Summary
Most lung adenocarcinoma-associated Epidermal Growth Factor Receptor (EGFR) mutations confer sensitivity to specific EGFR tyrosine kinase inhibitors. The finding that exon 19 insertion mutations are also sensitive to this class of drugs suggests that testing for these mutations should be performed and that these patients will benefit from treatment with tyrosine kinase inhibitors.
In this issue of Clinical Cancer Research, He and colleagues (1) demonstrate that a rare class of EGFR mutations, namely exon 19 insertions are associated with sensitivity to tyrosine kinase inhibitors providing rationale for testing of these mutations in lung adenocarcinoma patients.
Ninety percent of all lung adenocarcinoma-associated EGFR mutations are in frame deletions in exon 19 or a point mutation in exon 21 (L858R) and fall within the exons that encode the tyrosine kinase domain of the receptor (2–4). These mutations confer transforming ability to the receptor and lead to its constitutive activation (5). EGFR mutations are most frequently found in tumors from never-smokers and East-Asians. Retrospective and prospective studies have established that 70% of tumors harboring these mutations respond to treatment with the EGFR tyrosine kinase inhibitors, gefitinib or erlotinib (6).
The progression-free survival and response rate of patients with EGFR mutant tumors treated with an EGFR TKI is superior to standard chemotherapeutic regimens (7, 8). Despite the promising results, the development of acquired resistance to these therapies is almost inevitable. Drug resistance emerges most frequently as a result of a secondary mutation in exon 20 of EGFR that leads to substitution of a methionine for a threonine at position 790 [T790M, (9)]. The T790M mutation is almost always observed in conjunction with a sensitivity-conferring mutation and is only rarely found in tumors prior to treatment with an EGFR TKI.
The remaining ten percent of EGFR mutations found in lung adenocarcinomas include insertions in exon 20 (about 4% of EGFR mutations) and point mutations that modify codons G719 (to A, C or S; 3%) and L861 (to Q; 2%) (10, 11). Point mutations that alter these latter residues are also transforming and confer sensitivity to EGFR TKIs. In contrast, while exon 20 insertions are capable of transforming cells, erlotinib and gefitinib are not effective on these EGFR mutants in vitro or in the clinic (11). As these results demonstrate the clinical management of patients with EGFR mutant tumors depends upon the nature of the mutation present and therefore requires accurate and comprehensive mutation detection strategies.
The manuscript in this issue of Clinical Cancer Research represents the first effort to comprehensively characterize the frequency and sensitivity of exon 19 insertion mutations to EGFR TKIs. By retrospectively analyzing mutational data of non- small cell lung cancer the authors identified eight exon 19 insertions, representing 1% of all EGFR mutations. The authors also identified an additional four tumors from other centers. Like most EGFR mutations, exon 19 insertions are associated with adenocarcinoma histology and a null or limited smoking history. Three of the four patients with metastatic disease responded to TKI suggesting that EGFR exon 19 insertion mutations have a similar response rate to TKIs as exon 19 deletion mutations and the L858R, G719X and L861Q point mutations. It remains unclear whether these mutations have the same progression free and overall survival as the classic mutations.
Molecular modeling and crystallographic studies of EGFR have provided insight into the effect of different EGFR mutations on the structure of the tyrosine kinase domain. In particular, they have shed light on how the mutations may lead to constitutive activation of the kinase and affect sensitivity to TKIs. The tyrosine kinase domain of EGFR has two lobes: a smaller N-lobe and a larger C-lobe. The correct positioning of the C-helix (within the N-lobe) and the activation loop (within the C-lobe) are required for activation of the EGFR tyrosine kinase domain. In wild-type EGFR, ligand binding and receptor dimerization lead to the asymmetric interaction of the kinase domains of the two receptor dimers leading to correct positioning of both the C-helix and the activation loop thus favoring the active conformation of the kinase.
The L858 residue lies tucked in a hydrophobic pocket in the activation loop of the kinase when EGFR is in the inactive state. Substitution of leucine for arginine causes the activation loop to “flip out” destabilizing the inactive conformation and favoring the active conformation (12). Exon 19 deletion mutations occur in a protein strand (called the β3 strand) adjacent to the C-helix. Although crystal structures of these mutants have been elusive, it is postulated that reducing the length of this strand may favor the active conformation of the kinase. Interestingly, a number of different exon 19 deletion mutations are observed in lung cancers and the most common ones all lead to amino acid substitutions of residue L747. Similarly, the exon 19 insertion mutations all lead to substitution of residue L747. However, the exon 19 insertion mutations appear from these initial studies more uniform in length than the exon 19 deletions and always lead to substitution of L747 to proline. It is possible that exon 19 insertions also alter the position of the C-helix favoring the active conformation of the kinase. Similarly, exon 20 insertion mutations, which occur at the beginning of the C-helix also are likely to stabilize the active conformation of the kinase. Crystallographic studies of the exon 19 and 20 insertion and deletion mutations will be valuable to understand how these protein modifications affect sensitivity to TKIs.
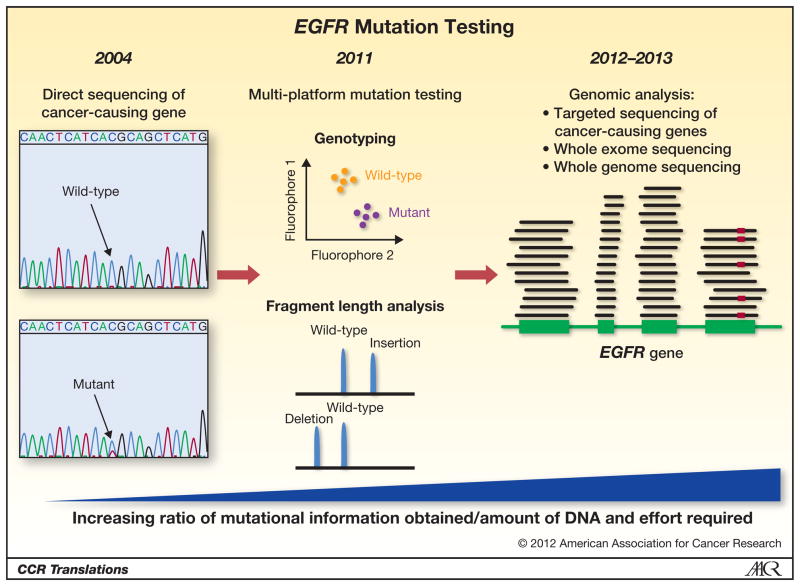
The sensitivity of EGFR exon 19 insertions to TKIs together with the low prevalence of these mutations in patients with lung adenocarcinomas brings up the important issue of whether to incorporate routine testing for them (and other rare but “actionable” mutations) in the clinic. Currently several institutions are pioneering the testing of all lung adenocarcinoma samples for known driver mutations most of which are point mutations in genes such as EGFR, KRAS, BRAF and PIK3CA. Compared to the identification of point mutations, detecting insertions and deletions is more complex. This is due to the fact that deletions and insertions can vary with respect to the size and nature of the mutation, characteristics that are difficult to distinguish with many genotyping platforms. Fragment length analysis can overcome these problems and is a useful complementary test to genotyping analyses. Clinically, multi-platform analyses that combine these techniques are increasingly important as we gain more information on rare mutations and correlate specific mutations with response to different targeted therapies (Figure 1). In the near future, these multi-platform analyses are likely to be replaced by targeted sequencing of cancer-related genes using new generation sequencing platforms and eventually whole exome or whole genome sequencing.
Figure 1. The evolution of EGFR mutation testing.
Direct sequencing of EGFR was initially used to detect mutations when they were first identified (left panel). More recently, as sequencing technologies have evolved, genotyping assays are being used to screen for point mutations complemented by fragment length analyses to accurately detect insertion and deletion mutations in EGFR and other genes (middle panel). Within the next 1–2 years these technologies will be replaced by high-throughput sequencing of cancer-related genes including EGFR (right panel), which will allow for the identification of all mutations in the gene using one assay. In the right panel, the EGFR gene is depicted in green. The horizontal lines represent sequencing reads that cover exonic regions of the gene. The red mark indicates the presence of a mutation in a read.
As molecular profiling of tumors enters mainstream practice, we will begin to identify a wider spectrum of mutations, some of them that confer sensitivity to clinically available therapies or drugs in trials. One limitation of current therapy of EGFR mutant lung cancer is that the existing inhibitors have been tailored to inhibit the wild type kinase. It is entirely possible that in the future we will have mutation specific inhibitors with differential activity based on the exact EGFR mutation identified. This exciting prospect of tailoring therapy to individual tumors poses challenges: Which assays should be used to ensure that all varieties of mutations are identified? How can we rapidly assess whether novel mutations are clinically significant and respond to specific drugs? Should we test for all mutations even though their presence may be extremely rare? As elegantly presented in this paper, gathering information on rare mutations may be valuable for patient care.
References
- 1.He M, Capelletti M, Nafa K, Yun CH, Arcila M, Miller V, et al. EGFR Exon 19 Insertions: A new family of Sensitizing EGFR Mutations in Lung Adenocarcinoma. Clincal Cancer Research. 2012 doi: 10.1158/1078-0432.CCR-11-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science (New York, NY) 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greulich H, Chen TH, Feng W, Janne PA, Alvarez JV, Zappaterra M, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS medicine. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nature reviews Cancer. 2010;10:760–74. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. The New England journal of medicine. 2009;361:947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 8.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. The lancet oncology. 2010;11:121–8. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 9.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS medicine. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitsudomi T, Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. The FEBS journal. 2010;277:301–8. doi: 10.1111/j.1742-4658.2009.07448.x. [DOI] [PubMed] [Google Scholar]
- 11.Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. The lancet oncology. 2011 doi: 10.1016/S1470-2045(11)70129-2. [DOI] [PubMed] [Google Scholar]
- 12.Kumar A, Petri ET, Halmos B, Boggon TJ. Structure and clinical relevance of the epidermal growth factor receptor in human cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2008;26:1742–51. doi: 10.1200/JCO.2007.12.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]