Abstract
BACKGROUND
Changes in DNA methylation may play an important role in the deleterious reproductive effects reported in association with exposure to environmental pollutants. In this pilot study, we identify candidate methylation changes associated with exposure to pollutants in women undergoing in vitro fertilization (IVF).
METHODS
Blood and urine were collected from women on the day of oocyte retrieval. Whole blood was analyzed for mercury and lead, and urine for cadmium using inductively coupled plasma mass spectrometry. Unconjugated bisphenol A (BPA) was analyzed in serum using high-performance liquid chromatography with Coularray detection. Participants were dichotomized as higher or lower exposure groups by median concentrations. Using the Illumina GoldenGate Methylation Cancer Panel I, DNA methylation in whole blood from 43 women was assessed at 1505 CpG sites for association with exposure levels of each pollutant. Candidate CpG sites were identified using a Diff Score >|13| (P< 0.05) and an absolute difference >10% which were confirmed using bisulfite pyrosequencing.
RESULTS
Methylation of the GSTM1/5 promoter was increased for women with higher mercury exposure (P= 0.04); however, no correlation was observed (r= 0.17, P= 0.27). Reduced methylation was detected in the COL1A2 promoter in women with higher exposure to lead (P= 0.004), and an inverse correlation was observed (r = − 0.45, P= 0.03). Lower methylation of a promoter CpG site at the TSP50 gene was detected in women with higher BPA exposure (P= 0.005), and again an inverse correlation was identified (r = − 0.51, P= 0.001).
CONCLUSIONS
Altered DNA methylation at various CpG sites was associated with exposure to mercury, lead or BPA, providing candidates to be investigated using a larger study sample, as the results may reflect an independently associated predictor (e.g. socioeconomic status, diet, genetic variants, altered blood cell composition). Further studies accommodating variations in these factors will be needed to confirm these associations and identify their underlying causes.
Keywords: bisphenol A (BPA), DNA methylation, IVF, lead (Pb), mercury (Hg)
Introduction
Exposures to mercury (Hg), lead (Pb), cadmium (Cd) and bisphenol A (BPA) are pervasive (CDC, 2010) and concern is growing that such exposures may adversely influence reproductive function among women (Buck Louis et al., 2006; Mendola et al., 2008). Substantial evidence suggests that Hg (ATSDR, 1999b), Pb (ATSDR, 2007) and Cd (ATSDR, 1999a; Thompson and Bannigan, 2008) alter fertility and reproductive outcomes in animals and humans at high doses. However, reproductive effects at persistent low doses of Hg, Pb and Cd, to which most humans are continually exposed through dietary, airborne and other so-called ‘background’ sources are less well understood (Younglai et al., 2005; Bloom et al., 2011a,c). Several investigators have reported adverse reproductive outcomes in association with higher female background exposures to Hg (Choy et al., 2002; Cole et al., 2006) and Pb (Borja-Aburto et al., 1999; Chang et al., 2006; Bloom et al., 2011c), whereas somewhat paradoxical increases in fecundity-related end-points during in vitro fertilization (IVF) have been reported by several groups in association with background exposures to Cd (Younglai et al., 2002; Al-Saleh et al., 2008; Bloom et al., 2010). Background exposures to the estrogenic plastic monomer BPA (Richter et al., 2007; Wetherill et al., 2007) have also been associated with reproductive dysfunction in animal studies (Hunt et al., 2003; Susiarjo et al., 2007; Cabaton et al., 2011), and concern is emerging about adverse reproductive effects in association with exposures among women (Takeuchi et al., 2004; Sugiura-Ogasawara et al., 2005; Mok-Lin et al., 2010; Bloom et al., 2011b; Fujimoto et al., 2011; Kandaraki et al., 2011); however, these hypotheses remain controversial (vom Saal et al., 2007; Eichenlaub-Ritter et al., 2008; Padmanabhan et al., 2008; Hengstler et al., 2011).
There is accumulating evidence that the reproductive effects of Hg, Pb, Cd and BPA may be mediated, at least in part, by epigenetic changes, specifically, alterations in the pattern of DNA methylation acquired during embryonic development or adulthood (Bollati and Baccarelli, 2010). The addition of methyl groups to sequences of cytosine–phosphate–guanine (CpGs), often found in clusters within a gene promoter region (CpG islands), typically reduces or prevents the expression of the associated gene. DNA methylation has been shown to vary with environmental exposures, and can be associated with altered gene expression depending on location. Assays of DNA methylation at LINE-1 and Alu elements, which are ubiquitous repetitive nucleic acid sequences found throughout the human genome, has been frequently used as surrogate markers for overall or ‘global’ methylation in studies of environmental exposures (Nelson et al., 2011).
By measuring methylation at LINE-1 and Alu sequences, investigators have previously reported alterations of genome-wide DNA methylation in human blood in association with exposure to organic pollutants (Rusiecki et al., 2008), traffic pollution (Baccarelli et al., 2009) and tobacco smoke (Breton et al., 2009). Exposure to Pb has been associated with reduced LINE-1, but not Alu, methylation in the peripheral blood (Wright et al., 2010). In contrast, low-dose Cd appears to up-regulate the expression of DNA methyltransferases, enzymes that facilitate the addition of methyl groups to nucleic acid, in vitro and therefore is proposed to increase DNA methylation (Jiang et al., 2008). In addition, research in rodents has shown that fetal BPA exposure is associated with changes in DNA methylation in various tissues (Dolinoy et al., 2007; Prins et al., 2008; Yaoi et al., 2008; Bromer et al., 2010), with a variation at the environmentally sensitive Agouti locus resulting in phenotypic changes (Dolinoy et al., 2007). Altered methylation patterns at genes associated with cellular communication have been reported for the prostate tissue sampled from male rats treated subcutaneously with low-dose BPA (Ho et al., 2006). In humans, breast epithelial cells showed many alterations in gene expression following treatment with low doses of BPA, for which a corresponding DNA methylation change was confirmed for one gene, LAMP3 (Weng et al., 2010). Similarly, such changes are seen in association with the consumption of the synthetic and potent estrogen diethylstilbestrol, which results in promoter methylation at estrogen responsive genes (Edwards and Myers, 2007), and increased rates of reproductive irregularities and cancers reported in subsequent generations are likely associated with such epigenetic modifications (Newbold, 2008).
While the animal literature is growing substantially, there remains a dearth of knowledge as to the extent of the association between environmental pollutants and DNA methylation changes in humans. In this pilot study, we begin to address this knowledge gap through the identification of candidate CpG sites demonstrating altered DNA methylation in association with Hg, Pb, Cd and BPA measured in a convenience sample of women undergoing IVF and participating in the Study of Metals and Assisted Reproductive Technologies (SMART). Although our experimental design is limited by the use of a mixed cell population derived from whole blood, the results of this preliminary study will be useful for the design of a future confirmatory investigation.
Materials and Methods
Samples
Sample selection and the clinical protocol for this study were previously described in detail (Bloom et al., 2010). In brief, 58 women undergoing a first completed IVF cycle at the University of California at San Francisco (UCSF) Center for Reproductive Health were recruited to the SMART, between 12 March 2007 and 29 April 2008. Women received baseline infertility evaluations and subsequently underwent gonadotrophin-induced ovarian stimulation according to clinic protocols. When at least two ovarian follicles exceeded 17 mm diameter on ultrasound, hCG was administered and oocytes were retrieved by transvaginal needle aspiration 36h later. On the day of oocyte collection, fasting whole blood specimens were collected from an intravenous port, immediately after insertion and prior to fluid administration, to facilitate conscious sedation during the procedure. The blood was collected into (1) one 6-ml lavender-topped Vacutainer® tube (Becton-Dickinson and Co., Franklin Lakes, NJ, USA), containing 10.8-mg K2-EDTA for trace element analysis, which was inverted several times and then allowed to stand for 10 min and (2) one serum separator Vacutainer® tube (Becton-Dickinson and Co., Franklin Lakes), which was allowed to stand for 20–30 min and then centrifuged at 700g for 10 min. Fasting urine specimens were also collected using a collection cup. Whole blood, serum and urine were aliquoted into 1.8-ml polypropylene cryovials and frozen immediately at −80°C. The study protocol was approved by the UCSF Committee for Human Research as well as the Institutional Review Boards of the New York State Department of Health (NYS DOH) and the University at Albany.
Exposure assessment
The analysis for blood Hg and Pb and urine Cd was previously described in detail (Bloom et al., 2010). In brief, blood and urine specimens were transported on dry ice to the Trace Elements Section of the Laboratory of Inorganic and Nuclear Chemistry, Wadsworth Center, NYS DOH (Albany, NY, USA), the principal reference laboratory for NYS. Sufficient blood volume was available for the determination of Hg and Pb in 50 women; however, a mandatory requirement by the State of California for the reporting of all blood Pb measurements with personal identifiers (California Occupational Blood Lead Registry, 2011) prompted 23 women to ‘opt-out’ of the blood Pb testing. Sufficient urine volume was available for Cd measurements in 55 women. An analytical method developed for the PerkinElmer Sciex ELAN DRC II inductively coupled plasma-mass spectrometer (PerkinElmer Life and Analytical Sciences, Shelton, CT, USA), equipped with Dynamic Reaction Cell technology (DRC-inductively coupled plasma mass spectrometry) was used to determine blood Hg and Pb (Palmer et al., 2006), in addition to urine Cd (Minnich et al., 2008). Limits of detection (LOD) were 0.20 μg/l for blood Hg, 0.17 μg/dL for blood Pb and 0.02 μg/l for urine Cd. For results that were below the method LOD, the instrument reported values (including some negative values) were used without censoring to preclude potential bias associated with the use of such procedures (Schisterman et al., 2006).
The analysis for BPA was previously described in detail (Bloom et al., 2011a,b,c). In brief, serum specimens were shipped on dry ice to the Endocrine Disruptors Laboratory at the University of Missouri (Columbia, MO, USA). Unconjugated serum BPA was measured in 44 women by high-performance liquid chromatography with an ESA Coularray 5600 detector (ESA Inc., Chelmsford, MA, USA), using known internal standards. The recovery averaged 89% for a 5-ng BPA/ml spike in human serum samples (MP Biomedicals, Solon, OH, USA). The LOD was 0.3 ng/ml and we extrapolated from the standard curve to generate values for those samples measured below the lowest on column standard equal to 0.05 ng. Where no evidence for the presence of BPA at any concentration existed, we assigned a value of zero.
DNA methylation
Whole blood specimens were shipped on dry ice to the Robinson Laboratory at the Department of Medical Genetics, University of British Columbia (Vancouver, Canada). DNA was extracted from 1 to 2 ml of frozen whole peripheral blood using conventional methods for 51 women. Samples were purified using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany), and bisulfite converted using the EZ DNA Methylation Kit (Zymo Research Corporation, Irvine, CA, USA). A total of 750 ng of each bisulfite converted sample was run on the Illumina GoldenGate Cancer Panel I bead array (Illumina, Inc., San Diego, CA, USA). This panel assays 1505 CpG sites throughout the genome that are associated with cell replication, cell differentiation, or oxidative stress and thus may have relevance for reproduction. A total of 43 specimens with the required quantity of DNA were successfully assayed; 2 specimens failed PCR quality controls and therefore Illumina results were reported for 41 women. Data was normalized to background intensity levels using BeadStudio software (Illumina, Inc.).
Candidate CpG sites were validated in all women using bisulfite pyrosequencing performed on the Pyromark ID machine and using the PyroGoldSQA reagent kit (Qiagen, Hilden, Germany). Assays for the selected candidate CpG sites GSTM1_P266, COL1A2_P407 and TSP50_P137 were designed using the Pyrosequencing Assay Design Software (primer sequences are listed in Supplementary data, Table S1). Each 15-µl PCR reaction contained 1 × PCR Buffer (Qiagen, Hilden, Germany), 3-mM Gibco dNTPs (Invitrogen, Carlsbad, CA, USA), 0.9-U HotStart Taq DNA polymerase (Qiagen, Hilden, Germany), 6 µM of each of the forward and reverse primers and ∼15 ng of bisulfite converted DNA. Cycling conditions were 95°C for 15 min, then 95°C for 30 s, annealing temperature (listed in Supplementary data, Table S1) for 30 s and 72°C for 30 s ×50, with a final extension of 72°C for 10 min. Global methylation of LINE-1 elements was done utilizing published primers (Bollati et al., 2007) and PCR cycling conditions for the commercially available LINE-1 assay (Qiagen, Hilden, Germany).
Genotyping
To further characterize DNA methylation in the GSTM1 promoter region, a deletion polymorphism encompassing the Illumina probes (GSTM1_P363 and GSTM1_P266) was genotyped using allelic discrimination with previously published primers and cycling conditions (Sata et al., 2003), and gel electrophoresis.
Statistical analysis
Concentrations of environmental pollutants were dichotomized into ‘higher exposure’ and ‘lower exposure’ groups by the median values for Hg (2.88 µg/l blood), Pb (0.73 µg/dl blood), Cd (0.38 µg/l urine) and BPA (2.39 µg/l serum). Higher exposure and lower exposure groups for environmental pollutants were evaluated by age above and below the mean, and by male factor versus female factor infertility using the χ2 or Fisher exact test as appropriate. BeadStudio software, v.3.1.3.0 (Illumina, Inc.) was used for difference analysis at CpG sites. Methylation at CpG sites for higher and lower exposure groups was compared for each pollutant, and candidate CpG sites were identified as those with both a Diff Score >13 (P< 0.05) and an absolute difference >10% between the means for each group. Those probes which contained single nucleotide polymorphisms (SNPs) and that failed quality control tests (detection P > 0.05) were omitted. Mann–Whitney U-tests were used to compare % DNA methylation as measured by pyrosequencing for each candidate CpG site and LINE-1 elements, between higher and lower pollutant-exposure groups. Pearson correlation coefficients were used to evaluate linear associations between the logarithm (i.e. to base 10) of each pollutant (after the addition of a constant equal to 1.00 to accommodate negative values). Statistical significance was defined as P< 0.05 for a two-tailed test. Statistical analyses were conducted using SPSS v.15.0 (IBM Co., Somers, NY USA), VassarStats (Vassar College, Poughkeepsie, NY USA) and SAS v.9.2 (SAS Institute, Cary, NC USA), and graphs were generated using R 2.11.0 (The R Project for Statistical Computing, Auckland, New Zealand).
Results
Global DNA methylation changes were investigated for associations with body burdens of Hg, Pb, Cd and BPA measured in women undergoing IVF treatment. Of the 43 women with epigenetic analysis, sufficient blood and/or urine was available for quantification of Hg in 43, Pb in 24, Cd in 42 and BPA in 35 women. The mean (SD) age was 36 (4.2) years, with a range of 28–44, most had never smoked cigarettes (83.7%), and a substantial proportion were Asian (30.2%). The primary infertility diagnoses were ‘unexplained’ (34.9%), ‘male factor’ (23.3%), ‘diminished ovarian reserve’ (18.6%) and ‘tubal factor’ (9.3%), and there was one (2.3%) diagnosis each of ‘endometriosis’ and ‘anovulation’, and four (9.3%) ‘other’ diagnoses. The majority of values were measured above the LODs for Hg (100% >LOD), Pb (100% >LOD), Cd (98% >LOD) and BPA (86% >LOD). Median (range) values were 2.9 µg/l blood (0.3–8.8) for Hg, 0.7 µg/dl blood (0.3–1.5) for Pb, 0.4 µg/l urine (0.02–1.2) for Cd, 0.3 µg/g for Cd normalized to urine creatinine (0.04–0.98) and 2.4 µg/l serum (0.0–67.4) for BPA. Women <36 years of age had a greater probability (P= 0.048) of ‘higher exposure’ to Hg (65.0%) than women >36 years of age (34.8%). No other differences were suggested for pollutant exposure by age or infertility diagnosis.
Using the Illumina GoldenGate Methylation Cancer Panel I, candidate CpGs were identified for Hg (2 CpG sites), Pb (1 CpG site) and BPA (1 CpG site); however, no candidates were identified for Cd (Supplementary data, Fig. S1). Each of the candidate CpG sites was validated using bisulfite pyrosequencing, the results of which were highly correlated with average_beta values provided by the Illumina platform (Supplementary data, Fig. S2). We report only pyrosequencing methylation values (%) for all subsequent analyses.
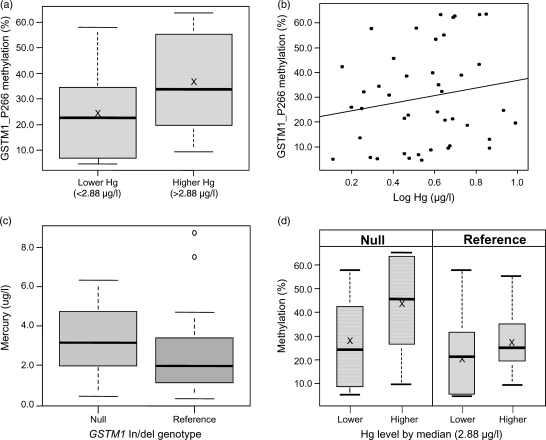
In a comparison of higher and lower Hg exposure groups, the GSTM1_P266 and GSTM1_P363 CpG sites, located within the promoter region upstream of the CpG island for the glutathione S-transferase M1 gene (GSTM1), were identified as candidate changes. However, the correlation between methylation values at these two CpG sites was highly significant (r = 0.93; P< 0.0001) and therefore we only followed up GSTM1_P266 using bisulfite pyrosequencing (Supplementary data, Fig. S3). Higher GSTM1_P266 CpG site methylation was measured for the higher exposure Hg group (mean = 35.8%) compared with the lower exposure Hg group (mean = 25.7%; P= 0.04; Fig. 1a). However, no significant correlation was detected (r = 0.17; P= 0.27) between Hg concentration and methylation at this site (Fig. 1b).
Figure 1.
(a) GSTM1_P266 methylation (%) compared between lower (n = 21) and higher (n = 22) mercury exposure groups (P= 0.04), ‘X’ indicates mean value; (b) correlation between GSTM1_P266 methylation (%) and the log of blood mercury concentrations (µg/l), (r= 0.17; P= 0.27); (c) mercury levels (µg/l) compared between women with GSTM1 null (n = 20) and reference (n = 23) genotypes (P= 0.36) and (d) GSTM1_P266 methylation (%) compared between higher and lower mercury exposure groups in women with null GSTM1 genotype (left panel; n = 11 and n = 9, respectively; P= 0.09) or reference GSTM1 genotype (right panel; n = 10 and n = 13, respectively; P= 0.50), ‘X’ indicates mean value.
The GSTM1 gene is located within a paralogous gene cluster on chromosome 1p13.3, which contains 5 GSTM family genes (Xu et al., 1998). As a result, the probes for both GSTM1_P266 and P363 hybridize with complete sequence identity to the promoter regions of both the GSTM1 and GSTM5 genes. In addition, there is a well-characterized deletion polymorphism encompassing the GSTM1 gene and its promoter (Xu et al., 1998) with a frequency of >50% in Caucasian populations (Board et al., 1990). We therefore genotyped this deletion polymorphism to further investigate the nature of the methylation change in this region associated with blood Hg concentrations. We detected no difference (P= 0.36) in Hg levels between women with null (homozygous deletion) and reference GSTM1 genotypes (Fig. 1c). The reference genotype can include both heterozygotes and homozygotes for the non-deleted allele, which unfortunately cannot be differentiated using this approach. When comparing DNA methylation of the GSTM1_P266 site in 20 women with the null genotype (measured methylation represents only the site within the GSTM5 promoter), we identified a trend (P= 0.09) towards lower methylation among the lower Hg-exposure group (mean = 27.2%) compared with the higher Hg-exposure group (mean = 44.4%) as demonstrated by Fig. 1d. In contrast, in 23 women with the reference genotype (measured methylation represents an average between the promoter regions of GSTM1 and GSTM5), there was no difference between low-exposure (mean = 21.2%) and high-exposure (mean = 27.6%) Hg groups (P= 0.50).
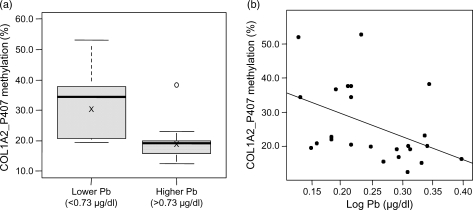
The candidate COL1A2_P407 CpG site within the promoter region of the collagen type 1 alpha-2 (COL1A2) gene was identified as associated with Pb concentrations. The methylation of COL1A2_P407 was higher (P= 0.004) in the lower exposure Pb group (mean = 31.7%) compared with the higher exposure Pb group (mean = 19.7%; Fig. 2a). Correspondingly, there was a statistically significant negative correlation between Pb exposure and methylation at this site (r= − 0.45; P= 0.03; Fig. 2b).
Figure 2.
(a) COL1A2_P407 methylation (%) compared between lower (n = 13) and higher (n = 11) lead exposure groups (P= 0.004), ‘X’ indicates mean value, and (b) correlation between COL1A2_P407 methylation (%) and the log of blood lead concentrations (µg/dl; r= − 0.45; P= 0.03).
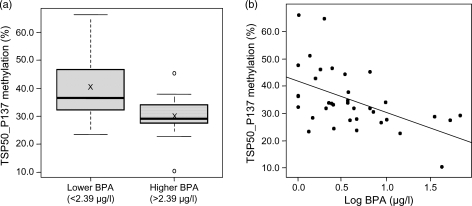
In comparison of women with lower and higher BPA concentrations, a methylation difference at the TSP50_P137 CpG site within the promoter CpG island of the testes-specific protease 50 (TSP50) gene was identified. DNA methylation of the TSP50_P137 site was higher (P= 0.005) among women in the lower exposure BPA group (mean = 40.3%) compared with the higher exposure BPA group (mean = 29.7%; Fig. 3a). Correspondingly, there was a moderate inverse correlation between BPA concentrations and methylation at this site (r= − 0.51; P= 0.001; Fig. 3b).
Figure 3.
(a) TSP50_P137 methylation (%) compared between lower (n = 17) and higher (n = 18) BPA exposure groups (P= 0.005), ‘X’ indicates mean value and (b) correlation between TSP50_P137 methylation (%) and the log of serum BPA concentrations (µg/l; r= − 0.51; P= 0.001).
No associations were identified for urine Cd concentrations and the epigenetic loci considered. Also, no difference was detected for LINE-1 methylation between higher exposure and lower exposure groups for Hg, Pb, Cd or BPA (Supplementary data, Fig. S4).
Discussion
This pilot study is the first, to our knowledge, to investigate DNA methylation changes in association with exposure to environmental pollutants in women undergoing IVF treatment. Candidate DNA methylation changes were identified in the gene promoters of GSTM1/5 (Hg exposure), COL1A2 (Pb exposure) and TSP50 (BPA exposure) in this study group.
We observed increased methylation at the CpG sites evaluated by the GSTM1_P266 Illumina probe in women within the higher Hg exposure group. GSTM1 is a member of the GSTM gene family, which encodes for enzymes that are involved in the metabolism of carcinogens and other hazardous environmental agents, and in the mediation of oxidative stress (Mannervik, 1985; Valko et al., 2007). However, interpretation of this association is complicated by the fact that this probe hybridizes to the promoter regions of both the GSTM1 and GSTM5 genes. Furthermore, there was no significant correlation (r = 0.17; P= 0.27) between Hg concentration and methylation at this site. The nature of this methylation change was further investigated by exploiting a deletion polymorphism, which encompasses the GSTM1 gene. We found that the trend of increased methylation with increased Hg exposure is limited to those women with a null GSTM1 genotype. Hence, the methylation effect is limited to only the GSTM5 promoter CpG site, suggesting down-regulation of this gene (Fig. 1d).
While GSTM1 remains the most extensively studied of the GSTM family, genes within this cluster share >85% sequence identity (Pearson et al., 1993); thus, the function of GSTM5 is expected to be similar. The GSTM1 null allele has been associated with increased susceptibility to cancers and poor clearance of hazardous agents (Strange et al., 1991; Van Poppel et al., 1992), and also has been suggested to play a role in human reproduction. The null allele was associated with recurrent pregnancy loss in Japanese women (Sata et al., 2003), and hypermethylation of the GSTM1 promoter was more frequently identified in the testes of infertile men (Dhillon et al., 2007). Increased DNA methylation and decreased gene expression of GSTM5 have also been associated with several types of cancer (Peng et al., 2009; Etcheverry et al., 2010; Bell et al., 2011), but it has not been studied in the context of human reproduction to our knowledge. It is, however, plausible that altered methylation of GSTM1 and GSTM5 might play a role in reproductive outcomes as a substantial literature underscores the importance of reactive oxygen species for human reproduction (Agarwal et al., 2005; Shkolnik et al., 2011).
In our study, COL1A2 promoter methylation was observed to negatively correlate with Pb exposure, and showed corresponding decreased methylation in the higher Pb exposure group. This result suggests that increased Pb may be associated with increased COL1A2 expression. The COL1A2 gene product is an important component of connective tissues, including the chorio-amniotic membranes and the uterine cervix (Romero et al., 2010). Interestingly, collagen genes, including COL1A2, are up-regulated in the placental tissue of pregnant tobacco smokers (Bruchova et al., 2010). Maternal COL1A2 polymorphisms have also been associated with preterm prelabour rupture of membranes, a leading cause of preterm birth (Romero et al., 2010). Environmentally induced alterations of collagen gene methylation, including COL1A2, might compromise human reproduction and a comprehensive investigation of this hypothesis is merited.
We detected decreased methylation within the TSP50 gene promoter in association with higher BPA exposure, and correspondingly there was a moderate negative correlation between methylation at this site and serum BPA levels. This result suggests that increased BPA exposure may be associated with increased expression of TSP50. The TSP50 gene encodes ‘testes-specific protease 50’, a product of unknown function that is expressed in the testes, and in which promoter CpG island hypomethylation is observed (Yuan et al., 1999). Increased expression of TSP50 due to aberrant loss of methylation has also been observed in female breast cancer tissue (Yuan et al., 1999; Shan et al., 2002). Our data suggests that TSP50 may be up-regulated with increasing BPA exposure due to a loss of methylation; however, little is known with regard to the function of this gene in women other than its initial identification in breast cancer tissue.
This study is a novel assessment of the effects of environmental exposures on DNA methylation in a sample of women undergoing IVF treatment, in which candidate changes at promoter CpG sites were identified for Hg (GSTM1/5), Pb (COL1A2) and BPA (TSP50). Whereas many prior human studies assess ‘global’ methylation (Nelson et al., 2011), we employed a strategy to identify specific functional epigenetic loci potentially related to human reproduction. In fact, we detected no associations using LINE-1, despite the aforementioned associations detected at specific sites, suggesting the utility of a function-specific approach to the assessment of environmental pollutants and epigenetic modification. A limitation of measuring methylation at individual CpGs is that it does not provide a comprehensive assessment of DNA methylation within a region of interest. While methodologies are being developed that can more comprehensively screen the genome, the more sites that are simultaneously evaluated the greater is the issue of false positives and negatives. As sites on the GoldenGate panel were selected on the basis of changes reported in association with cancer, these sites are more likely to be environmentally influenced than are random CpGs in the genome.
As previously reported, our study group shows higher median levels of Hg than those recorded for women in the general US population, whereas Cd and Pb levels are similar or lower (Bloom et al., 2010). Younger women demonstrated higher blood Hg levels than older in our study sample; however, no other differences were indicated for age and no difference in pollutant exposure were suggested between women diagnosed with infertility primarily due to a female factor versus a male factor. The range of serum BPA in our study group is somewhat higher than the levels reported from the limited number of previous studies assessing circulating concentrations (0.2–20 μg/l); however, these values are consistent with those reported for women during pregnancy (Vandenberg et al., 2010). Moreover, our assessment of BPA concentrations comprised only the unconjugated, or biologically active fraction in circulation (Matthews et al., 2001), providing a more specific evaluation of biologic activity than possible using total BPA concentrations. The use of polycarbonate (Sajiki et al., 1999) and polyvinylchloride (Calafat et al., 2009) products in scientific and medical equipment has raised concern regarding possible BPA contamination of biologic specimens. Our study protocol entailed collection of blood specimens using an intravenous port. However, this activity was conducted prior to connection to the intravenous line. Although no polycarbonate, epoxy resin or polyvinyl chloride containing supplies were employed, to our knowledge, during specimen collection, processing and storage during this study, we cannot conclusively rule out the possibility for BPA contamination by these means.
The pilot nature of our study presents several meaningful limitations that must be considered in the interpretation of these results. While our study cohort comprised women undergoing IVF, the limited sample size precludes an evaluation of the impact of epigenetic modifications on IVF outcomes; rather, this convenience sample afforded us the opportunity to identify candidate CpG sites so that a more comprehensive study may be conducted in the future. The small sample size also restricted the statistical power available to detect small differences in methylation between pollutant exposure groups. The selection of candidate CpG sites with >10% absolute difference between groups was done to address this limitation. Given the small number of candidate CpG sites identified and the hypothesis generating nature of this investigation, no correction for multiple comparisons was utilized in this study. This increases the likelihood of false positives within our data set, and therefore candidates will require validation in a larger study. In addition, our sample is heterogeneous with regard to infertility diagnosis with some women likely to be fertile (i.e. male factor infertility), whereas others were not; it is possible that such heterogeneity obscured associations between the environmental agents and epigenetic marks considered. Future investigations should endeavor to examine homogenous groups in terms of reproductive pathology, or the absence thereof. We also did not adjust for factors that might potentially alter methylation as well as the body burdens of Hg, Pb, Cd or BPA, and thus there is the possibility for unmeasured confounding. Furthermore, women in this study had been receiving reproductive medications, under various clinical protocols, for the purposes of IVF; the effect of such treatment on the body burden of environmental agents and the extent of methylation is unknown. Extrapolation of these study results to other groups should thus be made with caution.
A further limitation is that DNA methylation was assessed in whole peripheral blood and can vary by blood cell type, and so the methylation changes associated with pollutant exposures in this study may represent an alteration in blood cell composition, rather than a ‘true’ shift in methylation at the candidate loci (Wu et al., 2011). In an additional experiment using whole blood collected from 28 control women participating in an unrelated study (mean age 36 years; range 23–55), we evaluated associations between blood cell composition assessed by a Sysmex XE-2100 Automated Hematology Analyzer (Sysmex America, Inc. Mundelein, IL USA) and DNA methylation assessed by pyrosequencing at the candidate loci for which associations were detected (GTSM1_P266, COL1A2_P407, and TSP50_P137). No associations were identified between methylation at these sites and relative composition of neutrophils, leukocytes, monocytes, eosinophils or basophils when entered into multiple linear regression models (Supplementary data, Table S2). While these additional data lend credibility to the candidate loci identified, they do not eliminate the possibility that our results reflect a shift in blood cell composition given the mixed cell population we analyzed.
It is also important to note the possibility that the methylation changes are not a direct result of the exposure itself but of one or more unconsidered and co-associated factors, such as socioeconomic status, diet or genetic variants. In addition, if the DNA methylation changes in the peripheral blood do indeed reflect systemic environmental exposures relevant for effects on reproductive health, this does not necessarily reflect gene expression changes in the tissue of interest. Therefore, in future studies, the effects of these methylation changes on female reproductive tissues should be elucidated.
In conclusion, we conducted a novel assessment on the effects of environmental pollutants including Hg, Pb, Cd and BPA on both global and targeted DNA methylation in women undergoing IVF. Candidate CpG sites were identified for Hg (GSTM1/5), Pb (COL1A2) and BPA (TSP50). While limited by the use of whole blood specimens (i.e. a heterogeneous cell population), our findings raise the possibility that low level exposure to widely distributed environmental pollutants may be associated with altered DNA methylation. The potential impact of these study results on clinical reproductive outcomes and other aspects of human health merits consideration in a similar future study accommodating variations in the composition of whole blood specimens in a larger number of participants, and considering potential confounding factors.
Supplementary data
Supplementary data are available at http://humrep.oxfordjournals.org/.
Authors’ roles
M.S.B. and V.Y.F. conceived of and designed the study; V.Y.F. recruited study participants and collected data and biospecimens; P.J.P. and A.J.S. performed laboratory analysis for metals; F.S.v.S. and J.A.T. performed laboratory analysis for BPA; C.W.H. and W.P.R. performed epigenetic analysis; C.W.H., M.S.B., W.R., and D.K. completed data analysis and interpretation; C.W.H. drafted the article, which was revised critically for important intellectual content by M.S.B,. W.P.R., D.K., P.J.P., A.J.S., F.S.v.S., J.A.T., A.J.S. and V.Y.F.; C.W.H., M.S.B., W.P.R., D.K., P.J.P., A.J.S., F.S.v.S., J.A.T., A.J.S. and V.Y.F. approved of the final version of the manuscript for submission.
Funding
Institutional discretionary research funds available to M.S.B. and V.Y.F. were used to support this work. This research was also supported in part by Canadian Institutes for Health Research (CIHR) operating grant MOP-64220 provided to W.P.R. and by National Institute for Environmental Health Sciences (NIEHS) grant ES018764 provided to F.S.v.S.
Conflict of interest
F.S.v.S. served as a consultant for attorneys and has written reports concerning the published literature about the health effects of BPA in animals and humans. The other co-authors have no conflicts to declare.
Supplementary Material
Acknowledgements
We would like to thank the women in this study for their voluntary participation. In addition, we would like to thank Ruby Jiang and Dr Maria Peñaherrera for their assistance with sample preparation and processing in the Robinson Laboratory, Dr Michael Kobor for use of the Illumina GoldenGate array equipment and Dr Angela Devlin for use of the Biotage PyroMark™ MD system. We also acknowledge Giulia Conti, Julie D. Lamb and Gloria Cheng for their assistance in the coordination of study recruitment. We would also like to acknowledge the laboratory staff of the UCSF Center for Reproductive Health for their assistance in processing samples.
References
- Agarwal A, Gupta S, Sharma RK. Role of oxidative stress in female reproduction. Reprod Biol Endocrinol. 2005;3:28. doi: 10.1186/1477-7827-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Saleh I, Coskun S, Mashhour A, Shinwari N, El-Doush I, Billedo G, Jaroudi K, Al-Shahrani A, Al-Kabra M, El Din Mohamed G. Exposure to heavy metals (lead, cadmium and mercury) and its effect on the outcome of in-vitro fertilization treatment. Int J Hyg Environ Health. 2008;211:560–579. doi: 10.1016/j.ijheh.2007.09.005. [DOI] [PubMed] [Google Scholar]
- ATSDR. Toxicological Profile for Cadmium. Atlanta, GA: U.S. Agency for Toxic Substances and Disease Registry; 1999a. [PubMed] [Google Scholar]
- ATSDR. Toxicological Profile for Mercury. Atlanta, GA: U.S. Agency for Toxic Substances and Disease Registry; 1999b. [Google Scholar]
- ATSDR. Toxicological Profile for Lead. Atlanta, GA: U.S. Agency for Toxic Substances and Disease Registry; 2007. [PubMed] [Google Scholar]
- Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179:572–578. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A, Bell D, Weber RS, El-Naggar AK. CpG island methylation profiling in human salivary gland adenoid cystic carcinoma. Cancer. 2011;117:2898–2909. doi: 10.1002/cncr.25818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom MS, Parsons PJ, Steuerwald AJ, Schisterman EF, Browne RW, Kim K, Coccaro GA, Conti GC, Narayan N, Fujimoto VY. Toxic trace metals and human oocytes during in vitro fertilization (IVF) Reprod Toxicol. 2010;29:298–305. doi: 10.1016/j.reprotox.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom MS, Buck Louis GM, Sundaram R, Kostyniak PJ, Jain J. Associations between blood metals and fecundity among women residing in New York State. Reprod Toxicol. 2011a;31:158–163. doi: 10.1016/j.reprotox.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom MS, Kim D, Vom Saal FS, Taylor JA, Cheng G, Lamb JD, Fujimoto VY. Bisphenol A exposure reduces the estradiol response to gonadotropin stimulation during in vitro fertilization. Fertil Steril. 2011b;96:672–677. doi: 10.1016/j.fertnstert.2011.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom MS, Parsons PJ, Kim D, Steuerwald AJ, Vaccari S, Cheng G, Fujimoto VY. Toxic trace metals and embryo quality indicators during in vitro fertilization (IVF) Reprod Toxicol. 2011c;31:164–170. doi: 10.1016/j.reprotox.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Board P, Coggan M, Johnston P, Ross V, Suzuki T, Webb G. Genetic heterogeneity of the human glutathione transferases: a complex of gene families. Pharmacol Ther. 1990;48:357–369. doi: 10.1016/0163-7258(90)90054-6. [DOI] [PubMed] [Google Scholar]
- Bollati V, Baccarelli A. Environmental epigenetics. Heredity. 2010;105:105–112. doi: 10.1038/hdy.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, Byun HM, Jiang J, Marinelli B, Pesatori AC, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- Borja-Aburto VH, Hertz-Picciotto I, Lopez MR, Farias P, Rios C, Blanco J. Blood lead levels measured prospectively and risk of spontaneous abortion. Am J Epidemiol. 1999;150:590–597. doi: 10.1093/oxfordjournals.aje.a010057. [DOI] [PubMed] [Google Scholar]
- Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180:462–467. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromer JG, Zhou Y, Taylor MB, Doherty L, Taylor HS. Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. FASEB J. 2010;24:2273–2280. doi: 10.1096/fj.09-140533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruchova H, Vasikova A, Merkerova M, Milcova A, Topinka J, Balascak I, Pastorkova A, Sram RJ, Brdicka R. Effect of maternal tobacco smoke exposure on the placental transcriptome. Placenta. 2010;31:186–191. doi: 10.1016/j.placenta.2009.12.016. [DOI] [PubMed] [Google Scholar]
- Buck Louis GM, Lynch CD, Cooney MA. Environmental influences on female fecundity and fertility. Semin Reprod Med. 2006;24:147–155. doi: 10.1055/s-2006-944421. [DOI] [PubMed] [Google Scholar]
- Cabaton NJ, Wadia PR, Rubin BS, Zalko D, Schaeberle CM, Askenase MH, Gadbois JL, Tharp AP, Whitt GS, Sonnenschein C, et al. Perinatal exposure to environmentally relevant levels of bisphenol A decreases fertility and fecundity in CD-1 mice. Environ Health Perspect. 2011;119:547–552. doi: 10.1289/ehp.1002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calafat AM, Weuve J, Ye X, Jia LT, Hu H, Ringer S, Huttner K, Hauser R. Exposure to bisphenol A and other phenols in neonatal intensive care unit premature infants. Environ Health Perspect. 2009;117:639–644. doi: 10.1289/ehp.0800265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- California Occupational Blood Lead Registry. www.cdph.ca.gov/programs/olppp/Pages/Registry.aspx , accessed July 20th, 2011, State of California Department of Public Health. [Google Scholar]
- CDC. Atlanta, GA: U.S. Centers for Disease Control and Prevention; 2010. Fourth National Report on human exposure to environmental chemicals-updated tables, July 2010. [Google Scholar]
- Chang S-H, Cheng B-H, Lee S-L, Chuang H-Y, Yang C-Y, Sung F-C, Wu T-N. Low blood lead concentration in association with infertility in women. Environ Res. 2006;101:380–386. doi: 10.1016/j.envres.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Choy CMY, Lam CWK, Cheung LTF, Briton-Jones CM, Cheung LP, Haines CJ. Infertility, blood mercury concentrations and dietary seafood consumption: a case–control study. BJOG. 2002;109:1121–1125. doi: 10.1111/j.1471-0528.2002.02084.x. [DOI] [PubMed] [Google Scholar]
- Cole DC, Wainman B, Sanin LH, Weber JP, Muggah H, Ibrahim S. Environmental contaminant levels and fecundability among non-smoking couples. Reprod Toxicol. 2006;22:13–19. doi: 10.1016/j.reprotox.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Dhillon VS, Shahid M, Husain SA. Associations of MTHFR DNMT3b 4977 bp deletion in mtDNA and GSTM1 deletion, and aberrant CpG island hypermethylation of GSTM1 in non-obstructive infertility in Indian men. Mol Hum Reprod. 2007;13:213–222. doi: 10.1093/molehr/gal118. [DOI] [PubMed] [Google Scholar]
- Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TM, Myers JP. Environmental exposures and gene regulation in disease etiology. Environ Health Perspect. 2007;115:1264–1270. doi: 10.1289/ehp.9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenlaub-Ritter U, Vogt E, Cukurcam S, Sun F, Pacchierotti F, Parry J. Exposure of mouse oocytes to bisphenol A causes meiotic arrest but not aneuploidy. Mutat Res- Gen Toxicol Environ Mutag. 2008;651:82–92. doi: 10.1016/j.mrgentox.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Etcheverry A, Aubry M, de Tayrac M, Vauleon E, Boniface R, Guenot F, Saikali S, Hamlat A, Riffaud L, Menei P, et al. DNA methylation in glioblastoma: impact on gene expression and clinical outcome. BMC Genomics. 2010;11:701. doi: 10.1186/1471-2164-11-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto VY, Kim D, vom Saal FS, Lamb JD, Taylor JA, Bloom MS. Serum unconjugated bisphenol A concentrations in women may adversely influence oocyte quality during in vitro fertilization. Fertil Steril. 2011;95:1816–1819. doi: 10.1016/j.fertnstert.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Hengstler JG, Foth H, Gebel T, Kramer P-J, Lilienblum W, Schweinfurth H, Völkel W, Wollin K-M, Gundert-Remy U. Critical evaluation of key evidence on the human health hazards of exposure to bisphenol A. Crit Rev Toxicol. 2011;41:263–291. doi: 10.3109/10408444.2011.558487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SM, Tang WY, Belmonte De Frausto J, Prins GS. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66:5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PA, Koehler KE, Susiarjo M, Hodges CA, Ilagan A, Voigt RC, Thomas S, Thomas BF, Hassold TJ. Bisphenol A exposure causes meiotic aneuploidy in the female mouse. Curr Biol. 2003;13:546–553. doi: 10.1016/s0960-9822(03)00189-1. [DOI] [PubMed] [Google Scholar]
- Jiang G, Xu L, Song S, Zhu C, Wu Q, Zhang L, Wu L. Effects of long-term low-dose cadmium exposure on genomic DNA methylation in human embryo lung fibroblast cells. Toxicology. 2008;244:49–55. doi: 10.1016/j.tox.2007.10.028. [DOI] [PubMed] [Google Scholar]
- Kandaraki E, Chatzigeorgiou A, Livadas S, Palioura E, Economou F, Koutsilieris M, Palimeri S, Panidis D, Diamanti-Kandarakis E. Endocrine disruptors and Polycystic ovary syndrome (PCOS): elevated serum levels of bisphenol A in women with PCOS. J Clin Endocrinol Metab. 2011;96:E480–E484. doi: 10.1210/jc.2010-1658. [DOI] [PubMed] [Google Scholar]
- Mannervik B. The isoenzymes of glutathione transferase. Adv Enzymol Relat Areas Mol Biol. 1985;57:357–417. doi: 10.1002/9780470123034.ch5. [DOI] [PubMed] [Google Scholar]
- Matthews JB, Twomey K, Zacharewski TR. In vitro and in vivo interactions of bisphenol A and its metabolite, bisphenol A glucuronide, with estrogen receptors α and β. Chem Res Toxicol. 2001;14:149–157. doi: 10.1021/tx0001833. [DOI] [PubMed] [Google Scholar]
- Mendola P, Messer LC, Rappazzo K. Science linking environmental contaminant exposures with fertility and reproductive health impacts in the adult female. Fertil Steril. 2008;89:e81–e94. doi: 10.1016/j.fertnstert.2007.12.036. [DOI] [PubMed] [Google Scholar]
- Minnich MG, Miller DC, Parsons PJ. Determination of As, Cd, Pb, and Hg in urine using inductively coupled plasma mass spectrometry with the direct injection high efficiency nebulizer. Spectrochim Acta Part B At Spectrosc. 2008;63:389–395. [Google Scholar]
- Mok-Lin E, Ehrlich S, Williams PL, Petrozza J, Wright DL, Calafat AM, Ye X, Hauser R. Urinary bisphenol A concentrations and ovarian response among women undergoing IVF. Int J Androl. 2010;33:385–393. doi: 10.1111/j.1365-2605.2009.01014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson HH, Marsit CJ, Kelsey KT. ‘Global methylation’ in exposure biology and translational medical science. Environ Health Perspect. 2011;119:1523–1528. doi: 10.1289/ehp.1103423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbold RR. Prenatal exposure to diethylstilbestrol (DES) Fertil Steril. 2008;89:e55–e56. doi: 10.1016/j.fertnstert.2008.01.062. [DOI] [PubMed] [Google Scholar]
- Padmanabhan V, Siefert K, Ransom S, Johnson T, Pinkerton J, Anderson L, Tao L, Kannan K. Maternal bisphenol-A levels at delivery: a looming problem? J Perinatol. 2008;28:258–263. doi: 10.1038/sj.jp.7211913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CD, Lewis ME, Jr, Geraghty CM, Barbosa F, Jr, Parsons PJ. Determination of lead, cadmium and mercury in blood for assessment of environmental exposure: a comparison between inductively coupled plasma-mass spectrometry and atomic absorption spectrometry. Spectrochim Acta Part B At Spectrosc. 2006;61:980–990. [Google Scholar]
- Pearson WR, Vorachek WR, Xu SJ, Berger R, Hart I, Vannais D, Patterson D. Identification of class-mu glutathione transferase genes GSTM1-GSTM5 on human chromosome Ip13. Am J Hum Genet. 1993;53:220–233. [PMC free article] [PubMed] [Google Scholar]
- Peng DF, Razvi M, Chen H, Washington K, Roessner A, Schneider-Stock R, El-Rifai W. DNA hypermethylation regulates the expression of members of the Mu-class glutathione S-transferases and glutathione peroxidases in Barrett's adenocarcinoma. Gut. 2009;58:5–15. doi: 10.1136/gut.2007.146290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins GS, Tang WY, Belmonte J, Ho SM. Perinatal exposure to oestradiol and bisphenol A alters the prostate epigenome and increases susceptibility to carcinogenesis. Basic Clin Pharmacol Toxicol. 2008;102:134–138. doi: 10.1111/j.1742-7843.2007.00166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter CA, Birnbaum LS, Farabollini F, Newbold RR, Rubin BS, Talsness CE, Vandenbergh JG, Walser-Kuntz DR, vom Saal FS. In vivo effects of bisphenol A in laboratory rodent studies. Reprod Toxicol. 2007;24:199–224. doi: 10.1016/j.reprotox.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero R, Friel LA, Velez Edwards DR, Kusanovic JP, Hassan SS, Mazaki-Tovi S, Vaisbuch E, Kim CJ, Erez O, Chaiworapongsa T, et al. A genetic association study of maternal and fetal candidate genes that predispose to preterm prelabor rupture of membranes (PROM) Am J Obstet Gynecol. 2010;203:361.e1–361.e30. doi: 10.1016/j.ajog.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic inuit. Environ Health Perspect. 2008;116:1547–1552. doi: 10.1289/ehp.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajiki J, Takahashi K, Yonekubo J. Sensitive method for the determination of bisphenol-A in serum using two systems of high-performance liquid chromatography. J Chromatogr B Biomed Appl. 1999;736:255–261. doi: 10.1016/s0378-4347(99)00471-5. [DOI] [PubMed] [Google Scholar]
- Sata F, Yamada H, Kondo T, Gong Y, Tozaki S, Kobashi G, Kato EH, Fujimoto S, Kishi R. Glutathione S-transferase M1 and T1 polymorphisms and the risk of recurrent pregnancy loss. Mol Hum Reprod. 2003;9:165–169. doi: 10.1093/molehr/gag021. [DOI] [PubMed] [Google Scholar]
- Schisterman EF, Vexler A, Whitcomb BW, Liu A. The limitations due to exposure detection limits for regression models. Am J Epidemiol. 2006;163:374–383. doi: 10.1093/aje/kwj039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J, Yuan L, Xiao Q, Chiorazzi N, Budman D, Teichberg S, Xu HP. TSP50, a possible protease in human testes, is activated in breast cancer epithelial cells. Cancer Res. 2002;62:290–294. [PubMed] [Google Scholar]
- Shkolnik K, Tadmor A, Ben-Dor S, Nevo N, Galiani D, Dekel N. Reactive oxygen species are indispensable in ovulation. Proc Natl Acad Sci USA. 2011;108:1462–1467. doi: 10.1073/pnas.1017213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange RC, Matharoo B, Faulder GC, Jones P, Cotton W, Elder JB, Deakin M. The human glutathione S-transferase: a case–control study of the incidence of the GST1 0 phenotype in patients with adenocarcinoma. Carcinogenesis. 1991;12:25–28. doi: 10.1093/carcin/12.1.25. [DOI] [PubMed] [Google Scholar]
- Sugiura-Ogasawara M, Ozaki Y, Sonta SI, Makino T, Suzumori K. Exposure to bisphenol A is associated with recurrent miscarriage. Hum Reprod. 2005;20:2325–2329. doi: 10.1093/humrep/deh888. [DOI] [PubMed] [Google Scholar]
- Susiarjo M, Hassold TJ, Freeman E, Hunt PA. Bisphenol A exposure in utero disrupts early oogenesis in the mouse. PLoS Genet. 2007;3:0063–0070. doi: 10.1371/journal.pgen.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Tsutsumi O, Ikezuki Y, Takai Y, Taketani Y. Positive relationship between androgen and the endocrine disruptor, bisphenol A, in normal women and women with ovarian dysfunction. Endocr J. 2004;51:165–169. doi: 10.1507/endocrj.51.165. [DOI] [PubMed] [Google Scholar]
- Thompson J, Bannigan J. Cadmium: toxic effects on the reproductive system and the embryo. Reprod Toxicol. 2008;25:304–315. doi: 10.1016/j.reprotox.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Valko M, Leibfritz D, Moncol J, Cronin MTD, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Van Poppel G, De Vogel N, Van Balderen PJ, Kok FJ. Increased cytogenetic damage in smokers deficient in glutathione S-transferase isozyme. Carcinogenesis. 1992;13:303–305. doi: 10.1093/carcin/13.2.303. [DOI] [PubMed] [Google Scholar]
- Vandenberg LN, Chahoud I, Heindel JJ, Padmanabhan V, Paumgartten FJR, Schoenfelder G. Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Environ Health Perspect. 2010;118:1055–1070. doi: 10.1289/ehp.0901716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vom Saal FS, Akingbemi BT, Belcher SM, Birnbaum LS, Crain DA, Eriksen M, Farabollini F, Guillette LJ, Jr, Hauser R, Heindel JJ, et al. Chapel Hill bisphenol A expert panel consensus statement: integration of mechanisms, effects in animals and potential to impact human health at current levels of exposure. Reprod Toxicol. 2007;24:131–138. doi: 10.1016/j.reprotox.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng YI, Hsu PY, Liyanarachchi S, Liu J, Deatherage DE, Huang YW, Zuo T, Rodriguez B, Lin CH, Cheng AL, et al. Epigenetic influences of low-dose bisphenol A in primary human breast epithelial cells. Toxicol Appl Pharmacol. 2010;248:111–121. doi: 10.1016/j.taap.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetherill YB, Akingbemi BT, Kanno J, McLachlan JA, Nadal A, Sonnenschein C, Watson CS, Zoeller RT, Belcher SM. In vitro molecular mechanisms of bisphenol A action. Reprod Toxicol. 2007;24:178–198. doi: 10.1016/j.reprotox.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Wright RO, Schwartz J, Wright RJ, Bollati V, Tarantini L, Park SK, Hu H, Sparrow D, Vokonas P, Baccarelli A. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect. 2010;118:790–795. doi: 10.1289/ehp.0901429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HC, Delgado-Cruzata L, Flom JD, Kappil M, Ferris JS, Liao Y, Santella RM, Terry MB. Global methylation profles in DNA from different blood cell types. Epigenetics. 2011;6:76–85. doi: 10.4161/epi.6.1.13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SJ, Wang YP, Roe B, Pearson WR. Characterization of the human class Mu glutathione S-transferase gene cluster and the GSTM1 deletion. J Biol Chem. 1998;273:3517–3527. doi: 10.1074/jbc.273.6.3517. [DOI] [PubMed] [Google Scholar]
- Yaoi T, Itoh K, Nakamura K, Ogi H, Fujiwara Y, Fushiki S. Genome-wide analysis of epigenomic alterations in fetal mouse forebrain after exposure to low doses of bisphenol A. Biochem Biophys Res Commun. 2008;376:563–567. doi: 10.1016/j.bbrc.2008.09.028. [DOI] [PubMed] [Google Scholar]
- Younglai EV, Foster WG, Hughes EG, Trim K, Jarrell JF. Levels of environmental contaminants in human follicular fluid, serum, and seminal plasma of couples undergoing in vitro fertilization. Arch Environ Contam Toxicol. 2002;43:121–126. doi: 10.1007/s00244-001-0048-8. [DOI] [PubMed] [Google Scholar]
- Younglai EV, Holloway AC, Foster WG. Environmental and occupational factors affecting fertility and IVF success. Hum Reprod Update. 2005;11:43–57. doi: 10.1093/humupd/dmh055. [DOI] [PubMed] [Google Scholar]
- Yuan L, Shan J, De Risi D, Broome J, Lovecchio J, Gal D, Vinciguerra V, Hao-Peng X. Isolation of a novel gene, TSP50, by a hypomethylated DNA fragment in human breast cancer. Cancer Res. 1999;59:3215–3221. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.