Abstract
Viruses are the most abundant known infectious agents on the planet and are significant drivers of diversity in a variety of ecosystems. Although there have been numerous studies of viral communities, few have focused on viruses within the indigenous human microbiota. We analyzed 2 267 695 virome reads from viral particles and compared them with 263 516 bacterial 16S rRNA gene sequences from the saliva of five healthy human subjects over a 2- to 3-month period, in order to improve our understanding of the role viruses have in the complex oral ecosystem. Our data reveal viral communities in human saliva dominated by bacteriophages whose constituents are temporally distinct. The preponderance of shared homologs between the salivary viral communities in two unrelated subjects in the same household suggests that environmental factors are determinants of community membership. When comparing salivary viromes to those from human stool and the respiratory tract, each group was distinct, further indicating that habitat is of substantial importance in shaping human viromes. Compared with coexisting bacteria, there was concordance among certain predicted host–virus pairings such as Veillonella and Streptococcus, whereas there was discordance among others such as Actinomyces. We identified 122 728 virulence factor homologs, suggesting that salivary viruses may serve as reservoirs for pathogenic gene function in the oral environment. That the vast majority of human oral viruses are bacteriophages whose putative gene function signifies some have a prominent role in lysogeny, suggests these viruses may have an important role in helping shape the microbial diversity in the human oral cavity.
Keywords: saliva, bacteriophage, virus, microbiome, virome, metagenome
Introduction
The human oral cavity harbors a robust ecosystem inhabited by numerous different eukaryotes, bacteria, archaea and viruses (Lepp et al., 2004; Vianna et al., 2008; Nasidze et al., 2009; Bik et al., 2010; Ghannoum et al., 2010). Much is known about some of the bacterial inhabitants because of their involvement in common disease states, such as Streptococcus mutans and dental caries (Hamada and Slade, 1980). Chronic periodontitis is a disease with no single known etiological agent, but in which substantial alterations in the indigenous bacterial communities are found in the subgingival crevice (Jenkinson and Lamont, 2005; Ledder et al., 2007) and saliva (Mager et al., 2003; Sakamoto et al., 2004).
Viruses represent the most abundant infectious agents on the planet; moreover, viruses of bacteria are believed to exist wherever their bacterial hosts are present. Numerous studies describe viral communities in different habitats (Suttle, 2005; Gino et al., 2007; Andersson and Banfield, 2008); however, few have described these communities in humans (Breitbart et al., 2008; Nakamura et al., 2009; Willner et al., 2009, 2010; Reyes et al., 2010). We believe that there may be vast, uncharacterized communities of bacteriophages present in each of the ecological niches in humans. Because of their alternate lifestyles, involving either primarily lytic behavior—with the potential to eradicate certain bacteria, or primarily lysogenic behavior—with the potential to convey new function to their host (Canchaya et al., 2003), bacteriophages may have a substantial capacity to alter human bacterial communities (Kunin et al., 2008; Rohwer and Thurber, 2009), and as a result, may have a role in both health and in disease, such as chronic periodontitis (Gorski and Weber-Dabrowska, 2005).
One of the primary current approaches for characterizing microbial communities is broad-range 16S rRNA PCR amplification and amplicon sequencing (Bik et al., 2006; Ley et al., 2008; Costello et al., 2009). This approach provides information about community membership and phylogenetic relationships, but fails to reveal much about the functional potential of the community. Metagenomics (shotgun or community-wide sequencing) is an increasingly practical, alternative approach for community characterization, and assessment of functional potential that is broadly applicable, including to viral communities. There have been only a few published studies of viral community composition in humans (Breitbart et al., 2008; Nakamura et al., 2009). One study of respiratory viral communities demonstrated large differences between healthy subjects and those with cystic fibrosis (Willner et al., 2009). In the human gastrointestinal tract, viral populations are highly individual-specific, and are characterized by the presence of temperate bacteriophages with substantial genetic stability over time (Reyes et al., 2010). In human oropharyngeal samples, the presence of platelet-binding factors pblA and pblB in viruses implicates them as potential contributors to bacterial community virulence (Willner et al., 2010). We analyzed the salivary viromes of five periodontally healthy human subjects over a 60- to 90-day period to gain a broader appreciation for the viral inhabitants of the human oral cavity, their potential contribution to virulence and metabolism, and their relationship with oral bacteria.
Materials and methods
Human subject enrollment
All subjects were enrolled and donated three separate saliva samples over a 60- to 90-day period. The first and second saliva samples were collected on days 1 and 30 for all subjects, and the third collection occurred either on day 60 or 90 for each individual subject. Subject recruitment and enrollment were approved by the Stanford University Administrative Panel on Human Subjects in Medical Research. All subjects completed a questionnaire demonstrating their willingness to participate in the study. Five subjects were enrolled who had taken no antibiotics for at least 1 year prior to beginning the study, and who had no pre-existing medical conditions associated with significant immunosuppression. Subject no. 1 and no. 2 were members of the same household for the duration of this study. All subjects self-reported their health status. Each subject was subjected to a full baseline periodontal examination consisting of measurements of probing depths, clinical attachment loss, Gingival Index, Plaque Index and gingival irritation (Loe, 1967), and was found to have healthy oral tissues and no periodontitis (overall clinical attachment loss of <1 mm), with a diagnosis of slight localized gingivitis. A minimum of 3 ml of saliva was collected in the morning before breakfast prior to any oral hygiene practices, and the saliva was stored at −20 °C until further analysis. None of the subjects took antibiotics during the study.
Isolation and visualization of viruses
To visualize virus-like particles in human saliva, we modified an existing procedure commonly used to isolate viruses from environmental samples (Thurber et al., 2009). Saliva was filtered sequentially using 0.45 and 0.2-μm filters (VWR, Radnor, PA, USA) to remove cellular and other debris, stained using SYBR-gold and visualized by epifluorescence microscopy (Noble and Fuhrman, 1998). The concentration of the virus-like particles was estimated based on the average number of particles from at least four separate high-power fields. Viral concentrates also were visualized by electron microscopy (FEI Tecnai TF 30 He Polara) at a magnification ranging from 45K to 75K. To isolate human oral viruses for DNA preparation, the filtered fraction was purified on a cesium chloride gradient. Only the fraction with a density corresponding to most known bacteriophages (Murphy et al., 1995) was retained; further purified on Amicon YM-100 protein purification columns (Millipore Inc., Billerica, MA, USA); treated with DNase-I; and subjected to lysis and DNA purification using the Qiagen UltraSens virus kit (Qiagen, Valencia, CA, USA). The resulting DNA was amplified using Qiagen RepliG MDA (Qiagen) and fragmented bar-coded libraries were created as described (Dethlefsen and Relman, 2010), followed by sequencing using primer-A on a 454 Life Sciences Genome Sequencer FLX instrument using Titanium chemistry (Roche Applied Science, Indianapolis, IN, USA). Virome sequence data from this study is available through the Metagenomics Analysis Server at metagenomics.anl.gov.
Analysis of viral sequence data
Reads from each sequence data set were filtered to remove low-quality reads, which were defined as short reads (reads <100 nucleotides), reads with >10 homopolymer tracts and reads with ambiguous characters. The remaining reads were analyzed using a CLC Genomics workbench 3.65 (CLC bio USA, Cambridge, MA, USA) to construct assemblies based on 98% identity with a minimum of 20% total read overlap, consistent with criteria developed to discriminate between highly related bacteriophages (Breitbart et al., 2002). Because the shortest reads were 100 nucleotides, the minimum tolerable overlap was 20 nucleotides, and the average overlap was no less than 27 nucleotides, depending on the characteristics of each virome (Supplementary Table 1). Contigs were assigned to categories based on the presence of known homologs using blastX analysis of the NCBI NR database (E-score <10−3). Contigs were designated to the category ‘Hominid' if they had significant homology to Homo sapiens or Pan troglodytes. Those contigs assigned to the ‘virus' category were further assigned to a putative host taxonomy based on their blastX best hit. The same criteria were used to generate and assign contigs for a group of 11 fecal viromes (Reyes et al., 2010) and a pool of respiratory tract viromes (Willner et al., 2009).
Heatmaps were generated by creating a database of blastX best hits for all contigs across all subjects and time points, and depicted using Java Treeview (Saldanha, 2004). Heatmap data were normalized based on the total number of viral contigs for each virome. Principal-coordinates analysis was performed using Bray Curtis values using QIIME (Caporaso et al., 2010). Shared homologs present in each virome were analyzed by creating custom blast databases for each virome; comparing each database with all other viromes using blastN analysis (E-score <10−5); and normalizing the results to the size of the smaller virome. The metabolic potential of each virome was determined using blastX analysis of the SEED database using MG-Rast (E-score <10−5) (Meyer et al., 2008). Virulence factor homologs were identified using blastX analysis of the Virulence Factor Database (E-score <10−5) (Yang et al., 2008), and putative functions were assigned based on database annotation.
Viral contigs were analyzed using FGenesV (Softberry Inc., Mount Kisco, NY, USA) for open reading frame prediction, and individual Open reading frames were analyzed using blastX analysis against the NCBI non-redundant database (E-score <10−5). If the best hit was to a gene with no known function, lower level hits were used for the annotation as long as they had known putative function and still met the E-score cut-off (10−5). Veillonella dispar ATCC 17748 and its associated prophage were analyzed by using blastN analysis. Virome reads were mapped to the V. dispar ATCC 17748 genome using CLC Genomics workbench 3.65 (CLC bio USA), based on 90% identity with a minimum of 50% mapping overlap. Each virome also was analyzed to determine community structure, evenness, diversity and estimated number of genotypes using PHACCS (Angly et al., 2005) based on the Power Law, which represented the best fit.
Analysis of 16S rRNA sequences
We amplified the V1–V2–V3 region of the 16S rRNA gene from each specimen from each time point using primers that have been optimized for pyrosequencing (Dethlefsen and Relman, 2010). The forward primer consists of a 10:1:1 ratio of the following primers: 8FM-B 5′-CCCTGTGTGCCTTGGCAGTCTCAGCAAGAGTTTGATCMTGGCTCAG-3′ 8FT-B 5′-CCCTGTGTGCCTTGGCAGTCTCAGCAAGAGTTTGATTCTGGCTCAG-3′ and 8Fbif-B 5′-CCCTGTGTGCCTTGGCAGTCTCAGCAAGGGTTCGATTCTGGCTCAG-3′. This primer contains the 454 Life Sciences primer-B sequence and a two-base linker sequence ‘CA', and modifications of the broad-range 16S rRNA primer 8F 5′-AGAGTTTGATCMTGGCTCAG-3′. The reverse primer 515R-A 5′-CATCCCTGCGTGTCTCCGACTCAGNNNNNNNNNNGGTACCGCGGCKGCTGGCAC-3′ contains the 454 Life Sciences primer-A sequence, a unique 10-nt barcode for each subject sample (represented above by ‘N'), the broad-range bacterial 16S rRNA primer 515R (5′-TACCGCGGCKGCTGGCAC-3′) and a two-base linker sequence ‘CA'. PCR was performed in 50-μl reaction volumes using the Roche FastStart HiFi polymerase kit (Roche Applied Science). Each reaction consisted of 39.8 μl of H2O, 5 μl of HiFi buffer with MgCl2, 1 μl of dNTPs, 1.2 μl of forward primer, 1 μl of reverse primer, 1 μl of HiFi polymerase and 1 μl of salivary DNA template. The following cycling parameters were used: 3-min initial denaturation at 95 °C, followed by 25 cycles of denaturation (30 s at 95 °C), annealing (45 s at 51 °C) and extension (5 min at 72 °C), followed by a final extension (10 min at 72 °C). The products were approximately 550 bp in length and were gel-purified using the Qiagen QIAquick Gel Extraction kit (Qiagen), and further purified by Ampure bead purification (Beckman Coulter Genomics, Morrisville, NC, USA). The purified amplicons were quantified using PicoGreen (Invitrogen, Carlsbad, CA, USA) and were pooled in equimolar ratios. Pyrosequencing was performed using primer-A on a 454 Life Sciences Genome Sequencer FLX instrument with Titanium chemistry (Roche Applied Science). 16S rRNA sequence data from this study is available from the NCBI Sequence Read Archive under accession number SRA024393.1.
Sequences were processed in a manner similar to procedures described previously (Hamady et al., 2008). Sequences were removed from the analysis if they were <300 nt, had an uncorrectable barcode, contained any ambiguous characters or contained >10 homopolymers. Sequences were assigned to their respective samples based on their 10-nt barcode sequence and similar sequences were clustered into operational taxonomic units (OTUs) using a minimum identity of 97% using CD-Hit (Li and Godzik, 2006). To limit overestimation of the microbial diversity present, pyrosequencing noise was reduced using Pyronoise (Quince et al., 2009, #602). Representative sequences from each OTU were chosen and aligned using NAST (DeSantis et al., 2006b) based on the Greengenes database (DeSantis et al., 2006a). Phylogenetic trees were constructed using FastTree based on Kimura's two-parameter distances, and taxonomy was assigned to each OTU using the RDP classifier (Wang et al., 2007; Price et al., 2009). Shared OTUs were compared between each subject at each time point to generate heatmaps using Java Treeview (Saldanha, 2004). Principal-coordinates analysis was performed based on Beta Diversity using weighted Unifrac distances. Rarefaction analysis was performed based on species richness estimates of 10 000 iterations using EcoSim (Lee et al., 2005). Good's coverage was determined as the estimation of the number of singletons in the population (n), compared with the total number of sequences (N), using the equation (1−(n/N)) × 100 (Good, 1953). Beta Diversity was determined using Sorensen's similarity index (Magurran, 2004).
Comparison of bacterial and viral counts
Putative taxonomic assignments for viral hosts and for bacteria were compared by residual analysis using a Pearson's χ2-test of the proportion of viral counts to the null hypothesis that these counts are a multinomial sample from the observed vector of bacterial counts. The asymptotic normal distribution of residuals and a Bonferroni correction were used to assess the statistical significance of Pearson residuals. Rank analysis was performed using Kendall's tau measure of rank correlation. As all counts observed are whole numbers, to evaluate ties, small random uniform variables were added to counts of viruses and bacteria, and rank correlation statistics were calculated. This procedure was repeated 500 times and the mean rank statistic was referred to the normal distribution with appropriate variance to assess a P-value.
Results
Isolation and visualization of human salivary viral populations
To analyze viral communities in human saliva, we recruited five subjects with good overall periodontal health and obtained saliva at three time points over a 60- to 90-day period. Each sample was collected in the morning prior to breakfast or routine oral hygiene practices. Based on epifluorescence microscopy of filtered saliva, virus-like particles were present at an estimated concentration of 108 particles per milliliter of saliva in all five subjects (Supplementary Figure 1). A variety of virus-like morphologies were revealed by electron microscopy, including those with short tail stubs (Supplementary Figure 2a) among other less frequently identified types (Supplementary Figures 2b–d).
Salivary virus comparisons within and between subjects
To isolate viral populations, samples were filtered, purified on a cesium chloride gradient, and DNA was extracted and subjected to pyrosequencing. The resulting sequence reads were assembled into contigs (Supplementary Figure 3) using the stringent criterion of 98% identity over a minimum of 20% total read overlap (Supplementary Table 1) (Breitbart et al., 2002). We focused on contigs for identification of viruses rather than virome reads because the larger fragments allow more productive searches for homologous sequences, and create a further barrier to contaminating cellular elements. Also, a majority of the contigs were constructed with far greater than 20 × coverage, providing substantial confidence in the assembly process. There were minimal identifiable contaminating cellular elements, with the preponderance identified as clonal environmental contaminants (Supplementary Table 2). To identify the putative origin of each contig, each was subjected to blastX analysis using the NCBI NR database. The majority of the contigs had no known homologs (Breitbart et al., 2003; Bench et al., 2007); however, for each subject a substantial proportion had known viral homologs (Supplementary Figure 4). A number of the contigs had bacterial homologs; however, contigs with known viral homologs outnumbered them in each case. Relatively few contigs (ranging from 0 to 1%) had homology to viral sequences other than bacteriophages (Supplementary Figure 5), suggesting that bacteriophages constitute the majority of the human salivary double-stranded DNA viral population. Of those contigs with homology to eukaryotic viruses, most had homology to Torque Teno viruses (Hino and Miyata, 2007).
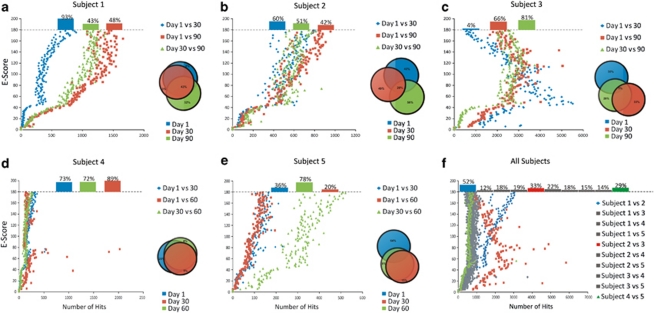
To determine whether viral community constituents are shared within a subject over time or between different subjects, we subjected each virome to blastN analysis in reference to the other viromes, to determine the presence of shared homologs. There was substantial conservation of homologs within all subjects across all time points, with the greatest conservation occurring in Subject no. 1 between day 1 and day 30, and across all time points for Subject no. 4 (Figures 1a–e). When comparing different subjects, there was extensive homology between the collective viromes of Subject no. 1 and no. 2, whereas each shared considerably less among the other subjects (Figure 1f). Subject no. 1 and no. 2 were members of the same household, which suggests that the extensive homology results from direct mixing or from shared environmental factors (Willner et al., 2009).
Figure 1.
Plots of shared homologs for intra-subject and inter-subject comparisons of viromes. Databases were created for the reads from each virome, and homologs between viromes were determined based on significant blastN hits (E-score <10−5). The number of significant hits per E-score is shown on the x-axis and E-scores are shown on the y-axis. For E-score values ⩾180 (the equivalent of 0), the proportion of significant hits is shown above the dashed line. (a–e) Intra-subject comparisons for subjects 1, 2, 3, 4, and 5, respectively. (f) Inter-subject comparisons. The insets with Venn diagrams show the overall percentage of shared homologs amongst each virome.
Viral contribution to metabolism and virulence
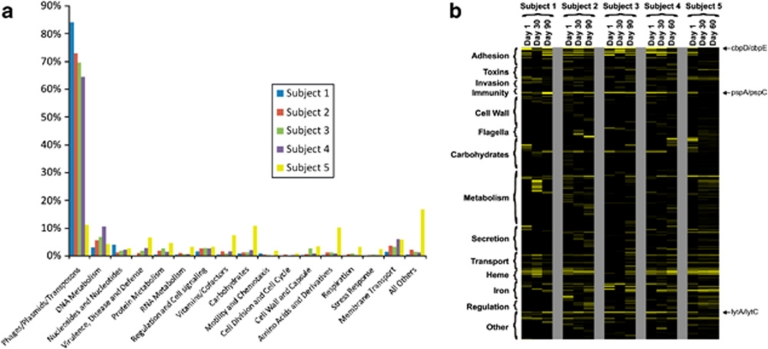
To determine whether viruses contribute to the metabolic potential in the human oral ecosystem, we subjected each virome to blastX analysis using the SEED database (Meyer et al., 2008). As would be expected for a viral population, the dominant features of the metabolic profile for each virome were nucleic acid metabolism and virulence (Figure 2a); however, sequences without SEED database homologs formed the majority of the viromes (Supplementary Figure 6). As might be expected, there were no complete metabolic pathways identified in these viromes.
Figure 2.
Analysis of metabolic potential and virulence factor homologs in viromes from each subject. (a) Percentage of viromes from each subject devoted to various putative metabolic categories based on reads with significant blastX homology to known entries in the SEED database (E-score <10−5). (b) Heatmap of virulence factor homologs present in viral contigs for each subject at all time points. Virulence factors were defined as those gene sequences that contribute substantially and may be either directly or indirectly involved in pathogenesis, and homologs were determined based on significant (E-score <10−5) blastX homology to the Virulence Factor Database (Yang et al., 2008). Putative functional categories are listed on the left.
We examined virome contigs to determine whether genes encoding virulence factors are present in the genome structure of salivary viruses using the Virulence Factor Database (Yang et al., 2008). The Virulence Factor Database was constructed with the purpose of identifying factors that contribute to disease processes, including more conventional virulence factors such as protein toxins, and less conventional virulence factors such as regulators and siderophores that are indirectly involved in pathogenesis, but are important for microorganisms to establish infection. Each subject presented a different profile of virulence factor homologs across all time points (Figure 2b); however, numerous virulence factor homologs are present in multiple contigs in each virome, suggesting that their presence may be of substantial importance to the viral community. Among the most commonly identified are pspA and pspC (involved in complement fixation and IgA degradation) (Dave et al., 2004; Ren et al., 2004), and cbpD and cbpE (involved in adhesion to the nasopharynx) (Gosink et al., 2000). Virulence factor homologs putatively involved in platelet binding, iron scavenging, lipopolysaccharide biosynthesis, cell wall antigenic variation and DNA methylation also were identified in viral contigs (Supplementary Figure 7 and Supplementary Table 3). The presence of these viral virulence factor homologs in salivary viruses suggests that these viruses may serve as a reservoir of pathogenic gene function in the human oral cavity.
Putative lysogenic viruses
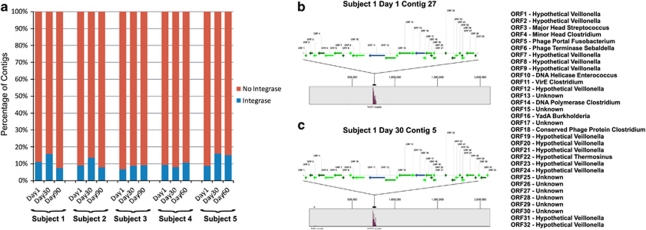
We analyzed viral contigs using blastX analysis to determine whether there were integrase homologs, which generally suggests the presence of lysogenic viruses. For each subject, we found that approximately 10% of the viral contigs had integrase homologs (Figure 3a), indicating that lysogenic viruses may be prominent in this human habitat. We also identified a viral contig in Subject no. 1, present on both day 1 and day 30, with a large number of homologs to the oral commensal V. dispar (Figures 3b and c). Indeed, through blastN analysis, nearly the entire contig is present in the genome of strain V. dispar ATCC 17748, with the exception of an ≈500-bp segment (Figures 3b and c). Mapping of virome reads to the genome of V. dispar ATCC 17748 reveals that the vast majority of the reads map to the region of the putative prophage rather than being evenly distributed over the length of the genome, which further substantiates that these phage are present in the salivary environment and are not the result of bacterial contamination. Interestingly, a small proportion of the reads on day 30 map to a separate genome region with a substantial number of hypothetical and putative phage genes, suggesting that Veillonella may have multiple phage present in the salivary environment simultaneously. We also found a putative streptococcal virus with an integrase homolog and an accompanying phage repressor homolog (Supplementary Figure 8a), further indicative of lysogenic viruses in the community. Similar to our finding of the putative Veillonella prophage, we found a putative Megasphaera phage in which many of the homologs also have synteny with the sequenced bacterial strain (Supplementary Figure 8b).
Figure 3.
Percentage of integrases in viral contigs and putative prophage assemblies from Subject no. 1. (a) Contigs with integrase homologs are shown in blue and contigs without integrase homologs are shown in red for each subject across all time points. Putative Veillonella phage assemblies from Subject no. 1 on day 1 (b) and day 30 (c). Putative virulence factor homologs virE and yadA are shown in blue, and all other open reading frames are shown in green. (b) Viral contig (27 429 nucleotides, 1421 reads, average coverage 22 × ) on day 1 and (c) viral contig (27 904 nucleotides, 1302 reads, average coverage 20 × ) on day 30. In panels b and c, the genome of V. dispar ATCC17748 is shown, and the proportion of virome reads mapping to different portions of the genome are shown in purple.
Host/virus community structure
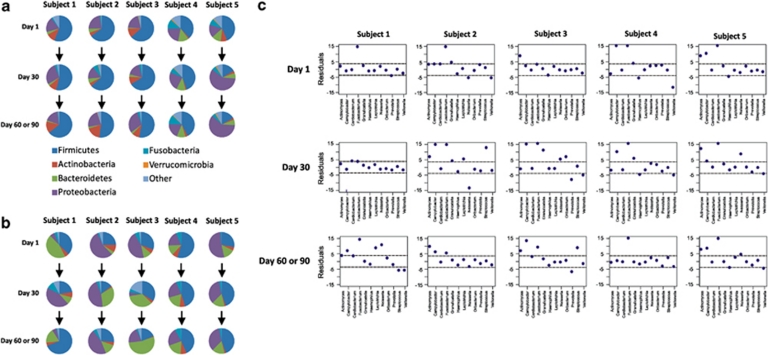
To improve our understanding of viral populations compared with their bacterial hosts in human saliva, we subjected each viral contig to blastX analysis using the NCBI NR database and used the best hit as an indicator of its host bacterium. At the phylum level, Firmicutes predominated across most all subjects and time points, followed by Proteobacteria and Actinobacteria (Figure 4a). We also sequenced the bacterial 16S rRNA genes from each subject (Supplementary Table 4) as a comparison data set. Whereas there was substantial conservation of viral taxonomy (Figure 4a), the bacterial phyla present were highly variable (Figure 4b) at a high taxonomic level. We also compared the populations of bacteria and viruses present at a lower taxonomic rank by testing whether the ranks of viruses and bacteria at the genus level were more concordant than would be expected by chance. We did not find significant concordant relationships using Kendall's tau distance for the majority of the time points in each subject (Table 1). After combining all time points within individual subjects to increase the power of detecting discordant relationships, we did find significant concordant relationships for all subjects (Table 1). Examination of individual genera using Pearson's χ2-test to measure residuals demonstrated that for most time points there was significant concordance between bacteria and virus for Streptococcus, Prevotella, Veillonella, Leptotrichia, Neisseria, Granulicatella and Cardiobacterium (Figure 4c). In many subjects, Actinomyces, Fusobacterium and Campylobacter presented the most discordant relationships (Figure 4c). There were few significant bacteria/virus relationships detected at the level of phylum (Table 1), probably because of the lack of power of a rank test on samples with limited categories (n=7). The presence of discordance for certain genera and concordance for others suggests that salivary viruses might have a different impact on their respective bacterial hosts.
Figure 4.
Taxonomic assignments and residual plots comparing viruses and their bacterial hosts for all subjects at all time points. Phylum-level taxonomic assignments for putative viral hosts based on blastX best hits of contigs against the NCBI NR database are shown in panel a, and assignments for bacteria based on 16S rRNA sequences are shown in panel b. Genus-level residual plots for taxonomic assignments comparing bacterial taxonomy with putative viral host taxonomy are shown in panel c. The dashed lines represent significant residuals with P-values <0.01.
Table 1. Significance values for bacteria and virus comparisons.
| Phylum | Genus | |
|---|---|---|
| Subject no. 1 | ||
| Day 1 | 0.707 | 0.806 |
| Day 30 | 0.260 | 0.133 |
| Day 90 | 0.260 | 0.193 |
| Subject no. 2 | ||
| Day 1 | 0.707 | 0.175 |
| Day 30 | 0.452 | 0.228 |
| Day 90 | 0.133 | 0.251 |
| Subject no. 3 | ||
| Day 1 | 0.806 | 0.251 |
| Day 30 | 0.260 | 0.276 |
| Day 90 | 0.260 | 0.152 |
| Subject no. 4 | ||
| Day 1 | 0.060 | 0.003a |
| Day 30 | 0.260 | 0.304 |
| Day 60 | 0.221 | 0.035a |
| Subject no. 5 | ||
| Day 1 | 0.260 | 0.210 |
| Day 30 | 0.260 | 0.251 |
| Day 60 | 0.260 | 0.029a |
| All subjectsb | ||
| Subject no. 1 | 0.260 | 0.021b |
| Subject no. 2 | 0.133 | 0.013b |
| Subject no. 3 | 0.133 | 0.023b |
| Subject no. 4 | 0.133 | 0.001b |
| Subject no. 5 | 0.260 | 0.017b |
P-values <0.05.
Combined time points within individual subjects.
We compared the viruses present in each subject at the genus level to determine whether they are conserved over time. For each subject, there is substantial conservation of viruses with similar putative host range (Supplementary Figure 9a). Beta diversity in the viromes was determined at the genus level, with an assigned value of 1 when viruses persist over time and 0 when the viruses are completely distinct at each time point. For all subjects, the beta diversity varied from 0.65 to 0.82, suggesting that viruses from the same hosts are generally conserved (Supplementary Figure 9a). Similar results were found for salivary bacteria (Supplementary Figure 9b).
We used PHACCS (Angly et al., 2005) to help decipher the ecology of viruses within the community, and found that there was a substantial number of estimated viral genotypes in human saliva (ranging from 293 to 2200) and a high level of evenness in the community (Supplementary Table 5). Examination of bacterial diversity through analysis of shared bacterial OTUs also revealed substantial diversity within and between subjects over time (Supplementary Figure 10). The high level of diversity for both viral and bacterial communities provides further evidence that the human oral cavity represents a complex and potentially dynamic ecosystem.
Comparisons with other human viromes
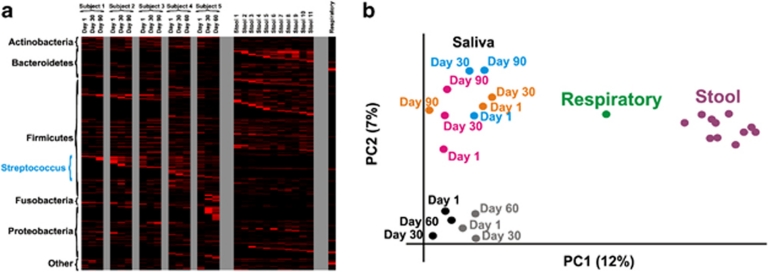
We analyzed viral blastX best hits to determine the diversity of viruses present in salivary communities (Figure 5a). Although there were numerous viral contigs with the same best hit among different subjects, many were unique to a specific subject and time point. Across all individuals, approximately 43% of the viral contigs shared the same best hit with another contig, suggesting that there are highly related yet distinct viruses that populate the human oral cavity. Similar results also were demonstrated for salivary bacterial communities (Supplementary Figures 9b and 11), indicating that shared genera are largely conserved over time in each subject. When compared with viromes from human stool (Reyes et al., 2010) and the respiratory tract (Willner et al., 2009), few of the contigs shared the same best hit, whereas most of the viral contigs were unique to their particular habitat (Figure 5a). Indeed, as demonstrated through principal-coordinates analysis, viromes from stool and the respiratory tract are distinct from those of saliva (Figure 5b). Even among the salivary viral communities, many are distinct to their individual host; however, for some time points, the viral communities are only partially reflective of their host environment.
Figure 5.
Heatmap of taxonomic assignments based on blastX best hits for viral contigs (a) and principal-coordinates analysis of viruses based on blastX best hits for viral contigs (b). At each heatmap time point, values are normalized by the total number of viral contigs. Principal-coordinates analysis was performed on Bray–Curtis values for viruses at all time points for each subject. Blue represents Subject no. 1, orange represents Subject no. 2, magenta represents Subject no. 3, gray represents Subject no. 4 and black represents Subject no. 5. A pooled respiratory virome is represented by green and 11 individual stool viromes are represented by purple.
Discussion
Viruses are critical determinants of bacterial community structure and function in all habitats so far examined. Our analysis of both bacterial and viral components of the microbial communities in human saliva over a 60- to 90-day period suggests that the same may be true for the human oral cavity. From the diverse morphologies of the virus-like particles present in human saliva (Supplementary Figure 2), to the estimated 108 virus-like particles per milliliter in each of our subjects (Supplementary Figure 1), our data reveal that there is a persistent community of double-stranded DNA viruses in saliva from healthy human subjects, with the most abundant virus types present identified almost exclusively as bacteriophages.
Our comparison of the viral and bacterial components of the human salivary microbiome also has uncovered important features of the interplay between human oral microbial communities. The substantial numbers of coexisting viral genotypes, whose specific membership is not static over time (Figure 5), demonstrates the complexity and robustness of the human oral ecosystem. While the putative host range of these viruses appears to be a fairly stable component of the ecosystem (Supplementary Figure 9a), the relative proportions of the salivary bacterial microbiota vary (Figure 4b). The presence of shared homologs, particularly between subjects residing in the same household (Figure 1d), suggests that environmental factors have a substantial role in determining the composition of the oral viral community; however, a more detailed analysis than the one presented here would be necessary to conclusively demonstrate such a phenomenon. While salivary viruses may have an important role in shaping the oral microbiome, it has yet to be determined whether they are representative of the viruses that likely are present in the oral bio-film.
For a portion of our analysis, we place greater emphasis on viral contigs rather than reads because in our stringent construction of contigs we noticed that for many contigs the average coverage far exceeded 20 × , and we were able to reproduce the same contigs from separate viromes in the same subject (Figures 3b and c), thereby providing us with substantial confidence in the construction process. Also, by focusing on the viral contigs, the longer stretches of sequence permitted more productive searches for homologous sequences in the NCBI NR database. Alternative methods to advancing the identification of novel bacteriophages from metagenome data include increasing the read length of sequences and cloning larger fragments prior to sequencing. One of the greatest limitations of virome analysis is the presence of contaminating cellular DNA. While our techniques produced data sets with limited cellular contamination (Supplementary Table 2) compared with other studies, the process of assembly creates a further barrier to contamination, because reads from larger bacterial and eukaryotic genomes are less likely to assemble than reads from smaller viral genomes. The vast majority of contigs with homology to eukaryotes were to redundant eukaryote DNA, with the exception of the few eukaryotic viruses found with homology to Torque Teno viruses.
We found that a substantial proportion of salivary viral sequences were homologous across all time points within individual subjects (Figures 1a–e). Although this finding might reflect the presence of the same viruses over time, it is more likely due to shared characteristics among different viruses, such as the numerous virulence factor homologs that are present in each subject across all time points (Figure 2b). The presence of shared profiles within subjects over time (Figure 5a and Supplementary Figure 11a), but not between subjects, suggests that there are inherent properties specific to each human environment that determine viral community composition. The effect of habitat on virome community membership is further exemplified by the distinct differences found when comparing salivary viromes with those from human stool and the respiratory tract (Figure 5b).
We base our identification of viral virulence factors on the Virulence Factor Database (Yang et al., 2008), which was constructed with the purpose of identifying factors that are involved in disease processes, which includes factors with both direct and indirect roles in pathogenesis. There was an extraordinary relative abundance of reads that were homologous to known virulence factors found in viral contigs from each subject across all time points (Figure 2b and Supplementary Table 3). Four of the most prevalent virulence factors (pspA, pspC, cbpD and cbpE) are putatively involved in immune evasion through breakdown of complement or IgA, and adhesion to the nasopharynx (Supplementary Table 3). The presence of these factors suggests that salivary viruses have the potential to have a role in the pathogenicity of their host bacteria.
We have just begun to explore the potential contributions of viruses to human ecosystems. Our analysis of viruses in human saliva has uncovered properties of viruses that differ from those previously found in analysis of human stool and respiratory viruses (Willner et al., 2009; Reyes et al., 2010). The vast majority of the human salivary viruses were identified as viruses of bacteria, with a substantial proportion of the population having integrase homologs (Figure 3a), suggesting a predominant role in lysogeny. That we found putative viruses of Veillonella (Figure 3b), Streptococcus (Supplementary Figure 8a) and Megasphaera (Supplementary Figure 8b), whose gene structure suggests they might also exist as prophages within their respective hosts, further supports the presence of lysogenic viruses in the community. Many of the viral contigs have homologs predicted to be involved in the pathogenic functions of bacteria. As such, these findings represent an intriguing feature of salivary viruses, where they may serve as reservoirs of pathogenic gene function in the human oral environment.
Acknowledgments
This work was supported by the Robert Wood Johnson Foundation; the UNCF-Merck Science Initiative; the Burroughs Wellcome Fund; and NIH 1K08AI085028 to DTP, the National Institutes of Health Director's Pioneer Award DP1OD000964 to DAR, and NSF DMS 0940077 to JS. DAR is supported by the Thomas C and Joan M Merigan Endowment at Stanford University. We thank Nafisi Ghori at the Stanford University High Resolution Electron Microscope Facility and Les Dethlefsen for contribution to this work.
Author contributions
Conceived and designed experiments: DTP and DAR. Performed experiments: DTP, MH, CD-L and RW. Analyzed data: DTP, JS, MH, FLR and DAR. Contributed reagents and performed examinations: PL and GCA. Wrote the paper: DTP and DAR.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- Angly F, Rodriguez-Brito B, Bangor D, McNairnie P, Breitbart M, Salamon P, et al. PHACCS, an online tool for estimating the structure and diversity of uncultured viral communities using metagenomic information. BMC Bioinformatics. 2005;6:41. doi: 10.1186/1471-2105-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bench SR, Hanson TE, Williamson KE, Ghosh D, Radosovich M, Wang K, et al. Metagenomic characterization of Chesapeake Bay virioplankton. Appl Environ Microbiol. 2007;73:7629–7641. doi: 10.1128/AEM.00938-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci USA. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B, et al. Viral diversity and dynamics in an infant gut. Res Microbiol. 2008;159:367–373. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead D, et al. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci USA. 2002;99:14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brussow H. Phage as agents of lateral gene transfer. Curr Opin Microbiol. 2003;6:417–424. doi: 10.1016/s1369-5274(03)00086-9. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave S, Carmicle S, Hammerschmidt S, Pangburn MK, McDaniel LS. Dual roles of PspC, a surface protein of Streptococcus pneumoniae, in binding human secretory IgA and factor H. J Immunol. 2004;173:471–477. doi: 10.4049/jimmunol.173.1.471. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Jr, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006b;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006a;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA. Microbes and Health Sackler Colloquium: incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2010;108 (Suppl 1:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gino E, Starosvetsky J, Armon R. Bacteriophage ecology in a small community sewer system related to their indicative role in sewage pollution of drinking water. Environ Microbiol. 2007;9:2407–2416. doi: 10.1111/j.1462-2920.2007.01355.x. [DOI] [PubMed] [Google Scholar]
- Good IJ. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- Gorski A, Weber-Dabrowska B. The potential role of endogenous bacteriophages in controlling invading pathogens. Cell Mol Life Sci. 2005;62:511–519. doi: 10.1007/s00018-004-4403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosink KK, Mann ER, Guglielmo C, Tuomanen EI, Masure HR. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect Immun. 2000;68:5690–5695. doi: 10.1128/iai.68.10.5690-5695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Slade HD. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev. 1980;44:331–384. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino S, Miyata H. Torque Teno virus (TTV): current status. Rev Med Virol. 2007;17:45–57. doi: 10.1002/rmv.524. [DOI] [PubMed] [Google Scholar]
- Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13:589–595. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Kunin V, He S, Warnecke F, Peterson SB, Garcia Martin H, Haynes M, et al. A bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res. 2008;18:293–297. doi: 10.1101/gr.6835308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledder RG, Gilbert P, Huws SA, Aarons L, Ashley MP, Hull PS, et al. Molecular analysis of the subgingival microbiota in health and disease. Appl Environ Microbiol. 2007;73:516–523. doi: 10.1128/AEM.01419-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SG, Kim CM, Hwang KS. Development of a software tool for in silico simulation of Escherichia coli using a visual programming environment. J Biotechnol. 2005;119:87–92. doi: 10.1016/j.jbiotec.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA. Methanogenic Archaea and human periodontal disease. Proc Natl Acad Sci USA. 2004;101:6176–6181. doi: 10.1073/pnas.0308766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Loe H. The gingival index, the plaque index and the retention index systems. J Periodontol. 1967;38 (Suppl:610–616. doi: 10.1902/jop.1967.38.6.610. [DOI] [PubMed] [Google Scholar]
- Mager DL, Haffajee AD, Socransky SS. Effects of periodontitis and smoking on the microbiota of oral mucous membranes and saliva in systemically healthy subjects. J Clin Periodontol. 2003;30:1031–1037. doi: 10.1046/j.0303-6979.2003.00418.x. [DOI] [PubMed] [Google Scholar]
- Magurran A. Measuring Biological Diversity. Blackwell Publishing: Oxford, UK; 2004. [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis AW, Martelli GP, et al. 1995Virus Taxonomy: Sixth Report of the International Committee on Taxonomy of VirusesVol. Supplement 10.Springer-Verlag: New York [Google Scholar]
- Nakamura S, Yang CS, Sakon N, Ueda M, Tougan T, Yamashita A, et al. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS One. 2009;4:e4219. doi: 10.1371/journal.pone.0004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasidze I, Li J, Quinque D, Tang K, Stoneking M. Global diversity in the human salivary microbiome. Genome Res. 2009;19:636–643. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble RT, Fuhrman JA. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microb Ecol. 1998;14:113–118. [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- Ren B, McCrory MA, Pass C, Bullard DC, Ballantyne CM, Xu Y, et al. The virulence function of Streptococcus pneumoniae surface protein A involves inhibition of complement activation and impairment of complement receptor-mediated protection. J Immunol. 2004;173:7506–7512. doi: 10.4049/jimmunol.173.12.7506. [DOI] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, et al. Viruses in the faecal microbiota of monozytotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- Sakamoto M, Huang Y, Ohnishi M, Umeda M, Ishikawa I, Benno Y. Changes in oral microbial profiles after periodontal treatment as determined by molecular analysis of 16S rRNA genes. J Med Microbiol. 2004;53:563–571. doi: 10.1099/jmm.0.45576-0. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Suttle CA. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. Laboratory procedures to generate viral metagenomes. Nat Protoc. 2009;4:470–483. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- Vianna ME, Holtgraewe S, Seyfarth I, Conrads G, Horz HP. Quantitative analysis of three hydrogenotrophic microbial groups, methanogenic archaea, sulfate-reducing bacteria, and acetogenic bacteria, within plaque biofilms associated with human periodontal disease. J Bacteriol. 2008;190:3779–3785. doi: 10.1128/JB.01861-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, et al. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS One. 2009;4:e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner D, Furlan M, Schmieder R, Grasis JA, Pride DT, Relman DA, et al. Microbes and Health Sackler Colloquium: metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc Natl Acad Sci USA. 2010;108 (Suppl 1:4547–4553. doi: 10.1073/pnas.1000089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Chen L, Sun L, Yu J, Jin Q. VFDB 2008 release: an enhanced web-based resource for comparative pathogenomics. Nucleic Acids Res. 2008;36:D539–D542. doi: 10.1093/nar/gkm951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.