Abstract
The biological functions of glycoconjugate glycans arise in the context of structural heterogeneity resulting from non-template driven biosynthetic reactions. Such heterogeneity is particularly apparent for the glycosaminoglycan (GAG) classes, of which heparan sulfate (HS) is of particular interest for its properties in binding to many classes of growth factors and growth factor receptors. The structures of HS chains vary according to spatial and temporal factors in biological systems as a mechanism where by the functions of the relatively limited number of associated proteoglycan core proteins is elaborated. Thus, there is a strong driver for the development of methods to discover functionally relevant structures in HS preparations for different sources. In the present work, a set of targeted tandem mass spectra were acquired in automated mode on HS oligosaccharides deriving from two different tissue sources. Statistical methods were used to determine the precursor and product ions, the abundances of which differentiate between the tissue sources. The results demonstrate considerable potential for using this approach to constrain the number of positional glycoform isomers present in different biological preparations toward the end of discovery of functionally relevant structures.
Keywords: Glycan, glycosaminoglycan, heparan sulfate, tandem MS, hierarchical clustering, electrospray, glycomics
Introduction
Heparan sulfate (HS) is a glycosaminoglycan (GAG) that consists of repeating disaccharides units containing variable modifications. HS is present on all animal cell surfaces and directly interacts with myriad of extracellular signaling molecules. It is required for embryonic development [1] and for the functioning of every adult physiological system [2]. The interactions between many families of growth factors and growth factor receptors are modulated depending on the structures of HS expressed on cell surfaces and extracellular matrices. Thus, it is not surprising that HS structural biochemistry is central to understanding of disease mechanisms including tumor growth, angiogenesis, amyloid deposition, tissue remodeling and repair, and host-pathogen interactions.
Biosynthesis of HS begins in the endoplasmic reticulum with the addition of a tetrasaccharide linker that is attached to a proteoglycan serine residue via a xylose monosaccharide. The saccharide chains are extended and subsequently modified by a series of enzymes in the Golgi apparatus. Nascent HS chains consist of repeating disaccharide units of glucuronic acid (GlcUA) and N-acetylglucosamine (GlcNAc) [4GlcAβ1-4GlcNAcβ1-] that undergo a series of modification reactions. The chains are first acted upon by N-deacetylase/N-sulfotransferase (NDST) enzymes that remove N-acetate groups and replace them with N-sulfate groups. Heparins, expressed in connective tissue mast cell granules, are a class of HS in which the NDSTs act to modify nearly all of the GlcNAc residues in the chain. In most other tissues, the NDSTs create HS chains with domains of high degree of N-sulfation and those with a high degree of N-acetylation and those with intermediate content. Following the actions of the NDSTs, the chains undergo O-sulfation and/or uronic acid epimerization. Such modifications occur at specific sites. The most common disaccharide of heparin is 4IdoUA(2S)α1-4GlcNS(6S)α1, where 2S = 2-O-sulfation and 6S = 6-O-sulfation. Addition of a sulfate to position 3 of the glucosamine is a rare but biologically significant modification. The general structure of HS from most tissues differs from that of heparin in that unsulfated and intermediate sulfation domains are present.
HS chains are heterogeneous by nature and expressed in a spatially and temporally regulated manner. Thus, biomedical investigations require HS structural information from samples isolated from specific tissues and disease states. The analytical challenge is to determine the structures of HS chains, given their inherent heterogeneity of composition with respect to biosynthetic modifications. For a given composition, it is possible for a set of positional isomers to be present, and this must be taken into consideration. Analytical strategies for HS analysis include (1) chemical or enzymatic partial hydrolysis of labeled or unlabeled HS [3, 4]; (2) separation using liquid chromatography modes including size exclusion, strong anion exchange, reverse-phase, reversed phase ion-pairing chromatography or capillary electrophoresis [5]; (3) comparative detection between the different conditions of hydrolysis. If each strategy adopted in the analysis of HS is a source of information concerning the structure of such compounds, the use of complementary strategies is necessary to obtain complete sequence information.
Two approaches used widely for the structural elucidation of HS are nuclear magnetic resonance (NMR) spectroscopy [6] and mass spectrometry (MS) [7–10]. The use of NMR provides detailed information regarding uronic acid isomers and positions of sulfate and acetate groups. The major limitation of this technique is the large amount of starting material required for analysis. Furthermore, NMR has been used primarily to characterize smaller oligomers, limited typically to hexasaccharides. Tandem MS has been proved to be a useful, highly reproducible and very sensitive means for the characterization of GAG disaccharides [11–17]. A typical approach to identify and quantify HS oligosaccharide isomer mixtures was developed using tandem MS and consisted of comparison of the product ion profiles of unknown isomer mixtures with those acquired from pure isomer standards [14, 18–22]. Selected fragment ions of the pure standards were observed to be diagnostic for the abundances of the isomeric compounds in the mixture. A correlation coefficient was calculated for each fragment ion of the standards and a system of equations allowed calculation of the proportion of each standard in the sample [14]. Pure oligosaccharide standards are available for chondroitin sulfate (CS) GAGs [14, 15, 19, 21–26] but unfortunately not for HS [13, 16]. In addition, because the compounds are typically present in isomeric mixtures when obtained from biological sources, such methods report the mixture percentages of the standard isomers and are incapable of determining complete sequence information. During a collisionally activated dissociation (CAD) tandem mass spectrometry experiment, the sulfate groups of HS chains tend to undergo dissociation at lower vibrational energies than do glycosidic bonds [27]. As a result, tandem MS experiments that involve significant vibrational excitation result in uninformative fragmentation of the sulfate groups. The extent to which this occurs is minimized as precursor ion charge states increase, and may also be minimized with the use of cations to pair with the negatively charged sulfate groups [27–29]. Tandem MS sequencing of HS oligosaccharides using CAD, electron capture dissociation, electron detachment dissociation or infrared multiphoton dissociation [29–33] is straightforward because of the competing fragmentation channels involved during the experiment. Post fragmentation, four types of fragment ions are observed in the tandem mass spectra of HS: loss of HSO4−, loss of neutral SO3, glycosidic bond cleavage and cross-ring cleavage [5]. Although glycosidic bond and cross-ring cleavages contain the most useful structural information, interpretation of the tandem MS is complicated by the presence of the accompanying SO3 losses. Under ideal conditions, complementary pairs of glycosidic bonds or cross-ring cleavages provide definitive structural information for the complete molecule; however, such pairs are often not observed. Furthermore, for most biological samples a mixture of HS isomers is present for a given oligosaccharide composition, and this complicates the product ion spectra.
In addition, the concept of direct sequencing is not applicable to HS samples that consist of mixtures of isomers. Although it is possible to reduce mixture complexity using multi-step chromatographic purification, this effort is only justified for a sample found to have significant biological activity. For the purpose of discovery and analysis of oligosaccharides of biological and/or therapeutic interest for HS/heparin preparations, it is necessary to interpret information produced directly from samples containing isomeric mixtures. Therefore, an appropriate method for interpretation of such data is essential.
In proteomics, tandem mass spectrometric data are used to search databases generated from genomic information. For such searches, homogeneity of the peptides is generally assumed in order to produce a sequence that matches the product ions observed. For glycomics analysis, product ion m/z and charge state values define monosaccharide compositions and the ion abundances correlate with the type of oligosaccharide isomer. Thus, it is necessary to consider product ion m/z values, charge states and abundances in interpreting the data. While the comparison of tandem MS data to databases or libraries using bioinformatics tools allows facile identification or sequencing of peptides [34–37], the process differs markedly for glycomics. Bioinformatics tools [38–42] as software of interpretation of MS and tandem MS data through different database are available for carbohydrates and GAGs [43, 44] but do not allow for the interpretation of isomeric mixtures. As a result, we have undertaken an effort to meet this need.
Two approaches commonly used to compare raw mass spectral data are clustering analysis and principal components analysis. Cluster analysis consists of dividing data into groups (clusters) in order to capture the natural structure of the data. One of the first references to clustering in the mass spectrometry field was for comparison of alkyl thiolesters and pharmaceutical products [45, 46]. The cluster analysis was used here to help the interpretation of tandem data and classify them in cluster representing the different classes of samples. Clustering analysis of tandem MS data was applied in the proteomics field for the following purposes: (1) to reduce the number of tandem mass spectra used in the identification of proteins by regrouping similar tandem data to decrease the redundancies of the analyses [47–49]; (2) to improve the understanding of fragmentation patterns (fragmentation vs. intensity) and open the area of improved protein identification algorithms [50] and (3) in label free quantification experiments for the discovery of biomarkers [51–54].
We have developed a novel fully automated approach based on the new interpretation of tandem MS data of HS oligosaccharides extracted from different organ tissues. We applied this approach to a set of data acquired using an automated CAD tandem MS acquisition parameters. Tandem MS data on 13 targeted precursor ions were acquired on HS oligosaccharides extracted from each of two bovine tissues (aorta, lung) automatically in quadruplicate. The size of the corresponding data set was approximately 1000 features (product ions), for which manual comparison was not feasible. A strategy based on software and bio informatics tools used commonly in the proteomics and biomarkers field was developed. Agglomerative hierarchical clustering (AHC) was used on the tandem MS data to demonstrate that sufficient information for differentiation of isomeric glycoforms in the four organs samples was present. The analysis was useful for recognition of fragmentation patterns corresponds to organ-specific HS structures.
Material and Methods
Preparation of the HS sample
Heparin lyase III from Flavobacterium heparinum was purchased from Ibex (Montreal, Canada). Heparan sulfate samples from bovine organs aorta (A) and lung (L) were a generous gift from Dr. Keiichi Yoshida and were prepared as described previously [55]. The extracted HS from bovine aorta and lung were depolymerized to completion using heparin lyase III as previously described [56]. Oligosaccharides were then fractionated using a Superdex Peptide 3.2/30 size exclusion column (GE Healthcare) equilibrated with 50 mM ammonium acetate using UV detection at 232 nm. The HS oligosaccharides present in fractions 15 from each tissue (Supplemental Figure 1) were analyzed using tandem mass spectrometry as described below.
Acquisition of mass spectral data
A ThermoFisher Scientific (San Jose, California) LTQ Orbitrap Discovery mass spectrometer coupled with a Triversa Nanomate system (Advion Biosystems, Inc., Ithaca, NY) was operated in negative ion mode. The Orbitrap ion optics were tuned to eliminate the fragmentation of the HS compounds during the ejection of ions from the ion trap and subsequent transmission to the C-trap. The temperature of the transfer line was set at 150°C. The Triversa Nanomate allowed automated acquisition of MS and tandem MS data.
Specific nomenclature
The following nomenclature is used throughout to define the compositions of HS oligoaccharides: [A,B,C,D,E] represents the number of units in the following composition: [ΔHexA, HexA, GlcN, SO3, Ac]. For example, the composition [1,2,3,4,1] is constituted by 1 unsaturated hexuronic acid, 2 hexuronic acid, 3 glucosamine, 4 sulfate groups and one acetate group. The Domon-Costello nomenclature [58] was used to describe the fragmentation pattern of HS compounds (Supplemental Figure 2). For the tandem MS product ions from GAG oligosaccharides, the number of sulfate (S) and acetate (Ac) groups is given in parenthesis after the product ion, i.e. Y2 (S, Ac).
Nano-Electrospray Conditions
Proper control of the electrospray source conditions was determined to be essential for generation of high quality tandem mass spectra from HS oligosaccharides [57]. In summary, if the ion charge state was higher than the number of sulfate groups of the oligosaccharides, the tandem mass spectra were rich in structural information. Otherwise, the tandem mass spectra contained abundant ions produced from the dissociation of sulfate groups. The charge state of the precursor ion is dependent on the nano-electrospray conditions. We observed that by dissolving the HS oligosaccharides 50% methanol, a combination of high ion charge state and stable nano-electrospray ions were obtained. The Triversa Nanomate method was programmed for 5 minutes of acquisition with the following parameters: pressure N2:0.7 PSI, voltage −1.3 kV. The nano-electrospray mass spectra showed the presence of 13 precursor ions common to all tissue types. They are represented here using the nomenclature described above: [1,0,1,1,1]−2 (m/z 228.5), [1,0,1,2,0]−3 (m/z 247.5), [1,1,2,3,1]−4 (m/z 238.0), [1,1,2,2,0]−3 (m/z 277.3), [1,1,2,2,1]−3 (m/z 291.0), [1,1,2,3,0]−3 (m/z 303.7), [1,1,2,3,1]−3 (m/z 317.7), [1,1,2,2,0]−2 (m/z 416.1), [1,1,2,2,1]−2 (m/z 437.1), [1,2,3,4,1]−5 (m/z 273.6), [1,2,3,3,1]−3 (m/z 317.7), [1,2,3,3,1]−4 (m/z 322.3), [1,2,3,4,1]−4 (m/z 342.3) and [1,2,3,3,1]−3 (m/z 430.1).
Tandem mass spectrometry acquisition
The method was as follows: one Orbitrap full scan (m/z 200–1,000 with RP 30,000 at m/z 400, 5×104 target value, 500 ms maximum injection time, 1 microscan) was followed by 4 CAD (RP 15,000 with 2×104 target value, 1,000 ms max inject time, 20 microscans, window of isolation 3, Energy 30) detected in the Orbitrap on the 1st, 2nd, 3rd and 4th most abundant ions detected in the corresponding full scan. Tandem and MS spectra were acquired in profile mode. A list of the 13 common precursor ions was obtained and used for the data acquisition. The exclusion time was 300s and the method was run for 5 min. The samples were acquired in quadruplicate technical replicates to allow statistical treatment. The designated precursor ions are shown in Supplemental Figure 1b,c.
Data Analysis
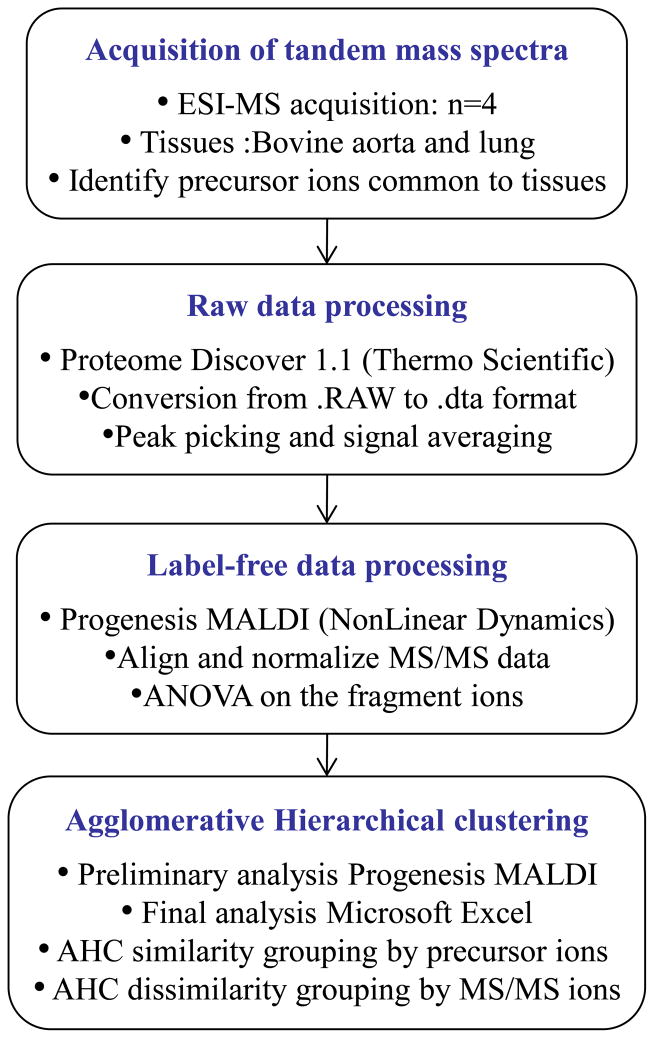
An overview of the data analysis approach is shown in Figure 1. The set of raw data files was processed using Proteome Discoverer version 1.1 (ThermoFisher Scientific) and converted to the Dta file format [59]. The Dta file names were changed manually to simplify the manipulation and of the content of the file: Type of Tissue (Aorta/Lung)_Replicate_(1/2/3/4)_Precursor ion.dta. The Dta file set was then analyzed using Progenesis MALDI version 1.2 (Nonlinear Dynamics, UK). Using Progenesis MALDI we performed spectral alignment and normalized the Dta mass lists across the complete data set of variance. The mass accuracy of the data was reduced to 0.1 Da via binning which did not affect the resulting clustering analysis. Each product ion present in tandem mass spectra from the different tissue type was considered to be an independent feature. Analysis of variance (ANOVA) performed on the product ions measured across complete data set with p-values <0.05 indicated significant changes across the different groups. A total of 78 daughter ions were considered statistically different, with p-values <10−16 (Supplemental Table 1).
Figure 1.
Workflow depicting tandem mass spectrometry data analysis.
Primary statistical analysis using agglomerative hierarchical clustering (AHC) was performed using Progenesis MALDI. AHC is one of the most common statistical tools used to define the degree of similarity/dissimilarity between objects and groups. It allows for iterative grouping or segmentation of objects, here precursor ions or product ions, into clusters to define the proximity of the objects. A distance measurement was defined for each AHC. Further statistical analyses using AHC and manual confirmation were performed using Microsoft Excel 2010. The data were clustered using both similarity and dissimilarity methods. Similarity clustering across precursors was performed to see if the precursors could be grouped dependent upon the origin of the tissue. Pearson’s correlation was used, and varied from 0 to 1 with 0 indicating the variables were uncorrelated. For dissimilarity, it was of interest to determine which product ions of the precursor ions differed most significantly in abundance. The clustering was realized using the Euclidean distance method.
Results and Discussion
Tandem MS on HS oligosaccharides
Interpretation of the tandem mass spectra of a single composition yields significant but incomplete structural information. The main reason is that a single composition generated by lyase III depolymerization yields a complex mixture of positional isomers. Additionally, the complexity of the MS2 data is increased by losses of labile sulfate groups from the precursor and product ions obtained during CAD fragmentation. The loss of sulfate from the precursor ions and product ions is dependent on the number of sulfate groups and the charge state of the ions involved. As the charge state of a precursor ion increases, the repulsion force between the sulfate group increases, thus favoring glycosidic bond dissociation or cross-ring cleavages [13, 16, 21, 60]. In our experience, when the charge state is lower than the number of sulfate groups in the precursor ion, the tandem mass spectrum is dominated by loss of SO3.
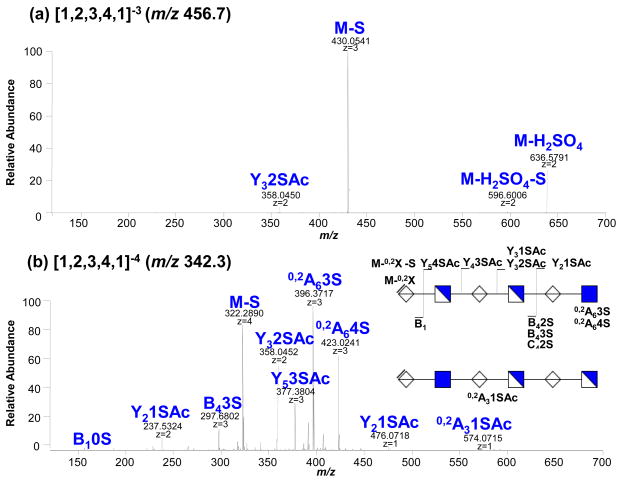
Representative tandem mass spectra of the compounds [1,2,3,4,1]3− (m/z 456.7) and [1,2,3,4,1]4− (m/z 342.3) obtained from bovine aorta tissue are shown in Figure 2. Each HS composition is represented by the numerical code [A, B, C, D, E] corresponding to the number of different of the following carbohydrates [ΔHexA, HexA, GlcN, SO3, Ac]. The tandem mass spectrum of the [1,2,3,4,1]−3 (m/z 456.7) (a) shows abundant product ions from loss of SO3 from the precursor and is therefore nearly devoid of structurally informative ions. In contrast, the tandem mass spectrum of [1,2,3,4,1]−4 (b) is rich in information and contains product ions that clearly identify the position of the acetate group. The product ions B1 and [M-0,2X] indicate that the first hexuronic acid at the non-reducing end does not carry a sulfate group. The complementary product ions Y2(1S,Ac)/B4(3S) and the product ions 0,2A6 (4S) unambiguously localize the acetate group to the first reducing end of the glucosamine residue. The product ion (m/z 574.0792) illustrated another complexity of the tandem data: it could be assigned as B3(1S) or 0,2A3(1S,Ac) product ions with the same error (0.8ppm). The ambiguity could be solved using MS3on this ion, which is not possible here due to low ion abundance. For either interpretation, the data indicate the presence of an isomer with acetate at the glucosamine in position 5, adjacent to the non-reducing end. The isomer with the acetate located at glucosamine 1 (reducing end) is dominant compared to the isomer with acetate at glucosamine in position 5. The relative intensity of the product ions indicating the acetyl group localized on glucosamine 5 is less than 5% while the product ions showing the presence of the acetate at glucosamine 1 ranges between 10% and 70%. A simple combinatorial calculation yields 378 possible isomers for the glycan composition [1,2,3,4,1]. The restriction posed by the predetermined location of the acetate group and by the lack of a sulfate group at the first hexuronic acid at the non-reducing end decreases this number to 70 possible combinations.
Figure 2.
Interpretation of tandem mass spectra of HS components: (a) [1,2,3,4,1]−3 (m/z) and (b) [1,2,3,4,1]4− (m/z) precursors obtained from bovine aorta tissue. Influence of the fragmentation pattern with the charge state and number of sulfates.
Conclusive determination of the location of the sulfate group on the chain is further complicated by the following: product ions may also be generated from sulfate losses from more highly sulfated precursors. This is indicated by the presence of the product ion 0,2A6(3S), [M-0,2X-S] and Y5 (3S) in the tandem mass spectrum of the compound [1,2,3,4,1]4− at 100%, 20% and 35% relative abundances (2b). Loss of sulfate from the product ions becomes more pronounced as the size of the oligosaccharide and the number of sulfates increase and modeling this mechanism is challenging. One way to counter the problem is to compare directly the pattern of fragmentation for an unknown precursor ion with that of a pure standard of known structure [13–16, 19, 22–26] and define the proportion of isomers present. Alternatively, the separation of individual isomers in the mixture may be performed to go further in the interpretation of the tandem mass spectrometry data; however, this is extremely difficult from a chromatography point-of-view and only justified for target oligosaccharides with known high value biological activities. It is therefore appropriate to develop an informatics approach to facilitate structure-function correlation from complex mixtures.
Agglomerative Hierarchical Clustering
Developing methods for global characterization of the differences in isomeric structures of HS preparations for the purpose of biomarker discovery [61–63] or for analysis of therapeutic HS preparations [64] is a primary focus for the present work. Regarding biomarker discovery, it is important to consider the need to screen a large number of compounds in a single experiment. Additionally, characterization of the isomerization of GAGs is crucial in determining their biological function [65–69]. The goal here, using a statistical classification approach with AHC, is to establish a classification-map of tandem MS data that will clearly afford detailed isomeric determination and classification of the HS oligosaccharides dependent upon tissue type and precursor ion m/z. The map would thus yield an exact representation of the tandem mass spectrometric data in terms of similarity of the precursor ions and would thus represent the dataset of features in both the MS and tandem MS space.
We performed two exploratory avenues of data processing using AHC. The first involved similarity clustering based upon precursor m/z to determine if it was feasible to differentiate HS obtained from lung and aorta and to determine the degree of similarity or difference in isoforms of identical m/z precursor ions. The second approach involved application of AHC using dissimilarity measurements on the tandem mass spectra ion series for each unique cluster obtained in the similarity space. The goal was to identify the product ions different in abundances between tissue types and to determine fine structure underlying these differences.
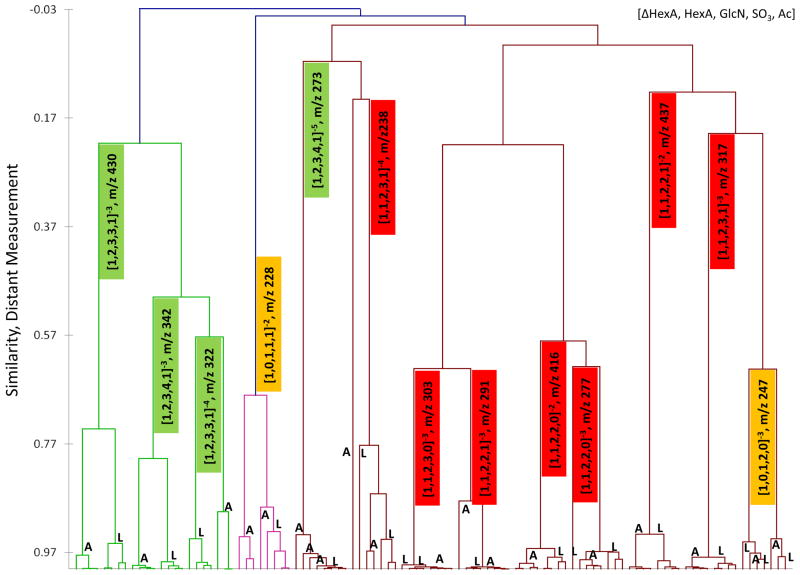
Agglomerative hierarchical clustering was performed on the tandem mass spectra obtained from the most prominent 13 precursor ions obtained from the lyase digest of HS obtained from bovine lung and aorta tissues. Results of similarity clustering of the 13 precursors are summarized in Figure 3 and demonstrate that all precursor ions clustered dependent upon the origin of the tissue and that precursors with the same m/z from lung and aorta clustered together. We observed that considerable variability in the distance measurement existed in the clustering of the precursors, ranging from 0.99, indicating near identical tandem mass spectra, to 0.573, indicating the presence of either novel ions in the tandem mass spectra or differences in ion intensities originating from the different tissues. A complete summation on our observations on similarity reflecting the degree of clustering of the precursors are presented in more detail below and summarized in Table 1.
Figure 3.
Agglomerative hierarchal clustering of heparan sulfate tandem mass spectra from aorta (A) and lung (L). The dendrogram illustrates by clustering (X axis) and distance measurement (Y axis) the degree of similarity of the tandem mass spectra originated from different precursor ions and tissues. The colored boxes indicate the size of the HS oligomer: dp2, yellow; dp4, red; dp6, green.
Table 1.
Overview of clustering for 13 precursor ions common to aorta and lung tissue in terms of and differentiation of glycoforms. DP = degree of polymerization.
| DP | Precursor m/z (z) | Error, ppm | Composition1 | Clustering2 | Differentiation3 | Notes |
|---|---|---|---|---|---|---|
| 2 | 247.5001 (2) | 0.9 | [1,0,1,2,0] | No | No | 3,6 |
| 4 | 238.0135 (4) | 0.3 | [1,1,2,3,1] | No | No | 6 |
| 6 | 273.621 (5) | 0.6 | [1,2,3,4,1] | No | No | 6 |
| 4 | 277.0313 (3) | 0.3 | [1,1,2,2,0] | Yes (0.967) | Yes | 7 |
| 4 | 291.0348 (3) | 0.4 | [1,1,2,2,1] | Yes (0.999) | No | 5 |
| 4 | 303.6836 (3) | 0.3 | [1,1,2,3,0] | Yes (0.999) | No | 5 |
| 4 | 317.6871(3) | −0.2 | [1,1,2,3,1] | Yes (0.969) | No | 7,5 |
| 4 | 416.0503 (2) | 0.3 | [1,1,2,2,0] | Yes (0.962) | No | 7 |
| 4 | 437.0556 (2) | −0.3 | [1,1,2,2,1] | Yes (0.932) | Yes | 7,5 |
| 2 | 228.5270 (2) | 0.2 | [1,0,1,1,1] | Yes (0.681) | No | 3 |
| 6 | 322.2885 (4) | −0.5 | [1,2,3,3,1] | Yes (0.573) | Yes | 7,5 |
| 6 | 342.2778 (4) | −0.1 | [1,2,3,4,1] | Yes (0.797) | Yes | 7 |
| 6 | 430.054 (3) | 0.1 | [1,2,3,3,1] | Yes (0.742) | Yes | 7 |
Composition based upon systematic HS naming scheme.
Clustering by similarity with distance measurement.
Differentiation of HS obtained from bovine aorta and lung tissue.
Co-isolation more than 15% of similar precursor ion and/or tandem mass spectra poor quality
Co-isolation around 5% of similar precursor ion
Lack of sensitivity in MS/MS, poor S/N
Quality tandem mass spectra and/or clean isolation
AHC confirms the quality of the tandem mass spectra
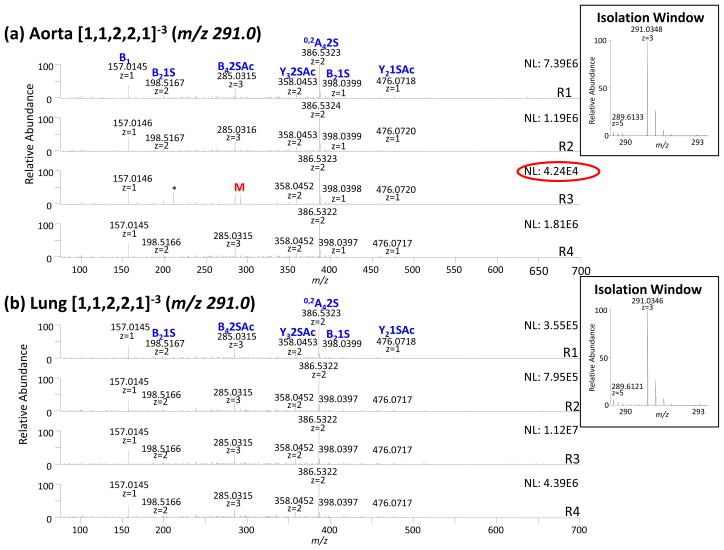
The reproducibility of tandem mass spectra is mirrored by the degree of clustering. We observed that if the replicate analyses of each precursor ion were reproducible, the tandem mass spectra produced from the same tissue were observed to cluster together. For the 13 precursor ions studied, 10 demonstrated tight clustering indicating acceptable reproducibility: [1,2,3,3,1]−4 (m/z 322.3), [1,2,3,3,1]−3 (m/z 430.1), [1,2,3,4,1]-4 (m/z 342.3), [1,1,2,2,0]−2 (m/z 416.1), [1,1,2,2,0]−3 (m/z 277.3), [1,1,2,3,0]−3 (m/z 303.7), [1,1,2,2,1]−2 (m/z 437.1), [1,1,2,3,1]−3 (m/z 317.7). We observed that two precursors ions [1,0,1,1,1]−2 (m/z 228.5) and [1,1,2,2,1]−3 (m/z 291.0) clustered well except for a single replicate. The ion intensity of this replicate in both cases was significantly lower than the other replicates. The tandem mass spectra of [1,1,2,2,1]−3 (m/z 291.0), shown in Figure 4, illustrates this observation. While the total ion current of aorta replicate 3 was on the order of 5E+4 counts, the other replicates were on the order of 1E+6 counts (4a). Furthermore, only aorta replicate 3 contains evidence of the precursor ion M (m/z 291.0).
Figure 4.
Tandem mass spectra of [1,1,2,2,1]−3 (m/z 291.0) obtained from bovine (a) aorta (b) lung tissues. Tandem MS data does not allow for differentiation of glycoforms specific to the tissue type. The product ions that differ in abundance significantly between aorta and lung are labeled with a star.
We observed three precursor ions [1,2,3,4,1]−5 (m/z 273.6), [1,0,1,2,0]−3 (m/z 247.5), and [1,1,2,3,1]−4 (m/z 238.0) which did not cluster ideally. In general for these compounds, the reproducibility of the tandem mass spectra was poor due to a lack of sensitivity in the tandem mass spectra (see for example Supplemental Figure 3) and low signal-to-noise ratio. The fact that the most intense ions were not constant from one replicate to the other perturbs the clustering analyses. This perturbation was also observed as higher P values in the original ANOVA analyses of these tandem MS clusters. Poor reproducibility in tandem mass spectra ultimately is reflected in ANOVA with P values > 0.05. The lack of reproducibility of the [1,0,1,2,0]−3 (m/z 247.5) resulted from co-isolation of precursor from the compound [1,1,2,4,0]−4 (m/z 247.59). Thus AHC proved to be an effective measure to indicate the reproducibility or the lack thereof of the tandem mass spectrometry data.
AHC across precursor ions quantify the degree of similarity of the tandem mass spectrometric data
AHC was performed across the precursor ions in order to determine which may be considered as good candidates for differentiation of HS obtained from aorta and lung tissues. For this type of clustering, distance measurement affords a relative-quantitative measure of the differences between the tandem mass spectra observed within this dataset and ranged from values of 0 to 1. A value of 1 indicates that the tandem mass spectra are nearly identical. In Table 1 the distance measurements are shown for the 10 precursor ions that demonstrated ideal clustering dependent upon their origin and yielded 3 distinct groups of clustering.
The first group contained 2 precursors, the component [1,1,2,3,0]−3 (m/z 303.7) and [1,1,2,2,1]−3 (m/z 291.0) which had a distance measurement equal to 0.99 indicating that the spectra were nearly identical. This is illustrated in Figure 4 which shows the tandem mass spectra of the precursor ion [1,1,2,2,1]−3 (m/z 291.0). Both components [1,1,2,2,1]−3 (m/z 291.0) and [1,1,2,3,0]−3 (m/z 303.7) (not shown) from both tissues were identical in terms of glycoform composition and were thus not good candidates to distinguish HS from the tissues.
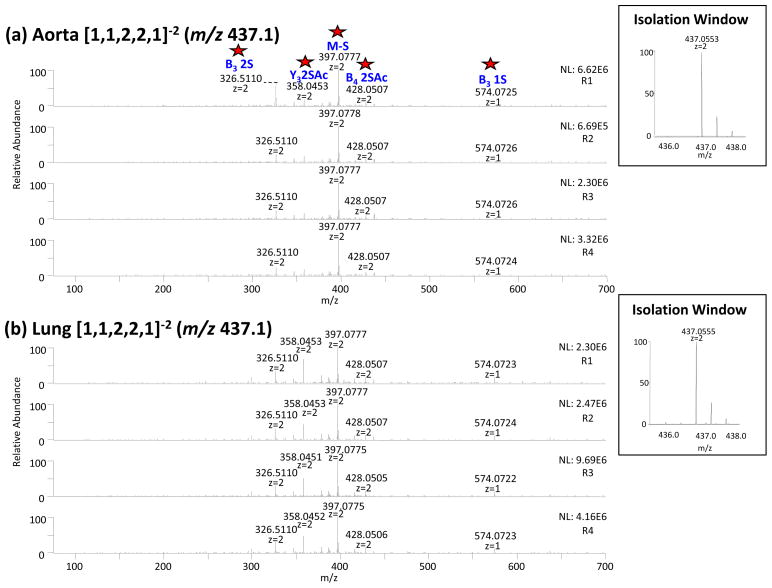
The second group was composed of 4 precursors, the components [1,1,2,2,0]−2 (m/z 416.1), [1,1,2,2,0]−3 (m/z 277.3), [1,1,2,2,1]−2 (m/z 437.1) and [1,1,2,3,1]−3 (m/z 317.7). Their distance measurements of the similarity clustering ranged between 0.969 and 0.937 indicating that a small degree of difference existed between the tandem mass spectra obtained from lung and aorta tissues. Manual comparison of the tandem mass spectra of the compounds [1,1,2,2,0]−2 (m/z 416.1), [1,1,2,2,0]−3 (m/z 277.3) and [1,1,2,3,1]−3 (m/z 317.7) did not indicate noteworthy differences. Tandem mass spectra of component [1,1,2,2,1]−2 (m/z 437.1) indicated some noticeable differences in the product ion intensities between the aorta and lung tissues including: 0,2X2 (1S) (m/z 300.0), B3 (2S) (m/z 326.5) and Y3 (2S,Ac) (m/z 358.0). These are illustrated in Figure 5. We also observed the co-isolation of the compound [1,3,4,5,0]−4 (m/z 436.0386) (−2.4 ppm) in the tandem mass spectra from the lung tissue (5b). The relative intensity of this component was less than 5% of the intensity of the target compound and contributed minimally to the tandem mass spectra, therefore it did not contribute to the intensity of the product ion Y3 2SAc (m/z 358.0). The component [1,1,2,2,1]−2 (m/z 437.1) is consistent with presence of different positional isomers in aorta vs. lung tissue. Of note is the fact that the product ion pattern for precursor ion [1,1,2,2,1]−3 (m/z 291.0) was similar for the lung and the aorta. Charge state influences product ion pattern, and it is therefore not surprising that the degree to which diagnostic product ions depends on charge state; however, the charge state distribution differs depending on the tissue type, consistent with the conclusion that tissue-specific isomers influence charge state.
Figure 5.
Tandem mass spectra of [1,1,2,2,1]−2 (m/z 437.5) obtained from bovine (a) aorta (b) lung tissues. Tandem MS data allowed for differentiation of glycoforms specific to the tissue type. The product ions that differ in abundance significantly between aorta and lung are labeled with a star.
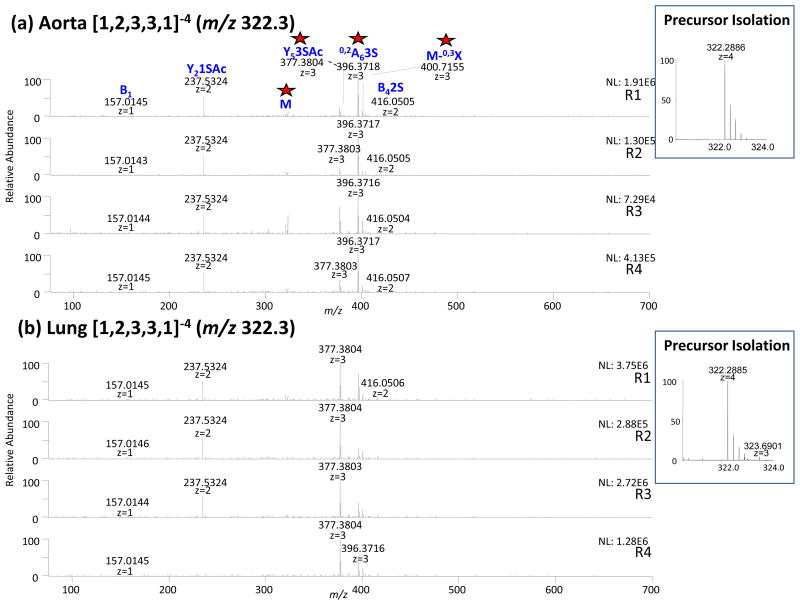
The third group contained 4 precursor ions, [1,2,3,3,1]−4 (m/z 322.3), [1,2,3,3,1]−3 (m/z 430.1), [1,2,3,4,1]−4 (m/z 342.3) and [1,0,1,1,1]−2 (m/z 228.5). These were present with distance measurements ranging from 0.573 to 0.797. These distance measurements were higher in value than for the compounds previously described because they arose due to the most abundant product ions, resulting from significantly different isomer composition rather than minor differences in the less abundant product ions. The tandem mass spectra of precursor ion [1,2,3,3,1]−4 (m/z 322.3) are presented in Figure 6 and indicate that the most intense product ion is Y5 (3S,Ac) (m/z 377.5) for the lung tissue and 0,2A6 (3S) (m/z 396.4) for the aorta tissue. The difference between the tandem MS data is due to intensity and not the appearance of new product ions. The verification of the window of isolation is important to be sure that the difference between tandem mass spectra from the 2 tissues is due to HS isomers specific to lung versus aorta and not from a co-isolation of unrelated compounds. Figure 6 indicates the precursor ion [1,2,3,3,1]−4 (m/z 322.3) are perfectly isolated for the aorta tissue. The window of isolation of the lung tissue contains a co-isolation of the component [0,2,2,3,1]−3 (m/z 323.6901) which accounted for less than 5% of the signal. The ions Y5(3S,Ac) (m/z 377.5) and 0,2A6(3S) (m/z 396.5) could not be assigned to any product ions of the structure [0,2,2,3,1]−3 (m/z 323.6901), consistent with the conclusion that the differences observed between the tandem mass spectra of the component [1,2,3,3,1]−4 (m/z 322.3) were significant and reflected the presence of different glycoforms in the both tissues. By contrast, the compound [1,1,2,3,1]−4 (m/z 228.5) clustered properly between lung and aorta tissue but this observation is due to a co-isolation of the compounds [1,1,2,3,0]−4 (m/z 227.5) at different intensity in the lung (<30%) and the aorta (>70%) tissue.
Figure 6.
Tandem mass spectra of [1,2,3,3,1]−4 (m/z 322.3) obtained from bovine (a) aorta (b) lung. Tandem MS data allowed for differentiation of glycoforms specific to the tissue type. The product ions that differ in abundance significantly between aorta and lung are labeled with a star.
The results summarized in Table 1 indicate the isolation of the compositions [1,2,3,4,1]−4 (m/z 342.3) and [1,2,3,3,1]−3 (m/z 430.1) were perfectly clean. This ideal clustering of both compositions from aorta and lung is consistent with the presence of different positional isomers in the lung versus aorta tissues. We observed (data not shown) that the most significant differences in intensity for the product ions for the aorta and lung tissues were 0,2A6 (3S) (m/z 396.4)//[M-S] (m/z 322.8) for the component [1,2,3,4,1]−4 (m/z 342.3) and B4 (2S) (m/z 407.0)//[M-S] (m/z 403.4) for the component [1,2,3,3,1] −3 (m/z 430.1). The observations are consistent with the conclusion of the existence of positional isoform differences between HS obtained from the lung and aorta tissues.
We hypothesized that the primary reason the components clustered together was due to a common structural core. Seven common product ions were clearly identified at different intensities between lung and aorta HS: B1 (m/z 157.0), Y5 (3S,Ac) (m/z 377.4), 0,2A6 (3S) (m/z 396.4), B4 (2S) (m/z 407.1), C4 (2S) (m/z 416.1), C5 (1S,Ac) (m/z 476.1), 0,2A3 (1S,Ac) (m/z 574.1). The presence of 0,2A6 (3S) (m/z 396.4) and the product ion 0,2A3 (1S,Ac) (m/z 574.1) clearly indicates the common HS core contains the acetate group and it could be identified for both those compounds at the first and fifth glucosamine residue of the oligosaccharide. While this core structure containing the sulfate compounds is plausible, our current study does not define this more precisely. To fully elucidate the structures of these compounds and characterize all possible compositional isoforms including position of sulfates that may give rise to diagnostic ions, it will be necessary to isolate the individual components by some means of separation. One possible solution to provide separation space that would afford sufficient resolution to solve this challenging problem is ion mobility mass spectrometry, an area of research that has been recently applied to the characterization of HS [10].
AHC by dissimilarity across product ions allows defining of structural information
AHC using dissimilarity measurements was performed on the tandem mass spectra ion series for each unique cluster obtained in the similarity space in order to determine which product ions were diagnostic for tissue type for each precursor m/z. The goals were to determine fine structure in homology and feature differences in product ion mass spectra from near neighbor clusters that would be indicative of structural homology and to determine the major differences in product ions resulting from different distributions of positional isomers of potential use as diagnostic marker ions. The distance measurement of AHC by dissimilarity is represented in this case as follows: a value of 1 indicates that the product ions differed significantly between tandem mass spectra of the same precursor ions while a value of 0 indicates homology.
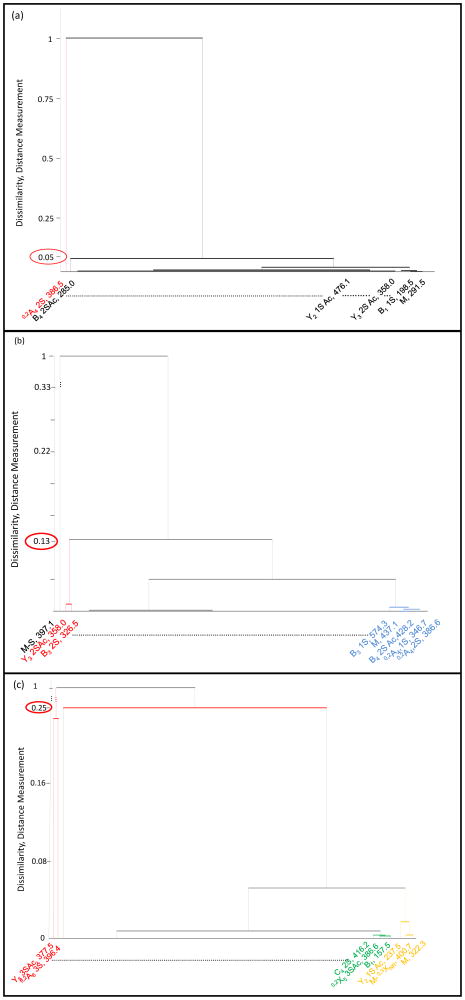
Example results of dissimilarity clustering are provided in Figure 7, which illustrates the clustering of the 3 general groups described above; (a) for precursor ion [1,1,2,2,1]3− (m/z 291.0), no major differences in product ion abundances between lung and aorta tissue, (b) for precursor ion [1,1,2,2,1]2− (m/z 437.5), slight differences in product ion abundances and (c))[1,2,3,3,1]4− (m/z 322.3), major differences in product ion abundances for HS oligosaccharides obtained from lung and aorta tissues. In Figure 7a depicting the tandem MS clustering of the component [1,1,2,2,1]−3 (m/z 291.0), we observed no major differences between the groups which was observed with a dissimilarity measurement of less than 0.05. The only feature that showed a difference, distance measurement equal to 1, was the most intense product ion in the tandem mass spectra, 0,2A4 (2S). We observed this perturbation in distance measurements consistently throughout the application of AHC when the product ion was the most intense in the tandem mass spectra.
Figure 7.
Agglomerative hierarchical clustering of product ions obtained from the components (a) [1,1,2,2,1]−3 (m/z 291.0), (b) [1,1,2,2,1]−2 (m/z 437.5) and (c)[1,2,3,3,1]−4 (m/z 322.3) obtained from bovine aorta and lung tissues. The product ion clusters indicate the presence of different positional isomers between the two tissues (aorta and lung).
In the second group, represented here in Figure 7b depicting the tandem MS clustering of the component [1,1,2,2,1]−2 (m/z 437.1), we observed some differences between the aorta and lung groups. Different clusters of product ions were observed. The first one included the most intense ion [M-S] (m/z 397.1) with a distance measurement equal to 1. The second cluster included the features Y3 (2S,Ac) (m/z 358.0) and B3 (2S) (m/z 326.5) with a distance measurement equal to 0.13. The last clustering with a distance measurement of < 0.13, is represented by the group of product ions B3(1S) (m/z 574.1), M (m/z 437.1), 0,2A4 (S,Ac) (m/z 346.7) and 0,2A4 (2,S) (m/z 386.6). The features Y3 (2S,Ac) (m/z 358.0) and B3 (2S) (m/z 326.5) constituted the diagnostic ions and represent the difference isomeric glycoforms present in the aorta and lung tissues. The distance measurement of 0.13 indicates a relatively moderate difference between the sets of tandem mass spectra.
In the third group, presented in Figure 7c depicting the tandem MS clustering of the component [1,2,3,3,1]−4 (m/z 322.3), major differences were observed between the aorta and lung groups. Here, two product ions Y5 (3S,Ac) (m/z 377.5) and 0,2A6 (3S) (m/z 396.4) clustered together with the distance measurement equal to 1. Both features clearly indicated the presence of different positional isoforms in HS from lung and aorta tissues and their variation of intensity between lung and aorta were significant. The second group also contained the first sub cluster C4 2S (m/z 416.2).
Conclusions
In the biological context, glycoconjugate glycans exist as mixtures of glycoforms and positional isomers. Thus, using assumptions relating to known activities of biosynthetic enzymes, the m/z value for a given HS oligosaccharide composition defines the range of positional isomers present in the mixture. As demonstrated here, the number of positional isomers may be constrained further using tandem mass spectrometric data.
For HS oligosaccharides, the number of positional isomers present for a given precursor ion composition is relatively large. The tandem mass spectrometric dissociation patterns for HS oligosaccharides is not entirely favorable to direct interpretation of structure in that uninformative losses of sulfate occur. Nonetheless, the tandem mass spectral profiles serve to differentiate HS from different tissues based on differences in product ion abundances. These observations are consistent with the conclusion that such patterns arise from different distributions of positional isomers in the tissue samples. We demonstrate that the use of AHC to analyze these data has three advantages. First, it provides a means of quality control for the tandem mass spectral data set. Second, it identifies the precursor ions for which significant differences in product ion abundances occur between the samples. Third, it identifies the product ions that have the most value for diagnostic purposes.
The level of fine structural detail produced by this AHC-based data analysis method is determined ultimately by the tandem MS parameters. In the present work, static infusion tandem MS was used. The advantage to this method was that the precursor ion charge states were higher than typically observed using on-line liquid chromatography MS. The disadvantage was that the lack of on-line separations contributed to the problem of co-isolation of different oligosaccharide compositions that produced similar m/z values. It is likely that the level of structural detail produced will increase through the use of LC/MS and or activated electron dissociation methods.
Even with the acknowledged limitations, the AHC method for analysis of tandem MS datasets shows great potential when applied to problems in glycomics. Complex positional isomer mixtures are typical in mixtures of released glycoconjugate glycans. The AHC method enables rapid analysis of large set of tandem MS data in a manner that permits discover of precursor and product ions that reflect the unique set of structures present in a given sample, and thus the underlying biological functions giving rise to these structures. As such, the method has significant potential value to the glycomics community.
Supplementary Material
Acknowledgments
This work was funded by NIH Grants P41RR10888, R01HL098950 and S10RR020946.
Abbreviations
- Ac
acetate
- AHC
agglomerative hierarchical clustering
- CAD
collisionally activated dissociation
- CS
chondroitin sulfate
- DP
degree of polymerization
- DS
dermatan sulfate
- GAG
glycosaminoglycan
- GlcUA
glucuronic acid
- GlcN
glucosamine
- GlcNAc
N-acetylglucosamine
- Hep
heparin
- HexA
hexuronic acid
- ΔHexA
Δ-4,5-unsaturated uronic acid
- HS
heparan sulfate
- IdoUA
iduronic acid
- MS
mass spectrometry
- NDST
N-deacetylase/N-sulfotransferase
- NMR
nuclear magnetic resonance
- NS
designates N-sulfation of GlcN
- 2S
designates 2-O-sulfation of IdoUA
- 6S
designates 6-O-sulfation of GlcN
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Perrimon N, Bernfield M. Specificities of heparan sulphate proteoglycans in developmental processes. Nature. 2000;404:725–728. doi: 10.1038/35008000. [DOI] [PubMed] [Google Scholar]
- 2.Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 3.Powell AK, Yates EA, Fernig DG, Turnbull JE. Interactions of heparin/heparan sulfate with proteins: Appraisal of structural factors and experimental approaches. Glycobiology. 2004;14:17R–30R. doi: 10.1093/glycob/cwh051. [DOI] [PubMed] [Google Scholar]
- 4.Prabhakar V, Capila I, Sasisekharan R. The structural elucidation of glycosaminoglycans. Methods Mol Biol. 2009;534:147–156. doi: 10.1007/978-1-59745-022-5_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaia J. On-line separations combined with MS for analysis of glycosaminoglycans. Mass Spectrom Rev. 2009;28:254–272. doi: 10.1002/mas.20200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pomin VH, Sharp JS, Li XY, Wang LC, Prestegard JH. Characterization of Glycosaminoglycans by N-15 NMR Spectroscopy and in Vivo Isotopic Labeling. Analytical Chemistry. 2010;82:4078–4088. doi: 10.1021/ac1001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ly M, Laremore T, Linhardt R. Proteoglycomics: Recent Progress and Future Challenges. OMICS; May 7, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaia J. Mass spectrometry and glycomics. OMICS. 2010;14:401–418. doi: 10.1089/omi.2009.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawasaki N, Itoh S, Hashii N, Takakura D, Qin Y, Huang XY, Yamaguchi T. The Significance of Glycosylation Analysis in Development of Biopharmaceuticals. Biological & Pharmaceutical Bulletin. 2009;32:796–800. doi: 10.1248/bpb.32.796. [DOI] [PubMed] [Google Scholar]
- 10.Schenauer MR, Meissen JK, Seo Y, Ames JB, Leary JA. Heparan Sulfate Separation, Sequencing, and Isomeric Differentiation: Ion Mobility Spectrometry Reveals Specific Iduronic and Glucuronic Acid-Containing Hexasaccharides. Anal Chem. 2009;81:10179–10185. doi: 10.1021/ac902186h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saad OM, Leary JA. Heparin sequencing using enzymatic digestion and ESI-MSn with HOST: a heparin/HS oligosaccharide sequencing tool. Anal Chem. 2005;77:5902–5911. doi: 10.1021/ac050793d. [DOI] [PubMed] [Google Scholar]
- 12.Saad OM, Myers RA, Castleton DL, Leary JA. Analysis of hyaluronan content in chondroitin sulfate preparations by using selective enzymatic digestion and electrospray ionization mass spectrometry. Anal Biochem. 2005;344:232–239. doi: 10.1016/j.ab.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 13.Saad OM, Leary JA. Compositional analysis and quantification of heparin and heparan sulfate by electrospray ionization ion trap mass spectrometry. Anal Chem. 2003;75:2985–2995. doi: 10.1021/ac0340455. [DOI] [PubMed] [Google Scholar]
- 14.Desaire H, Sirich TL, Leary JA. Evidence of block and randomly sequenced chondroitin polysaccharides: sequential enzymatic digestion and quantification using ion trap tandem mass spectrometry. Anal Chem. 2001;73:3513–3520. doi: 10.1021/ac010385j. [DOI] [PubMed] [Google Scholar]
- 15.Desaire H, Leary JA. Detection and quantification of the sulfated disaccharides in chondroitin sulfate by electrospray tandem mass spectrometry. J Am Soc Mass Spectrom. 2000;11:916–920. doi: 10.1016/S1044-0305(00)00168-9. [DOI] [PubMed] [Google Scholar]
- 16.Behr JR, Matsumoto Y, White FM, Sasisekharan R. Quantification of isomers from a mixture of twelve heparin and heparan sulfate disaccharides using tandem mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:2553–2562. doi: 10.1002/rcm.2079. [DOI] [PubMed] [Google Scholar]
- 17.Shi X, Zaia J. Organ-specific heparan sulfate structural phenotypes. J Biol Chem. 2009;284:11806–11814. doi: 10.1074/jbc.M809637200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowman M, Zaia J. Novel tags for the stable isotopic labeling of carbohydrates and quantitative analysis by mass spectrometry. Anal Chem. 2007;76:5777–5784. doi: 10.1021/ac070581b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller MJ, Costello CE, Malmstrom A, Zaia J. A tandem mass spectrometric approach to determination of chondroitin/dermatan sulfate oligosaccharide glycoforms. Glycobiology. 2006;16:502–513. doi: 10.1093/glycob/cwj093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hitchcock AM, Costello CE, Zaia J. Glycoform quantification of chondroitin/dermatan sulfate using an LC/MS/MS platform. Biochemistry. 2006;45:2350–2361. doi: 10.1021/bi052100t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hitchcock AM, Costello CE, Zaia J. Glycoform quantification of chondroitin/dermatan sulfate using a liquid chromatography-tandem mass spectrometry platform. Biochemistry. 2006;45:2350–2361. doi: 10.1021/bi052100t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bielik AM, Zaia J. Extraction of chondroitin/dermatan sulfate glycosaminoglycans from connective tissue for mass spectrometric analysis. Methods Mol Biol. 2010;600:215–225. doi: 10.1007/978-1-60761-454-8_15. [DOI] [PubMed] [Google Scholar]
- 23.Zaia J, McClellan JE, Costello CE. Tandem mass spectrometric determination of the 4S/6S sulfation sequence in chondroitin sulfate oligosaccharides. Anal Chem. 2001;73:6030–6039. doi: 10.1021/ac015577t. [DOI] [PubMed] [Google Scholar]
- 24.McClellan JE, Costello CE, O’Connor PB, Zaia J. Influence of charge state on product ion mass spectra and the determination of 4S/6S sulfation sequence of chondroitin sulfate oligosaccharides. Anal Chem. 2002;74:3760–3771. doi: 10.1021/ac025506+. [DOI] [PubMed] [Google Scholar]
- 25.Hitchcock AM, Yates KE, Shortkroff S, Costello CE, Zaia J. Optimized extraction of glycosaminoglycans from normal and osteoarthritic cartilage for glycomics profiling. Glycobiology. 2007;17:25–35. doi: 10.1093/glycob/cwl046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hitchcock AM, Yates KE, Costello CE, Zaia J. Comparative glycomics of connective tissue glycosaminoglycans. Proteomics. 2008;8:1384–1397. doi: 10.1002/pmic.200700787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naggar EF, Costello CE, Zaia J. Competing fragmentation processes in tandem mass spectra of heparin-like glycosaminoglycans. J Am Soc Mass Spectrom. 2004;15:1534–1544. doi: 10.1016/j.jasms.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 28.Zaia J, Li XQ, Chan SY, Costello CE. Tandem mass spectrometric strategies for determination of sulfation positions and uronic acid epimerization in chondroitin sulfate oligosaccharides. J Am Soc Mass Spectrom. 2003;14:1270–1281. doi: 10.1016/S1044-0305(03)00541-5. [DOI] [PubMed] [Google Scholar]
- 29.Wolff JJ, Laremore TN, Busch AM, Linhardt RJ, Amster IJ. Influence of charge state and sodium cationization on the electron detachment dissociation and infrared multiphoton dissociation of glycosaminoglycan oligosaccharides. J Am Soc Mass Spectrom. 2008;19:790–798. doi: 10.1016/j.jasms.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolff JJ, Laremore TN, Aslam H, Linhardt RJ, Amster IJ. Electron-induced dissociation of glycosaminoglycan tetrasaccharides. J Am Soc Mass Spectrom. 2008;19:1449–1458. doi: 10.1016/j.jasms.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolff JJ, Laremore TN, Busch AM, Linhardt RJ, Amster IJ. Electron detachment dissociation of dermatan sulfate oligosaccharides. J Am Soc Mass Spectrom. 2008;19:294–304. doi: 10.1016/j.jasms.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolff JJ, Laremore TN, Leach Ii FE, Linhardt RJ, Amster IJ. Electron capture dissociation, electron detachment dissociation and infrared multiphoton dissociation of sucrose octasulfate. Eur J Mass Spectrom (Chichester, Eng) 2009;15:275–281. doi: 10.1255/ejms.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolff JJ, Leach FE, 3rd, Laremore TN, Kaplan DA, Easterling ML, Linhardt RJ, Amster IJ. Negative electron transfer dissociation of glycosaminoglycans. Anal Chem. 2010;82:3460–3466. doi: 10.1021/ac100554a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lam H, Deutsch EW, Eddes JS, Eng JK, King N, Stein SE, Aebersold R. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics. 2007;7:655–667. doi: 10.1002/pmic.200600625. [DOI] [PubMed] [Google Scholar]
- 35.Lam H, Deutsch EW, Eddes JS, Eng JK, Stein SE, Aebersold R. Building consensus spectral libraries for peptide identification in proteomics. Nat Methods. 2008;5:873–875. doi: 10.1038/nmeth.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deutsch EW, Lam H, Aebersold R. Data analysis and bioinformatics tools for tandem mass spectrometry in proteomics. Physiol Genomics. 2008;33:18–25. doi: 10.1152/physiolgenomics.00298.2007. [DOI] [PubMed] [Google Scholar]
- 37.Bandeira N, Tsur D, Frank A, Pevzner PA. Protein identification by spectral networks analysis. Proc Natl Acad Sci U S A. 2007;104:6140–6145. doi: 10.1073/pnas.0701130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Apte A, Meitei NS. Bioinformatics in glycomics: glycan characterization with mass spectrometric data using SimGlycan. Methods Mol Biol. 2010;600:269–281. doi: 10.1007/978-1-60761-454-8_19. [DOI] [PubMed] [Google Scholar]
- 39.Souady J, Dadimov D, Kirsch S, Bindila L, Peter-Katalinic J, Vakhrushev SY. Software utilities for the interpretation of mass spectrometric data of glycoconjugates: application to glycosphingolipids of human serum. Rapid Commun Mass Spectrom. 2010;24:1039–1048. doi: 10.1002/rcm.4479. [DOI] [PubMed] [Google Scholar]
- 40.Vakhrushev SY, Dadimov D, Peter-Katalinic J. Software platform for high-throughput glycomics. Anal Chem. 2009;81:3252–3260. doi: 10.1021/ac802408f. [DOI] [PubMed] [Google Scholar]
- 41.von der Lieth CW, Lutteke T, Frank M. The role of informatics in glycobiology research with special emphasis on automatic interpretation of MS spectra. Biochimica et biophysica acta. 2006;1760:568–577. doi: 10.1016/j.bbagen.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 42.Gao HY. Generation of asparagine-linked glycan structure databases and their use. J Am Soc Mass Spectrom. 2009;20:1739–1742. doi: 10.1016/j.jasms.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 43.Tissot B, Ceroni A, Powell AK, Morris HR, Yates EA, Turnbull JE, Gallagher JT, Dell A, Haslam SM. Software tool for the structural determination of glycosaminoglycans by mass spectrometry. Anal Chem. 2008;80:9204–9212. doi: 10.1021/ac8013753. [DOI] [PubMed] [Google Scholar]
- 44.Tissot B, Haslam SM, Morris HR, Turnbull JE, Powell AK, Zhi ZL, Gallagher JT, Robinson CJ, Dell A. New mass spectrometry tools for glycosaminoglycans analysis. Glycobiology. 2006;16:1110–1110. [Google Scholar]
- 45.Chu KC. Applications of artificial intelligence to chemistry. Use of pattern recognition and cluster analysis to determine the pharmacological activity of some organic compounds. Anal Chem. 1974;46:1181–1187. doi: 10.1021/ac60345a030. [DOI] [PubMed] [Google Scholar]
- 46.Heller RSC, Chu CLKC. Interpretation of Mass Spectormetry Data Using Cluster Analysis-Alkyl Thiolesters. Analytical Chemistry. 1974;46:951. [Google Scholar]
- 47.Frank AM, Bandeira N, Shen Z, Tanner S, Briggs SP, Smith RD, Pevzner PA. Clustering millions of tandem mass spectra. J Proteome Res. 2008;7:113–122. doi: 10.1021/pr070361e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tabb DL, MacCoss MJ, Wu CC, Anderson SD, Yates JR., 3rd Similarity among tandem mass spectra from proteomic experiments: detection, significance, and utility. Anal Chem. 2003;75:2470–2477. doi: 10.1021/ac026424o. [DOI] [PubMed] [Google Scholar]
- 49.Beer I, Barnea E, Ziv T, Admon A. Improving large-scale proteomics by clustering of mass spectrometry data. Proteomics. 2004;4:950–960. doi: 10.1002/pmic.200300652. [DOI] [PubMed] [Google Scholar]
- 50.Huang Y, Tseng GC, Yuan S, Pasa-Tolic L, Lipton MS, Smith RD, Wysocki VH. A data-mining scheme for identifying peptide structural motifs responsible for different MS/MS fragmentation intensity patterns. J Proteome Res. 2008;7:70–79. doi: 10.1021/pr070106u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang H, Yi EC, Li XJ, Mallick P, Kelly-Spratt KS, Masselon CD, Camp DG, 2nd, Smith RD, Kemp CJ, Aebersold R. High throughput quantitative analysis of serum proteins using glycopeptide capture and liquid chromatography mass spectrometry. Mol Cell Proteomics. 2005;4:144–155. doi: 10.1074/mcp.M400090-MCP200. [DOI] [PubMed] [Google Scholar]
- 52.Hong YJ, Wang XD, Shen D, Zeng S. Discrimination analysis of mass spectrometry proteomics for ovarian cancer detection. Acta Pharmacol Sin. 2008;29:1240–1246. doi: 10.1111/j.1745-7254.2008.00861.x. [DOI] [PubMed] [Google Scholar]
- 53.Pan J, Chen HQ, Sun YH, Zhang JH, Luo XY. Comparative proteomic analysis of non-small-cell lung cancer and normal controls using serum label-free quantitative shotgun technology. Lung. 2008;186:255–261. doi: 10.1007/s00408-008-9093-7. [DOI] [PubMed] [Google Scholar]
- 54.Akella LM, Rejtar T, Orazine C, Hincapie M, Hancock WS. CLUE-TIPS, clustering methods for pattern analysis of LC-MS data. J Proteome Res. 2009;8:4732–4742. doi: 10.1021/pr900427q. [DOI] [PubMed] [Google Scholar]
- 55.Maccarana M, Sakura Y, Tawada A, Yoshida K, Lindahl U. Domain structure of heparan sulfates from bovine organs. J Biol Chem. 1996;271:17804–17810. doi: 10.1074/jbc.271.30.17804. [DOI] [PubMed] [Google Scholar]
- 56.Staples GO, Shi X, Zaia J. Extended N-sulfated domains reside at the nonreducing end of heparan sulfate chains. J Biol Chem. 2010;285:18336–18343. doi: 10.1074/jbc.M110.101592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zaia J, Costello CE. Tandem mass spectrometry of sulfated heparin-like glycosaminoglycan oligosaccharides. Anal Chem. 2003;75:2445–2455. doi: 10.1021/ac0263418. [DOI] [PubMed] [Google Scholar]
- 58.Domon B, Costello CE. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate Journal. 1988;5:397. [Google Scholar]
- 59.Ducret A, Van Oostveen I, Eng JK, Yates JR, 3rd, Aebersold R. High throughput protein characterization by automated reverse-phase chromatography/electrospray tandem mass spectrometry. Protein Sci. 1998;7:706–719. doi: 10.1002/pro.5560070320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saad OM, Leary JA. Delineating mechanisms of dissociation for isomeric heparin disaccharides using isotope labeling and ion trap tandem mass spectrometry. J Am Soc Mass Spectrom. 2004;15:1274–1286. doi: 10.1016/j.jasms.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 61.Guo CH, Koo CY, Bay BH, Tan PH, Yip GW. Comparison of the effects of differentially sulphated bovine kidney- and porcine intestine-derived heparan sulphate on breast carcinoma cellular behaviour. Int J Oncol. 2007;31:1415–1423. [PubMed] [Google Scholar]
- 62.Tomatsu S, Montano AM, Oguma T, Dung VC, Oikawa H, de Carvalho TG, Gutierrez ML, Yamaguchi S, Suzuki Y, Fukushi M, Sakura N, Barrera L, Kida K, Kubota M, Orii T. Dermatan sulfate and heparan sulfate as a biomarker for mucopolysaccharidosis I. J Inherit Metab Dis. 2010;33:141–150. doi: 10.1007/s10545-009-9036-3. [DOI] [PubMed] [Google Scholar]
- 63.Tokuyama Y, Takahashi T, Okumura N, Nonaka K, Kawaguchi Y, Yamaguchi K, Osada S, Gazdar A, Yoshida K. Aberrant methylation of heparan sulfate glucosamine 3-O-sulfotransferase 2 genes as a biomarker in colorectal cancer. Anticancer Res. 2010;30:4811–4818. [PubMed] [Google Scholar]
- 64.Zang Q, Keire DA, Wood RD, Buhse LF, Moore CM, Nasr M, Al-Hakim A, Trehy ML, Welsh WJ. Determination of galactosamine impurities in heparin samples by multivariate regression analysis of their (1)H NMR spectra. Anal Bioanal Chem. 2011;399:635–649. doi: 10.1007/s00216-010-4268-5. [DOI] [PubMed] [Google Scholar]
- 65.Theocharis AD, Tsara ME, Papageorgacopoulou N, Karavias DD, Theocharis DA. Pancreatic carcinoma is characterized by elevated content of hyaluronan and chondroitin sulfate with altered disaccharide composition. Biochim Biophys Acta. 2000;1502:201–206. doi: 10.1016/s0925-4439(00)00051-x. [DOI] [PubMed] [Google Scholar]
- 66.Theocharis AD, Theocharis DA, De Luca G, Hjerpe A, Karamanos NK. Compositional and structural alterations of chondroitin and dermatan sulfates during the progression of atherosclerosis and aneurysmal dilatation of the human abdominal aorta. Biochimie. 2002;84:667–674. doi: 10.1016/s0300-9084(02)01428-1. [DOI] [PubMed] [Google Scholar]
- 67.Theocharis AD, Skandalis SS, Tzanakakis GN, Karamanos NK. Proteoglycans in health and disease: novel roles for proteoglycans in malignancy and their pharmacological targeting. FEBS J. 2010;277:3904–3923. doi: 10.1111/j.1742-4658.2010.07800.x. [DOI] [PubMed] [Google Scholar]
- 68.Nadanaka S, Kitagawa H. Heparan sulphate biosynthesis and disease. Journal of Biochemistry. 2008;144:7–14. doi: 10.1093/jb/mvn040. [DOI] [PubMed] [Google Scholar]
- 69.Karangelis DE, Kanakis I, Asimakopoulou AP, Karousou E, Passi A, Theocharis AD, Triposkiadis F, Tsilimingas NB, Karamanos NK. Glycosaminoglycans as key molecules in atherosclerosis: the role of versican and hyaluronan. Curr Med Chem. 2010;17:4018–4026. doi: 10.2174/092986710793205354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.