Abstract
Objectives
Systemic lupus erythematosus (SLE) is a chronic multisystem genetically complex autoimmune disease characterised by the production of autoantibodies to nuclear and cellular antigens, tissue inflammation and organ damage. Genome-wide association studies have shown that variants within the major histocompatibility complex (MHC) region on chromosome 6 confer the greatest genetic risk for SLE in European and Chinese populations. However, the causal variants remain elusive due to tight linkage disequilibrium across disease-associated MHC haplotypes, the highly polymorphic nature of many MHC genes and the heterogeneity of the SLE phenotype.
Methods
A high-density case-control single nucleotide polymorphism (SNP) study of the MHC region was undertaken in SLE cohorts of Spanish and Filipino ancestry using a custom Illumina chip in order to fine-map association signals in these haplotypically diverse populations. In addition, comparative analyses were performed between these two datasets and a northern European UK SLE cohort. A total of 1433 cases and 1458 matched controls were examined.
Results
Using this transancestral SNP mapping approach, novel independent loci were identified within the MHC region in UK, Spanish and Filipino patients with SLE with some evidence of interaction. These loci include HLA-DPB1, HLA-G and MSH5 which are independent of each other and HLA-DRB1 alleles. Furthermore, the established SLE-associated HLA-DRB1*15 signal was refined to an interval encompassing HLA-DRB1 and HLA-DQA1. Increased frequencies of MHC region risk alleles and haplotypes were found in the Filipino population compared with Europeans, suggesting that the greater disease burden in non-European SLE may be due in part to this phenomenon.
Conclusion
These data highlight the usefulness of mapping disease susceptibility loci using a transancestral approach, particularly in a region as complex as the MHC, and offer a springboard for further fine-mapping, resequencing and transcriptomic analysis.
Introduction
Systemic lupus erythematosus (SLE) is a chronic multisystem autoimmune disease characterised by the production of autoantibodies to nuclear and cellular antigens, tissue inflammation and organ damage. There is a strong but complex genetic component to SLE susceptibility, whereby many polymorphisms each with a small or modest effect contribute to disease susceptibility. Genome-wide association studies have shown that variants within the major histocompatibility complex (MHC) region on chromosome 6 confer the greatest genetic risk for SLE in European and Chinese populations.1–3 The extended MHC spans almost 8 Mb and is divided into five subregions: extended class I (telomeric), class I, class III, class II and extended class II (centromeric). One of the most complex regions of the genome, this locus harbours two copy variable regions (the HLA-DRB genes in class II and the RCCX module containing complement component C4 in class III), some of the most polymorphic genes in the genome and conserved haplotypes where linkage disequilibrium (LD) extends over 2 Mb in some instances. The region has been the subject of extensive study given the importance of MHC alleles in the pathogenesis of tissue incompatibility, drug sensitivity, autoimmune, infectious and inflammatory diseases.
In European SLE cohorts, well-established associations are observed with highly conserved and extended haplotypes bearing the class II alleles HLA-DRB1*03:01 and HLA-DRB1*15:01.4 More recent high-density single nucleotide polymorphism (SNP) genotyping studies have demonstrated multiple independent signals across the MHC in northern European cohorts.5 6 However, the causal variants remain elusive due to tight LD across disease-associated MHC haplotypes, the highly polymorphic nature of associated variants and the heterogeneity of the SLE phenotype. Fine-mapping studies across the MHC region in other European and non-European SLE populations are lacking. The haplotypic diversity consequent on differing ancestry and environment demonstrated by these populations at the MHC region as well as non-MHC loci should allow further refinement of known association signals together with the identification of novel susceptibility variants. Given these known haplotypic differences, we undertook a high-density case-control SNP study of the MHC region in SLE cohorts of Spanish and Filipino ancestry using a custom Illumina chip in order to fine-map established association signals and potentially uncover novel susceptibility loci. In addition, we have performed comparative analyses between these two datasets and a northern European UK SLE cohort. In total we examined 1433 cases and 1458 matched controls.
Methods
Spanish cohort
The cohort comprised 464 cases and 468 controls. All cases were recruited from rheumatology clinics throughout Spain. Control samples were obtained from the Blood Bank Units of the hospitals where the cases originated.
Filipino cohort
The cohort comprised 335 SLE probands and 247 unrelated controls. We also included 26 trios (father, mother and affected child) to allow checks for Mendelian inheritance. All probands attended the Rheumatology and Clinical Immunology Clinics at the University of Santo Tomas Hospital, Manila, Philippines. Unrelated controls were recruited from spouses and acquaintances of the probands.
UK cohort
The cohort comprised 632 SLE probands and 742 unrelated controls from a previous study.5
All SLE probands fulfilled the American College of Rheumatology criteria for the classification of SLE.7 Written consent was obtained from all study participants.
Sample collection
DNA was obtained from whole blood using phenol-chloroform extraction. Native genomic DNA was used for the Spanish study. For the Filipino cohort, 100 ng (5 μl at 20 ng/μl) native DNA was whole genome amplified using the Qiagen REPLI-g Midi Kit (Cat. No. 150045) according to the manufacturer's written instructions.
Custom iSelect Illumina SNP array and genotyping
The samples were genotyped at the Feinstein Institute, USA using a custom Illumina iSelect chip comprising 10 788 SNPs: 6045 SNPs within the MHC region (29–33.5 Mb) and 4743 SNPs informative for major and European ancestry (see online supplement for SNP selection criteria).8 9
HLA genotyping
All HLA typing was performed using Luminex One Lambda SSO. Four-digit genotyping for HLA-B, HLA-DRB1 and HLA-DQB1 was performed in 82%, 99% and 44% of the Spanish cohort, respectively, following quality control (QC) at Hospital Virgen del Rocío, Seville, Spain and Hospital Virgen de las Nieves, Granada, Spain.
In order to assess LD relationships, four-digit HLA-DRB1 typing was performed in a subset of the Filipino cohort of known genotype for the top SNP, rs9271366, where DNA was available (n=89). Four-digit HLA-DRB1 typing was performed in 606 of 632 UK cases of SLE (96%). The Filipino and UK typing was performed at the Anthony Nolan Trust, London, UK. Two-digit HLA-DRB1 data were obtained for 694 of the 742 UK controls (92%) from the 1958 British birth cohort.
Quality control (QC) filters
All QC analyses except principal components analyses were performed using PLINK.10 Samples and SNPs were put forward for analysis if they met the following quality control filters: SNPs greater than 95% genotyping efficiency, minor allele frequency (MAF) >1% (failed Spanish n=271, Filipino n=758), non-deviation from Hardy-Weinberg equilibrium in controls on the basis of a false discovery rate of 0.05 (failed Spanish n=61, Filipino n=21). SNPs were excluded if they showed >10% Mendel error rate in the post-QC Filipino trios (n=7). Samples required >95% genotyping efficiency (failed Spanish n=50, Filipino n=50) and PI-Hat scores >0.2 on identity-by-descent analysis using ancestry informative markers (AIMs) in order to exclude cryptic relatedness and duplicate samples (failed Spanish n=27, Filipino n=3). In order to correct for population stratification, samples were excluded if they were outliers on principal components analysis using post-QC AIMs (performed using EIGENSTRAT and defined as >4 SDs from the mean)11 (failed Spanish n=62, Filipino n=88). The genomic inflation factor (λGC) was calculated using the post-QC AIMs after correction for population stratification (Spanish λGC=1.04 and Filipino λGC=1.09).
UK SLE cohort imputation
Genotypes were imputed using IMPUTE12 on the initial set of directly genotyped SNPs (n=1230) up to the Wellcome Trust Case-Control Consortium 2 (WTCCC2) study (n=7119). The WTCCC2 data were used as reference genotypes in the imputation, with dbsnp build 126 defining the genome map.13 No reference haplotypes were used in the imputation. Of the 7119 imputed SNPs in the UK SLE cohort, 3314 overlapped with the 6045 MHC SNPs genotyped in the Spanish and Filipino cohorts in this study and were used for analysis.
Statistical analyses
Single marker association analyses using logistic regression and stepwise logistic regression analyses were performed using PLINK and SNPTEST.14 We took the genotypes for the most associated SNP as a covariate and conditioned on this in the search for other independently associated SNPs in each dataset. If this analysis yielded further SNPs that passed our threshold of significance (see below), we added the top SNP to further stepwise logistic regression models and continued the process until no further SNPs passed our threshold of significance. Haplotypic association analyses were performed using PLINK and R statistical package. Data for SNP rs409558 in the Spanish and Filipino cohorts were meta-analysed using the standard inverse variance method. We performed tests of heterogeneity using the Breslow-Day test in PLINK. p values are represented following adjustment for the first principal component in each dataset or following adjustment for the first principal component and additional SNPs as covariates in SLR analyses in each dataset. A significance threshold of p=7×10−5 was set, given that genome-wide significance thresholds based on haplotype structure are typically in the range of 5–7×10−8 and that the MHC region constitutes approximately 1/1000th of the genome. The LD structure of the MHC region has been shown to be similar to that of the genome in general, but there appears to be greater LD between haplotype blocks in the MHC region so our significance threshold is likely to be conservative. In the Spanish cohort, separate logistic regression and conditional logistic regression analyses were performed for HLA-DRB1 alleles in order to assess relative predispositional effects. In order to account for multiple testing, Bonferonni-corrected p values were used as follows: HLA-DRB1, p=0.0023 (0.05/22 alleles tested). We examined LD relationships between SNPs and HLA alleles in each cohort by calculating the correlation coefficient (r2) using the Tagger algorithm in Haploview.15
Results
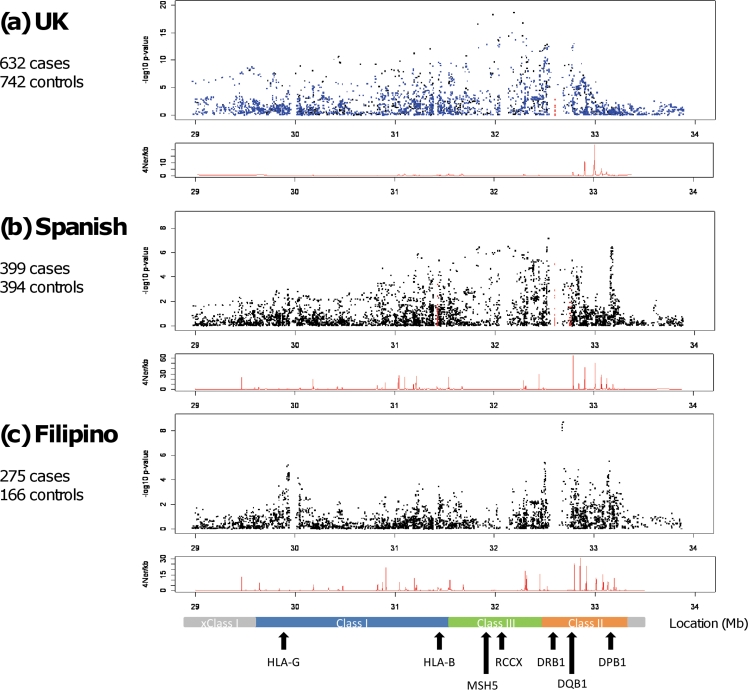
In all three datasets under study there was significant SNP association across the entire MHC region (figure 1). The UK SLE data confirmed previously published reports in northern European cohorts demonstrating principal SNP association within the class II and class III regions of the MHC.5 6 16 The most significantly associated SNP was rs1269852, located intergenic TNXB-ATF6B in the class III region of the MHC. Stepwise logistic regression demonstrated independent association at additional MHC loci including SNPs tagging the HLA-DRB1*1501 haplotype in class II, as well as class I SNPs located between HLA-B and HLA-C and 5′ PSORS1C1 (table 1).
Figure 1.
High-density transancestral single nucleotide polymorphism (SNP) mapping of the major histocompatibility complex (MHC) region in UK, Spanish and Filipino systemic lupus erythematosus (SLE). The panels show MHC region association plots for (A) UK, (B) Spanish and (C) Filipino SLE cohorts where genomic position (Mb) is shown on the horizontal axis with –log10 p values on the vertical axis. The black squares represent genotyped SNPs and the blue squares indicate imputed SNPs in the UK cohort. The red squares represent classically typed HLA alleles in the UK (HLA-DRB1 only) and Spanish (HLA-B, HLA-DRB1 and HLA-DQB1) cohorts. The panels beneath each association plot demonstrate the recombination rate for each cohort calculated using control haplotypes only generated with the program rhomap.27 A scaled map of the MHC region with relevant genes is shown in the bottom panel. RCCX represents the copy variable RCCX module containing the complement C4 gene (R=RP1/STK19, C=C4A/C4B, C=CYP21A2/CYP21A1P, X=TNXA/TNXB).
Table 1.
Primary and secondary single marker association in the major histocompatibility complex region in UK, Spanish and Filipino systemic lupus erythematosus
| SNP ID | Position | F_U | Associated allele | OR (95% CI) | p | Other disease | Location (LD)* | Gene expression¶ |
|---|---|---|---|---|---|---|---|---|
| UK SLE | ||||||||
| rs1269852 | 32188169 | 0.11 | C | 2.68 (2.16 to 3.33) | 2.48×10−19 | TNXB-ATF6B (760 kb) | ||
| rs3906272 | 31370903 | 0.04 | A | 2.84 (2.05 to 3.92) | 2.95×10−10 | HLA-C-HLA-B (22 kb) | ||
| rs3129868 | 32512355 | 0.11 | T | 1.67 (1.29 to 2.14) | 8.65×10−5 | BTNL2 – DRA (1501 tag SNP) (375 kb) | ||
| rs4713419 | 31101215 | 0.10 | C | 0.40 (0.25 to 0.63) | 7.55×10−5 | MUC21 – PSORS1C1 (9 kb) | HLA-C‡ | |
| Spanish SLE | ||||||||
| rs3130490 | 31847099 | 0.04 | A | 2.96 (1.95 to 4.51) | 3.94×10−7 | C6orf27 (620 kb) | ||
| rs3129768 | 32703061 | 0.13 | C | 1.91 (1.44 to 2.53) | 7.57×10−6 | MS | BTNL2-DRA (1501 tag SNP) (182 kb) | |
| rs3117213 | 33172583 | 0.18 | A | 1.76 (1.37 to 2.25) | 7.18×10−6 | ACPA+ RA CBD, SSc, PBC, Hep B | DPB1-DPB2 (29 kb) | |
| rs409558 | 31816126 | 0.20 | G | 0.57 (0.43 to 0.77) | 2.2×10−4 | MSH5 (20 kb) | MSH5§ | |
| Filipino SLE | ||||||||
| rs9271366 | 32694832 | 0.34 | G | 2.46 (1.83 to 3.30) | 1.97×10−9 | MS | DRB1-DQA1 (1502 tag SNP) (87 kb) | |
| rs2571391 | 30031817 | 0.16 | C | 0.36 (0.22 to 0.59) | 6.06×10−5 | MS | HLA-G-HLA-H (300 kb) | |
| rs2507987 | 31451012 | 0.48 | A | 0.48 (0.35 to 0.66) | 6.80×10−6 | HLA-B-MICA (6 kb) | ||
| rs409558 | 31816126 | 0.30 | G | 0.56 (0.37 to 0.86) | 7.47×10−3 | MSH5 (100 kb) | MSH5§ | |
| rs2071351 | 33151908 | 0.17 | A | 0.48 (0.28 to 0.83) | 8.44×10−3 | DPB1 (19 kb) | HLA-DPB1† | |
Stepwise logistic regression association results are shown in italics.
Meta-analysis of Spanish and Filipino data for the MSH5 SNP rs409558 revealed a locus-wide level of significance at p=1.92×10−5.
LD surrounding each SNP calculated in control population of each cohort using r2 cut-off >0.8.
All HapMap populations, CEU, YRI, CHB and JPT.
CEU and YRI only.
CEU only.
Data from GEO database (http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE6536).28
ACPA+RA, anti-citrullinated protein antibody positive rheumatoid arthritis; CBD, chronic beryllium disease; F_U, frequency in unaffected controls; Hep B, hepatitis B; LD, linkage disequilibrium; MS, multiple sclerosis; p, corrected/conditional p value (see Methods); PBC, primary biliary cirrhosis; SLE, systemic lupus erythematosus; SNP, single nucleotide polymorphism; SSc, systemic sclerosis; T1D, type 1 diabetes.
Association of MHC class II and class III variants with Spanish SLE
In the Spanish cohort, 399 cases and 394 controls were put forward for analysis following QC measures. Logistic regression analysis of 4924 post-QC SNPs showed that the peak signals in this southern European SLE cohort also arise from the class II and class III regions of the MHC (figure 1, table 1 and table S1 in online supplement). The most significantly associated SNP, rs9268832, was located in the class II pseudogene HLA-DRB9 (OR 1.80, CI 1.45 to 2.23, p=7.64×10−8) and showed moderate/weak LD with HLA-DRB1 alleles (figure S1 in online supplement). Serial stepwise logistic regression revealed a number of independent signals around HLA-DPB1 (best SNP rs3117213) as well as risk and protective signals in and surrounding MSH5 (best risk SNP rs3130490; best protective SNP rs409558). Interestingly, the SNPs with the best OR in this Spanish dataset were the aforementioned variants in and around the class III genes MSH5/C6orf27. The most associated of these SNPs was rs3130490 (OR 3.08, CI 2.03 to 4.66, p=1.04×10−7; (table 1, figure 2 and figure S2 in online supplement). This SNP showed strong LD with the top UK MHC SNP rs1269852 (r2=0.97). Thus, the primary MHC signal in Spanish SLE replicated that observed in the previously published UK dataset.5 In contrast to the UK data where the SNP rs3130490 showed strong LD with HLA-DRB1*03:01 (r2=0.71), the Spanish signal showed only moderate LD with HLA-DRB1*03:01 (r2=0.23), suggesting that variants in the class III region of the MHC may play a more important role than previously recognised. Conditioning on rs3130490 also revealed a number of potentially independent signals in the Spanish cohort, the best of which was the class II SNP rs3129768 located between HLA-DRB1 and HLA-DQA1 (OR 1.91, CI 1.44 to 2.53, p=7.57×10−6). This SNP showed moderate LD with HLA-DRB1*15:01 (r2=0.62). Again this contrasts with our northern European data where one of the main secondary association signals was observed with variants in strong LD with HLA-DRB1*15:01 (r2=0.93). Further stepwise logistic regression revealed association with the previously mentioned SNPs rs3117213 (HLA-DPB1) and rs409558 (MSH5) (table 1). LD analysis revealed that the association underlying rs9268832 probably represents a composite effect of rs3130490 and rs3129768, resulting in its greater statistical significance (figure S1 in online supplement).
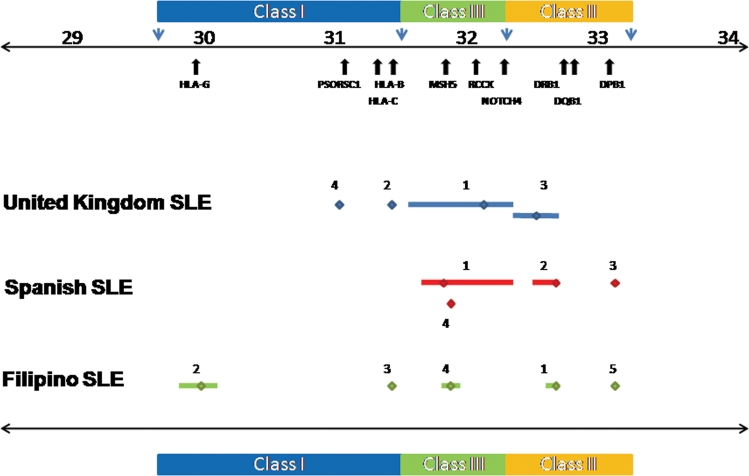
Figure 2.
Primary and secondary major histocompatibility complex (MHC) region association signals in UK, Spanish and Filipino systemic lupus erythematosus (SLE). This figure shows the primary and secondary association signals in UK, Spanish and Filipino SLE denoted in blue, red and green, respectively. The primary signals are labelled 1 and the secondary signals obtained by stepwise logistic regression are labelled 2–5 and correspond to the single nucleotide polymorphism (SNPs) shown in table 1. An indication of linkage disequilibrium (LD) surrounding each marker (calculated in the control population of each cohort using r2 cut-off >0.8) is shown by the bars flanking each marker. LD is <30 kb where flanking bars are absent. The genomic position is shown above the plot together with the positions of the relevant MHC region genes.
MHC region SNPs show association independent of HLA-DRB1 alleles in Spanish SLE
Analysis of HLA-DRB1 alleles alone demonstrated principal association with HLA-DRB1*03:01 (OR 1.89, CI 1.43 to 2.48, p=5.53×10−6) (table S2 in online supplement). Conditioning on HLA-DRB1*03:01 in order to assess relative predispositional effects, we found association with HLA-DRB1*15:01 (OR 1.83, CI 1.31 to 2.55; p=0.00045). Conditioning on these top two HLA-DRB1 alleles, we found association with HLA-DRB1*08:01 (OR 3.52, CI 1.55 to 8.01; p=0.0027). No other HLA-DRB1 alleles showed significant disease association following further stepwise logistic regression. Next we used the three principally associated HLA-DRB1 alleles as covariates in a serial stepwise logistic regression in the entire SNP dataset. We found that all the aforementioned SNPs showed some evidence of association independent of HLA-DRB1 alleles except rs3129768 (table S3 in online supplement). Similar results were obtained when conditioning the UK SNP data for HLA-DRB1*03:01 and HLA-DRB1*15:01 (table S4 in online supplement).
Major role for MHC class II and class I variants in Filipino SLE
Following QC measures, 275 cases and 166 controls were put forward for analysis in the Filipino SLE cohort. The overall pattern of association showed that, of the 3704 post-QC SNPs, the major signal arises from the class II region of the MHC and therefore differs from that observed in European SLE cohorts where principal associations are seen in class II and class III (figure 1, table 1 and table S5 in online supplement). The top SNP, rs9271366, was located between HLA-DRB1 and HLA-DQA1 (OR 2.46, CI 1.83 to 3.30, p=1.97×10−9). Furthermore, HLA-DRB1 typing in a subset of this cohort showed that the most highly associated SNP, rs9271366, was a perfect proxy for HLA-DRB1*15:02 in the Filipino population (r2=1) and suggests a major role for variants on this haplotype in Filipino SLE (table S6 in online supplement). These data are consistent with the known high allele frequency of HLA-DRB1*15:02 in Filipino reference and other Pacific rim populations where the allele frequency ranges from 37% to 48%.17 18 Furthermore, the association of HLA-DRB1*15:01 is well established in East Asian SLE cohorts from Japan and Korea, while the association of HLA-DRB1*15:02 with SLE has been reported in South East Asians from Thailand.19–21
SLR analyses on the top SNP (rs9271366) revealed independent signals in the class I region of the MHC between HLA-G and HLA-A (best SNP rs2571391: OR 0.36, CI 0.22 to 0.59, p=6.06×10−5). Further stepwise logistic regression revealed additional independent signals that replicate those observed in the Spanish cohort: MSH5 (best SNP rs409558) and HLA-DPB1 (best SNP rs2071351) (table 1 and figure S3 in online supplement). Meta-analysis of Spanish and Filipino data for the MSH5 SNP rs409558 revealed a locus-wide significance level at p=1.92×10−5 (ORmeta 0.58, CImeta 0.33 to 0.83).
Effect size of SNP rs9271366 is significantly greater in Filipino SLE than in European SLE
The most associated Filipino MHC SNP, rs9271366, which acts as a surrogate marker for HLA-DRB1*15:02 in this population, tags HLA-DRB1*15:01 in populations of European ancestry (r2=0.94 (UK controls); r2=0.77 (Spanish controls)) and, as such, shows disease association in European SLE with ORs of approximately 1.4 (table S7 in online supplement). The frequency of HLA-DRB1*15:02 is low in European populations (1–2%). The SNP rs9271366 also tags the SNPs demonstrating disease association following primary conditional analysis in the UK (r2 with rs3129868=0.98) and Spanish (r2 with rs3129768=0.77) cohorts because these SNPs (rs3129868 and rs3129768) are also in LD with HLA-DRB1*15:01 (table 1). As the effect size of the SNP rs9271366 is significantly greater in the Filipino SLE cohort than in the Europeans (Filipino OR 2.46, European OR ∼1.4, Breslow-Day p=5.25×10−4, table S7 in online supplement), it is interesting to speculate that this genetic variant or variants in LD may predispose to a more severe disease phenotype such as renal involvement, as is often observed in non-European populations.22–24
Refinement of SLE-associated HLA-DRB1*15 signal
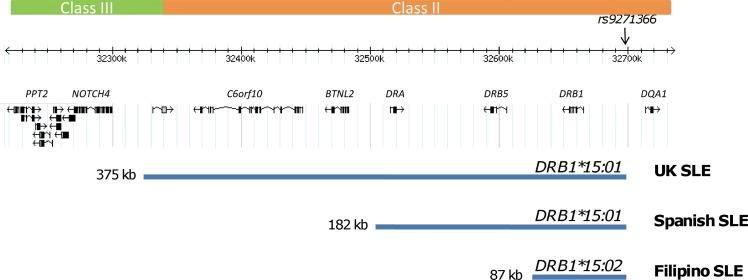
Previous attempts to fine-map the SLE-associated HLA-DRB1*1501 haplotypic signal could only delimit the region to approximately 500 kb of the MHC class II region in European-Americans using microsatellite typing.25 When the SNP rs9271366 was used as a surrogate marker for HLA-DRB1*15 haplotypes, we found that the LD surrounding this SNP varied in the different populations studied from a 375 kb region in UK SLE to a 182 kb region in Spanish SLE and to an 87 kb region encompassing HLA-DRB1 and the intergenic interval between HLA-DRB1 and HLA-DQA1 in Filipino SLE (figure 3). Hence, the transancestral mapping approach used in this study allowed refinement of the SLE-associated HLA-DRB1*15 signal.
Figure 3.
Transancestral fine-mapping of the HLA-DRB1*15 signal in UK, Spanish and Filipino systemic lupus erythematosus (SLE). The frequencies of HLA-DRB1*15 alleles show geographical variability. For example, in Europeans the common allele is HLA-DRB1*15:01, in Pacific and South East Asians it is HLA-DRB1*15:02, while in African populations it is HLA-DRB1*15:03 (http://allelefrequencies.net/). It is well established that haplotypes harbouring HLA-DRB1*15 alleles show primary or secondary association with SLE and data from this study support this view. However, the identity of causal variation has remained elusive due to the strong linkage disequilibrium present on the common disease-associated HLA-DRB1*15:01 haplotype in northern Europeans. Using the single nucleotide polymorphism (SNP) rs9271366 as a surrogate marker for the common HLA-DRB1*15 allele in each population studied, we have refined the disease-associated region from 375 kb in northern Europeans to 87 kb in the Filipino population. The latter region encompasses the HLA-DRB1 gene itself as well as part of the intergenic interval between HLA-DRB1 and HLA-DQA1.
Evidence for genetic interaction between the top independent MHC region SNPs in SLE
Haplotypic analyses were performed on the top three independent SNPs from each cohort to look for evidence of interaction. The top SNP was chosen from each cohort, together with the next two independently associated SNPs which were selected following stepwise logistic regression (table 2). Despite the relatively modest cohort sizes, these analyses suggested evidence of genetic interaction (non-additive effects) in all three populations studied (table S8 in online supplement). For example, in the Spanish cohort, a multiple logistic regression model was fitted using the top three independent SNPs (rs3130490, rs3129768 and rs3117213) as explanatory variables and interaction terms were tested for. Interestingly, the best model (difference in Akaike Information Criterion=4.8, difference in Bayesian Information Criterion=6.8) had an rs3130490*rs3129768 interaction where the effect on the OR was positive (5.1 (95% CI 1.12 to 23.08), p=0.03), plus an independent additive term for rs3117213 (see table S9 and figure S4 in online supplement).
Table 2.
Haplotypic association using the top three SNPs following serial step-wise logistic regression in each cohort is shown (see table 1 and table s8 in the online supplement for further details).
| SNP1 | SNP2 | SNP3 | HAPLOTYPE | F_A | F_U | F_Uexp | OR | 95% CI | p | pperm | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UKrisk | rs1269852 | rs3906272 | rs3129868 | CAT | <0.001 | <0.001 | 0.0004 | NA | NA | NA | NA |
| UKprot | rs1269852 | rs3906272 | rs3129868 | GGG | 0.524 | 0.729 | 0.7604 | 0.37 | 0.31 to 0.44 | 3.72×10−28 | 1×10−4 |
| Spanishrisk | rs3130490 | rs3129768 | rs3117213 | ACA | 0.004 | <0.001 | 0.001 | NA | NA | NA | NA |
| Spanishprot | rs3130490 | rs3129768 | rs3117213 | CAC | 0.506 | 0.688 | 0.6849 | 0.443 | 0.36 to 0.55 | 2.22×10−13 | 1×10−4 |
| Filipinorisk | rs9271366 | rs2571391 | rs2507987 | GAT | 0.273 | 0.092 | 0.1485 | 3.45 | 2.24 to 5.33 | 5.69×10−11 | 1×10−4 |
| Filipinoprot | rs9271366 | rs2571391 | rs2507987 | ACA | 0.005 | 0.031 | 0.0507 | NA | NA | NA | NA |
F_A, haplotypic frequency in affected SLE cases; F_U, haplotypic frequency in unaffected controls; F_Uexp, expected haplotypic frequency calculated from minor allele frequency of SNPs in controls; NA, not applicable (unable to perform statistical analyses as frequencies too low); p, p value; pperm, permuted p value (10000 permutations); prot, haplotype comprising all three protective alleles; risk, haplotype comprising all three risk alleles; SLE, systemic lupus erythematosus; SNP, single nucleotide polymorphism.
Increased frequency of MHC region risk alleles and haplotypes in Filipino SLE compared with European SLE
Next we examined haplotypic frequency and association using the top three independent SNPs in all three cohorts studied (table 2). We found that the haplotype harbouring the risk alleles of the top three SNPs was rare in European SLE cohorts. However, in Filipino SLE, the risk haplotype was common while the protective haplotype was rare (risk OR 3.45, CI 2.24 to 5.33, p=5.69×10−11; protective OR 0.002, CI 1×10−4 to 0.05, p=2.10×10−4). Thus, the population frequency of the top ranked risk alleles and risk haplotypes increases from the UK and Spain to the Philippines (risk haplotype frequencycases 0, 0.004 and 0.273, respectively), suggesting that the greater disease burden in non-European SLE populations may be due in part to this phenomenon. Furthermore, the frequency of protective haplotypes in each population decreases through the same gradient (protective haplotype frequencycases 0.525, 0.505 and 0.005, respectively).
Discussion
We present the results of the first high-density transancestral mapping study of the MHC region in SLE using cohorts from the UK, Spain and the Philippines. Despite the modest sample sizes, we have identified and replicated new independent loci with evidence of interaction across this complex region, some of which appear to be SLE-specific (MSH5) while others suggest shared mechanisms across autoimmune/inflammatory diseases (HLA-DPB1, HLA-G, HLA-B/C) (box 1). In particular, we were able to demonstrate a considerable effect from MHC variants in Filipino SLE using single marker and haplotypic analyses due to the high frequency of disease-associated variants in this cohort. There are no accurate prevalence data for SLE in the Philippines and most parts of Asia, even in the most recent literature. In general, published prevalence rates for SLE in Asia are broadly similar to those observed in Europeans and range between 30 and 50 per 100 000.22 24 26 Interestingly, prevalence rates appear to be higher in Asian migrant populations.24
Box 1 Major histocompatibility complex region susceptibility genes and haplotypes in systemic lupus erythematosus.
-
▶
HLA-DPB1: For the first time we report an association with single nucleotide polymorphisms (SNPs) in the region of HLA-DPB1 and systemic lupus erythematosus (SLE). The most associated SNP in this region is rs3117213, located between the pseudogenes HLA-DPA2 and COL11A2P. This SNP has previously shown association with ACPA-positive rheumatoid arthritis (RA) in northern European cohorts and is independent of the known RA HLA-DRB1 risk alleles.29 30 The association of HLA-DPB1 alleles with chronic beryllium disease is well established.31 Recent studies have also detected associations in the HLA-DPB1 region with chronic hepatitis B infection, Wegener's granulomatosis, primary biliary cirrhosis, Graves' disease, Takayasu's arteritis, juvenile idiopathic arthritis, systemic sclerosis and multiple sclerosis in European and non-European populations.29 32–36 The association of this region with these autoimmune, infectious and inflammatory diseases suggests that it is likely to represent a shared autoimmune/inflammatory locus.
-
▶
MSH5: The MSH5 (MutS homologue 5) gene, located in the major histocompatibility complex (MHC) class III region, comprises 26 exons and spans 25 kb.37 MSH5 belongs to a family of proteins involved in DNA mismatch repair and meiotic recombination. Recent evidence also suggests a role for human MSH5 in promoting ionising radiation-induced apoptosis.38
-
▶
HLA-G: The principal signal after stepwise logistic regression analysis on the top Filipino SNP rs9271366 is observed in the class I region of the MHC between HLA-G and HLA-H. HLA-H is transcribed but not translated. HLA-G is a non-classical HLA class I molecule that has been implicated in immune tolerance mechanisms and autoimmune disease.39 Unlike other classical HLA molecules, HLA-G exhibits limited nucleotide and protein polymorphism and shows marked tissue restriction in that the molecule is primarily expressed on cytotrophoblastic cells of the placenta where it is thought to mediate maternal-fetal tolerance40 41 (http://hla.alleles.org/proteins/class1.html). HLA-G expression can be induced in tumours, transplanted tissue and plaques from patients with multiple sclerosis. HLA-G is able to suppress immune responses through binding inhibitory receptors such as ILT2, ILT4 and KIR2DL4 which are expressed on a variety of immune cells including NK cells (KIR2DL4), CD4+ and CD8+ T cells, B cells, monocytes and dendritic cells.41 Recent studies in multiple sclerosis and SLE have suggested HLA-G as a putative disease susceptibility gene.39 A 14 bp insertion located in the 3′UTR of the gene has been associated with lower levels of HLA-G mRNA. Interestingly, the HLA-G*01:01:02 allele which is the most common allele carrying the 14 bp insertion shows strong linkage disequilibrium with the SLE-associated HLA-A*01-HLA-B*08-HLA-DRB1*03-HLA-DQA1*05-HLA-DQB1*02 haplotype in European populations.42
-
▶
RCCX module: The MHC class III risk haplotype, tagged by rs1269852 and rs3130490 showing primary association in the Spanish cohort, spans the copy variable RCCX module containing complement component C4. A similar haplotype, also extending across the RCCX module, harbours the most highly associated SNPs in our northern European SLE cohort.5 Despite the haplotypic diversity observed in the southern European Spanish population compared with northern Europeans, we have not been able to fine-map this signal using SNPs. The reasons underlying the conservation of this haplotype remain unclear but include selective pressures and co-regulation of gene expression.
-
▶
HLA-DRB1*15 alleles: The HLA-DRB1*15:01 and HLA-DRB1*15:02 alleles, which show association in European and Filipino SLE respectively, differ by only one amino acid—a valine (DRB1*15:01) to glycine (DRB1*15:02) substitution at position 86 of the amino acid sequence in pocket P1 of the peptide-binding cleft.43 Haplotypes harbouring HLA-DRB1*15:01 alleles confer risk for SLE and multiple sclerosis but also demonstrate protection in diseases such as type 1 diabetes and IgA deficiency.4 5 The mechanisms underlying these phenomena are unknown.
-
▶
HLA-C / HLA-B region: Data from the Filipino and UK SLE cohorts under study reveal association in the class I region of the MHC encompassing HLA-C/HLA-B/MICA. A recent genome-wide association study comparing HIV-1 controllers and progressors found that the same region harbours the major genetic determinants of HIV-1 control. Further analysis implicated specific HLA-B allele peptide groove amino acids, in addition to an independent HLA-C effect in the control of HIV infection.44
The primary class III risk haplotype tagged by rs1269852 and rs3130490 in Europeans displays extended LD such that it encompasses most of the MHC class III region. Stepwise logistic regression analyses demonstrated an identical protective haplotype encompassing the class III gene MSH5 alone in Filipino and Spanish SLE. These data suggest that dysregulation of MSH5 may underlie some of the risk attributable to the conserved class III risk haplotype. Further fine-mapping will be required to elucidate the nature of this signal. The class III variants that confer the greatest risk in SLE cohorts of European ancestry are rare in Filipino and Han Chinese SLE where MAF are approximately 0.001. These data suggest that either these variants are not important in SLE cohorts of south-east Asian ancestry or that different class III SNPs that are uncommon in Europeans and hence not typed in this study show association in these populations.
Two recent genome-wide association scans in SLE case-control cohorts of Chinese ancestry have shown that the most highly associated SNPs were located in the class II region of the MHC, between HLA-DRA and HLA-DQA2.2 3 We observed a similar pattern of association in the Filipino cohort under study. The most highly associated SNP in the Filipino cohort, rs9271366, is a surrogate marker for HLA-DRB1*15:02. This SNP showed the greatest overall association in the Hong Kong Chinese genome-wide association study and is ranked 8 of the top 13 MHC SNPs in the Han Chinese genome-wide association study in SLE, implicating HLA-DRB1*15:02 haplotypes in SLE susceptibility in these populations as well.
Using transancestral SNP mapping of the MHC region in SLE, we have successfully refined the established SLE-associated HLA-DRB1*15 signal to an interval encompassing HLA-DRB1 and HLA-DQA1. We have identified and replicated association in the genes MSH5 and HLA-DPB1 in Filipino and Spanish SLE cohorts, and also demonstrated association at HLA-G in Filipino SLE. These signals are independent of each other and HLA-DRB1 alleles and show some evidence of genetic interaction. These data highlight the usefulness of mapping disease susceptibility loci using a transancestral approach, particularly in a region as complex as the MHC, and offer a springboard for further fine-mapping, resequencing and transcriptomic analysis.
Supplementary Material
Acknowledgments
The authors would like to thank all study participants.
Footnotes
International MHC and Autoimmunity Genetics Network (IMAGEN): John D Rioux, Philippe Goyette, Timothy J Vyse, Lennart Hammarström, Michelle M A Fernando, Todd Green, Philip L De Jager, Sylvain Foisy, Joanne Wang, Paul I W de Bakker, Stephen Leslie, Gilean McVean, Leonid Padyukov, Lars Alfredsson, Vito Annese, David A Hafler, Qiang Pan-Hammarström, Ritva Matell, Stephen J Sawcer, Alastair D Compston, Bruce A C Cree, Daniel B Mirel, Mark J Daly, Tim W Behrens, Lars Klareskog, Peter K Gregersen, Jorge R Oksenberg and Stephen L Hauser.
Clinicians who provided access to SLE samples: Norberto Ortego Centeno, Department of Internal Medicine, Hospital San Cecilio, Granada, Juan Jimenez Alonso, Department of Internal Medicine, Hospital Virgen de las Nieves, Granada, Enrique de Ramon Garrido, Department of Internal Medicine, Hospital Carlos Haya, Malaga, Maria Teresa Camps Garcia, Department of Internal Medicine, Hospital Carlos Haya, Malaga, Julio Sanchez Roman, Department of Internal Medicine, Hospital Virgen del Rocio, Seville, Spain.
A full list of the investigators who contributed to the generation of the WTCCC data is available from www.wtccc.org.uk.
Funding MMAF and LB were funded through an Arthritis Research UK grant (18239). The IMAGEN consortium was supported by grant AI067152 from the National Institutes of Allergy and Infectious Diseases. This study makes use of data generated by the Wellcome Trust Case-Control Consortium. Funding for the project was provided by the Wellcome Trust under awards 076113 and 085475. We acknowledge the use of DNA from the British 1958 Birth Cohort collection (D Strachan, S Ring, W McArdle and M Pembrey) funded by the Medical Research Council grant G0000934 and Wellcome Trust grant 068545/Z/02.
Competing interests None.
Ethics approval This study was approved by the London Research Ethics Committee, UK (Ref: 06/MRE02/9) and the Comité de Ética del CSIC, Granada, Spain.
Provenance and peer review Not commissioned; externally peer reviewed.
References
- 1.Gateva V, Sandling JK, Hom G, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet 2009;41:1228–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han JW, Zheng HF, Cui Y, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet 2009;41:1234–7 [DOI] [PubMed] [Google Scholar]
- 3.Yang W, Shen N, Ye DQ, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet 2010;6:e1000841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernando MM, Stevens CR, Walsh EC, et al. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet 2008;4:e1000024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rioux JD, Goyette P, Vyse TJ, et al. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci USA 2009;106:18680–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barcellos LF, May SL, Ramsay PP, et al. High-density SNP screening of the major histocompatibility complex in systemic lupus erythematosus demonstrates strong evidence for independent susceptibility regions. PLoS Genet 2009;5:e1000696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan EM, Cohen AS, Fries JF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1982;25:1271–7 [DOI] [PubMed] [Google Scholar]
- 8.Tian C, Plenge RM, Ransom M, et al. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet 2008;4:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian C, Kosoy R, Nassir R, et al. European population genetic substructure: further definition of ancestry informative markers for distinguishing among diverse European ethnic groups. Mol Med 2009;15:371–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–9 [DOI] [PubMed] [Google Scholar]
- 12.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009;5:e1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burton PR, Clayton DG, Cardon LR, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet 2007;39:1329–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marchini J, Howie B, Myers S, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007;39:906–13 [DOI] [PubMed] [Google Scholar]
- 15.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–5 [DOI] [PubMed] [Google Scholar]
- 16.Harley JB, Alarcón-Riquelme ME, Criswell LA, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet 2008;40:204–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mack SJ, Bugawan TL, Moonsamy PV, et al. Evolution of Pacific/Asian populations inferred from HLA class II allele frequency distributions. Tissue Antigens 2000;55:383–400 [DOI] [PubMed] [Google Scholar]
- 18.Bugawan TL, Klitz W, Alejandrino M, et al. The association of specific HLA class I and II alleles with type 1 diabetes among Filipinos. Tissue Antigens 2002;59:452–69 [DOI] [PubMed] [Google Scholar]
- 19.Hashimoto H, Nishimura Y, Dong RP, et al. HLA antigens in Japanese patients with systemic lupus erythematosus. Scand J Rheumatol 1994;23:191–6 [DOI] [PubMed] [Google Scholar]
- 20.Hong GH, Kim HY, Takeuchi F, et al. Association of complement C4 and HLA-DR alleles with systemic lupus erythematosus in Koreans. J Rheumatol 1994;21:442–7 [PubMed] [Google Scholar]
- 21.Sirikong M, Tsuchiya N, Chandanayingyong D, et al. Association of HLA-DRB1*1502-DQB1*0501 haplotype with susceptibility to systemic lupus erythematosus in Thais. Tissue Antigens 2002;59:113–17 [DOI] [PubMed] [Google Scholar]
- 22.Osio-Salido E, Manapat-Reyes H. Epidemiology of systemic lupus erythematosus in Asia. Lupus 2010;19:1365–73 [DOI] [PubMed] [Google Scholar]
- 23.Navarra SV, Ishimori MI, Uy EA, et al. Studies of Filipino patients with systemic lupus erythematosus: autoantibody profile of first-degree relatives. Lupus 2011;20:537–43 [DOI] [PubMed] [Google Scholar]
- 24.Thumboo J, Wee H-L. Systemic lupus erythematosus in Asia: is it more common and more severe? APLAR J Rheumatol 2006;9:320–6 [Google Scholar]
- 25.Graham RR, Ortmann WA, Langefeld CD, et al. Visualizing human leukocyte antigen class II risk haplotypes in human systemic lupus erythematosus. Am J Hum Genet 2002;71:543–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Danchenko N, Satia JA, Anthony MS. Epidemiology of systemic lupus erythematosus: a comparison of worldwide disease burden. Lupus 2006;15:308–18 [DOI] [PubMed] [Google Scholar]
- 27.Auton A, McVean G. Recombination rate estimation in the presence of hotspots. Genome Res 2007;17:1219–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stranger BE, Forrest MS, Dunning M, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 2007;315:848–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding B, Padyukov L, Lundström E, et al. Different patterns of associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in the extended major histocompatibility complex region. Arthritis Rheum 2009;60:30–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orozco G, Barton A, Eyre S, et al. HLA-DPB1-COL11A2 and three additional xMHC loci are independently associated with RA in a UK cohort. Genes Immun 2011;12:169–75 [DOI] [PubMed] [Google Scholar]
- 31.Dai S, Murphy GA, Crawford F, et al. Crystal structure of HLA-DP2 and implications for chronic beryllium disease. Proc Natl Acad Sci USA 2010;107:7425–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu XM, Wang C, Zhang KN, et al. Association of susceptibility to multiple sclerosis in Southern Han Chinese with HLA-DRB1, -DPB1 alleles and DRB1-DPB1 haplotypes: distinct from other populations. Mult Scler 2009;15:1422–30 [DOI] [PubMed] [Google Scholar]
- 33.Zhou X, Lee JE, Arnett FC, et al. HLA-DPB1 and DPB2 are genetic loci for systemic sclerosis: a genome-wide association study in Koreans with replication in North Americans. Arthritis Rheum 2009;60:3807–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wieczorek S, Holle JU, Epplen JT. Recent progress in the genetics of Wegener's granulomatosis and Churg-Strauss syndrome. Curr Opin Rheumatol 2010;22:8–14 [DOI] [PubMed] [Google Scholar]
- 35.Kamatani Y, Wattanapokayakit S, Ochi H, et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet 2009;41:591–5 [DOI] [PubMed] [Google Scholar]
- 36.Radstake TR, Gorlova O, Rueda B, et al. Genome-wide association study of systemic sclerosis identifies CD247 as a new susceptibility locus. Nat Genet 2010;42:426–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Her C, Doggett NA. Cloning, structural characterization, and chromosomal localization of the human orthologue of Saccharomyces cerevisiae MSH5 gene. Genomics 1998;52:50–61 [DOI] [PubMed] [Google Scholar]
- 38.Tompkins JD, Wu X, Chu YL, et al. Evidence for a direct involvement of hMSH5 in promoting ionizing radiation induced apoptosis. Exp Cell Res 2009;315:2420–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larsen MH, Hviid TV. Human leukocyte antigen-G polymorphism in relation to expression, function, and disease. Hum Immunol 2009;70:1026–34 [DOI] [PubMed] [Google Scholar]
- 40.Apps R, Gardner L, Moffett A. A critical look at HLA-G. Trends Immunol 2008;29:313–21 [DOI] [PubMed] [Google Scholar]
- 41.Carosella ED, Favier B, Rouas-Freiss N, et al. Beyond the increasing complexity of the immunomodulatory HLA-G molecule. Blood 2008;111:4862–70 [DOI] [PubMed] [Google Scholar]
- 42.Kolte AM, Steffensen R, Nielsen HS, et al. Study of the structure and impact of human leukocyte antigen (HLA)-G-A, HLA-G-B, and HLA-G-DRB1 haplotypes in families with recurrent miscarriage. Hum Immunol 2010;71:482–8 [DOI] [PubMed] [Google Scholar]
- 43.Marsh SG, Bodmer JG. HLA class II nucleotide sequences, 1992. Immunogenetics 1993;37:79–94 [DOI] [PubMed] [Google Scholar]
- 44.Pereyra F, Jia X, McLaren PJ, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 2010;330:1551–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.