Abstract
Numerous mobile genetic elements (MGE) are associated with the human gut microbiota and collectively referred to as the gut mobile metagenome. The role of this flexible gene pool in development and functioning of the gut microbial community remains largely unexplored, yet recent evidence suggests that at least some MGE comprising this fraction of the gut microbiome reflect the co-evolution of host and microbe in the gastro-intestinal tract. In conjunction, the high level of novel gene content typical of MGE coupled with their predicted high diversity, suggests that the mobile metagenome constitutes an immense and largely unexplored gene-space likely to encode many novel activities with potential biotechnological or pharmaceutical value, as well as being important to the development and functioning of the gut microbiota. Of the various types of MGE that comprise the gut mobile metagenome, plasmids are of particular importance since these elements are often capable of autonomous transfer between disparate bacterial species, and are known to encode accessory functions that increase bacterial fitness in a given environment facilitating bacterial adaptation. In this article current knowledge regarding plasmids resident in the human gut mobile metagenome is reviewed, and available strategies to access and characterize this portion of the gut microbiome are described. The relative merits of these methods and their present as well as prospective impact on our understanding of the human gut microbiota is discussed.
Keywords: gut microbiome, horizontal gene transfer, host-microbe interactions, mobile genetic elements, mobile metagenome, plasmid isolation, TRACA
Introduction
Higher eukaryotic organisms have evolved in a biosphere dominated by prokaryotic cells (estimated to number ~1030),1 and live in intimate association with prokaryotic microbes that colonise various body sites to form host-associated ecosystems.2-8 Such communities develop and evolve under complex selective pressures, and unlike microbial ecosystems that reside in environments such as the soil and sea, host-associated ecosystems are not only subject to pressures resulting from competition between constituent members, but are also subject to selective pressure at the level of the metazoan host through effects on host fitness.2,4-6,8
The co-evolution of host and microbe in this context and the general complexity of many microbial ecosystems frequently results in the development of host-associated microbial communities with emergent properties that directly benefit the metazoan host.2,3,7-14 Modern humans are no exception, and we play host to numerous microbial communities with the most densely populated being found in the gastrointestinal tract (GIT).2,9,15-20 The adult human gut microbiota reaches population densities of ~1013–1014 individual cells in the distal colon which are dominated by bacteria derived from an estimated 150–800 species (with ~70–80% presently being uncultured). These belong primarily to the Firmicutes, Bacteroidetes, Actinobacteria and Proteobacteria divisions, with Bacteroidetes and Firmicutes co-dominating in this community.2,9,15-20
This microbial ecosystem plays an intimate role in the development and well-being of the human host, and undertakes a range of functions from which we benefit,2,3,7-14,20-29 including the salvage of energy from the diet through fermentation of components recalcitrant to host digestive mechanisms, protection against pathogens through competitive exclusion, and maturation of the immune system.2,8,19-24Conversely, bacterial residents of the gut obtain many benefits from the human host such as nutrients (in the form of mucus and dietary components), as well as a relatively stable environment.2,8 Recently, detailed investigation of the mechanisms underlying host-microbe interactions in key cultivatable members of the gut microbiota, as well as metagenomic approaches which permit access to the wider uncultivated fraction of this community, have further illuminated the intimate association of host and microbe in the GIT. This includes the ability of gut microbes to direct the development of aspects of host physiology, modulate host immune responses, maintain epithelial homeostasis, and the integration of microbial metabolic outputs with host metabolism.7-14,25-29 Overall, there is increasing evidence and awareness that constituents of the gut microbiota (and other human microbiomes) enter into symbiotic relationships with the human host, and play an intimate role in our development and wellbeing.2-8
This understanding has not only made the gut microbiota and other human microbiomes the focus of a significant global research effort, but has also prompted many researchers to begin viewing humans and other animals as gestalt entities composed of both prokaryotic and eukaryotic cells, which engage in a complex network of interactions.4,5,8,30 This has resulted in the development of new ecological and evolutionary models to describe, explain and predict the co-evolution of host and microbe,4-6,8,30 including the hologenome theory of evolution in which the overall unit of selection in evolution consists of the total genetic content of an eukaryotic host plus that of its associated prokaryotic partners, which are collectively referred to as the “holobiont.”4,5 However, host-associated communities such as the human gut microbiota in turn play host to a wide range of mobile genetic elements (MGE) and bacteriophages, which collectively may be referred to as the mobile metagenome.20,30-48 The role of this flexible gene pool in the development and functioning of the gut microbial community remains largely unexplored, yet there is emerging evidence that the gut mobile metagenome also reflects the co-evolution of bacteria with the human host in the GIT.20,30,32,48-50 Some MGE may therefore be unique to or enriched within this ecosystem (Table 1). Therefore, the human gut mobile metagenome is likely to encode genes important for survival and persistence in the GIT as well as activities involved in host-microbe and microbe-microbe interactions in this community.30-33
Table 1. Evidence for co-evolution of gut-associated MGE with the human host and the role of the gut mobile metagenome in community function, development and host-microbe interaction.
| Study | MGE examined | Summary |
|---|---|---|
| Kurokawa et al. 200720 |
Conjugative transposons |
CTn-1549 like family of elements designated CTnRINT observed to be enriched in the gut microbiomes of Japanese and American individuals examined. In conjunction a high level of recombinases and integrases also observed in gut microbiomes. |
| Jones et al. 201032 |
Plasmids |
Plasmids or plasmid families enriched and potentially unique to the human gut microbiome identified, with homologous sequences detected in gut microbiomes of geographically isolated hosts (America, Europe, Japan). Enrichment of some functions encoded by plasmids also observed. In particular plasmid pTRACA22 was noted as widely distributed and potentially unique to the human gut microbiome, with RelBE type toxin-antitoxin addiction modules (TAMs) enriched in terms of relative abundance in human gut microbiomes compared with other data sets examined. |
| Jones 201030 |
Plasmids |
Further evidence for wide distribution of pTRACA22 like plasmids and enrichment of pTRACA22 type RelBE TAMs provided from analysis of the METAHIT9 data set. Functions of HGT and the gut mobile metagenome considered from host perspective and within the hologenome theory of evolution.4,5 |
| Zaneveld et al. 201049 |
Plasmids |
Plasmids infecting gut-associated bacteria exhibit the same patterns of habitat-associated gene convergence observed for bacterial chromosomes. |
| Claesson et al. 200641 Li et al. 200740 Corr et al. 2007141 |
Plasmids |
Lactobacilli commonly harbor large megaplasmids encoding genes proposed to be involved in adaptation to the gut environment. Genes associated with the probiotic or protective effect of some species also appear to be encoded by megaplasmids. |
| Hehemann et al. 201050 |
HGT, putative plasmids |
Gain of functional capacity in the Japanese gut microbiome attributed to plasmid-mediated HGT of genes conferring ability to utilize seaweed glycans, from marine Bacteroides spp to gut commensal Bacteroides spp |
| Ebdon et al. 200647 |
Bacteriophage |
Isolation and characterization of bacteriophage infecting Bacteroides sp GB-124, indicated these phage were specific to the human gut, and carriage in the general population is high. |
| Reyes et al. 201048 |
Bacteriophage/virus-like particles |
Demonstrated large inter-individual variation in gut virome composition, but relative intra-individual temporal stability of gut viromes. Functions associated with a wide range of processes in anaerobic gut bacteria found to also be encoded by gut viromes. A dominance of temperate phages observed and a general lack of predator-prey dynamics in host-virus interactions suggested, which is in contrast to other microbial ecosystems. The latter may indicate host-level selection for a stable gut ecosystem. |
| Lepage et al. 2008204 |
Bacteriophage |
Distinct increase in concentrations of virus-like particles, predominantly bacteriophage, observed in patients with Crohns disease compared with healthy controls. Bacteriophage hypothesized to play a role in the dysbiosis of the gut microbiota observed in Crohn disease by destabilizing the community. |
| Lozupone et al. 200859 Xu et al. 200760 |
General HGT | Habitat associated gene convergence of glycoside hydrolases and glycosyltransferases in gut-associated bacteria and archaea largely generated through HGT. |
Furthermore, the high level of novel gene content typical of MGE, coupled with their predicted high level of diversity, suggests that the mobile metagenome constitutes an immense gene-space likely to encode many novel activities relevant to development and functioning of the gut microbiota, and with potential biotechnological or pharmaceutical value.30-33,46,51,52 Current estimates predict the diversity of MGE to be exceedingly high, with phage alone estimated to outnumber bacteria by an order of magnitude in many environments,53 and by around 2–5 times in the human gut microbiota.46 As such, this vast genetic resource likely accounts for a significant proportion of the “biological dark matter” believed to be extant in the biosphere, yet we remain relatively ignorant of the true diversity, abundance and functions encoded by many types of MGE.30,32,48,51,52
MGE are also well documented to promote and facilitate gene flow between distinct and disparate bacterial species driving bacterial adaptation and evolution, and the development of new functional pathways.30,31,54-61 There is increasing appreciation for the role of horizontal gene transfer (HGT) in the development of the gut microbiota, and stabilization of important activities of this community.8,20,30,31,59-62 Although this may be achieved through recruitment of member species with overlapping metabolic and functional profiles, HGT in the gut microbiota has been implicated as a major factor in the development of a functionally stable community, underlying the dissemination of gene sets encoding for key community activities to a wide range of community members.59,60,62-65 This is thought to generate functional redundancy among community members, which guards against loss of key activities.5,8,30,31,62 In addition, the observation that MGE can facilitate the transfer of genetic material across kingdom boundaries,66-68 including transfer from bacteria to mammalian cells,69 is of particular significance to host-associated microbial ecosystems such as the human gut microbiota.
Of the numerous types of MGE that will comprise the mobile metagenome of a microbial community, plasmids are of particular significance. Plasmids are frequently capable of mediating their own transfer between distinct species of bacteria, and often encode a range of accessory functions advantageous to their bacterial hosts under particular environmental conditions.30-33,52,54-58; they can also act as vehicles for the dissemination of other MGE such as transposons and integrons.30-32,58,70,71 In addition, natural plasmids often form the basis for the development of new molecular tools for organisms lacking basic genetic systems for their manipulation and functional dissection. Ultimately the development of strategies and detailed elucidation of mechanisms underlying host-microbe interactions in the gut microbiota will require the development of molecular tools which permit the detailed characterization of species. Plasmids and other MGE associated with the gut microbiota will likely provide a source of raw genetic material to develop such tools, as has been described for a range of species recently.64,72-76 In this article current knowledge regarding plasmids resident in the human gut mobile metagenome is reviewed, and available strategies to access and characterize this portion of the gut microbiome are discussed.
Overview of Plasmids Associated with Gut Microbes and Encoded Functions
Plasmid carriage is prevalent among cultivatable members of the gut microbiota, which until recently constituted the only accessible fraction of this community.38-44,64,70,72,74-90 Members of the Bacteroidetes, Enterobacteriacea and lactobacilli are frequently found to harbor multiple elements within the same cell,40-44,64,72,74,77-84,89 while plasmid carriage by isolates of common gut-associated Bifidobacterial species has generally been reported to be less common, with plasmid-free isolates encountered at a higher frequency.79,85-88 In contrast, there is a general paucity of data regarding plasmid carriage by numerically dominant members of the gut microbiota belonging to the Firmicutes division (predominantly Clostridia), which co-dominate with members of the Bacteroidetes division in this ecosystem.16-18 The general lack of data regarding clostridial plasmids from this population is likely a reflection of the difficulty in culturing the relevant members of gut Firmicutes, as well as limitations of classical endogenous and exogenous methods for plasmid isolation (discussed in detail in subsequent sections).31.33.58
More recently, genomic and metagenomic analyses have been adapted for study of gut-associated plasmids and have provided evidence for the co-evolution of plasmids infecting members of the gut microbiota with the human host (summarized in Table 1).30-32,49,50 In particular, Zanveld et al. (2010) have demonstrated that in the gut-associated bacterial species they analyzed, the gene content of plasmids exhibited the same general patterns of habitat-associated gene convergence found in the corresponding bacterial chromosomes.49 Closely related bacteria exhibited low levels of inter-species gene convergence moving toward greater levels in both chromosomes and plasmids with increasing phylogenetic distance.49 This is proposed to demonstrate increased specialization among closely related species (likely driven by competition for similar resources), but long-term convergence of functions at higher phylogenetic levels (indicative of host level selection).8,30,49 In essence, these findings and those summarized in Table 1, support the hypothesis that plasmids, and the mobile metagenome in general, are likely to encode functions symptomatic of challenges and activities important to life in the human gut.30-33 These findings also predict the existence of plasmid-encoded functions, or plasmid families, that are enriched or unique to the gut microbiota. Evidence for this has recently been provided in a metagenomic-based analysis of plasmids and encoded functions in the gut microbiome.32 In this study, plasmid pTRACA22 was identified as potentially unique or enriched in the gut mobile metagenome, when compared with murine or environmental metagenomic data sets.32 Sequences with high homology to pTRACA22 were detected in metagenomic datasets derived from the gut microbiomes of geographically isolated individuals indicating a long-term association with this community, and were not detected in any environmental or murine data sets analyzed.32 Furthermore, some functions encoded by the pTRACA22 element were found to be enriched in the human gut microbiome relative to other environments, including a novel RelBE type toxin-antitoxin addiction module.32
The enriched pTRACA22 RelBE module has now also been confirmed to be a functional and expressed gene system in the surrogate E. coli host (by RT-PCR), and preliminary PCR surveys with primers specific to the pTRACA22 RelBE genes has indicated that this module may be present on a large proportion of plasmids isolated from the gut environment (28/42 plasmids tested; B. Jones, unpublished data). Although it is presently unclear what role these pTRACA22 type RelBE modules may play in the gut microbiota (if any), they have been associated with numerous functions in bacteria relevant to life in the GIT, including survival under adverse conditions and exposure to antibiotics, resistance to bacteriophage predation, regulation of gene expression, and effects on human cells.30,32,91-101 This highlights how analysis of the mobile metagenome can identify novel functions with the potential to be important in community function and development, as well as access to activities of potential biotechnological or pharmaceutical or clinical interest. However, studies characterizing plasmid-encoded functions in the human gut microbiome have typically focused on clinically relevant traits relevant to treatment or pathogenesis of infectious disease, but factors mediating survival and persistence among gut commensals are now receiving more attention.
Antibiotic resistance
Many studies of plasmids and other MGE resident in cultivatable members of the gut microbiota have focused on those conferring antibiotic resistance, and harbored by easily cultivated members of this community that constitute prominent and problematic opportunistic pathogens, such as strains of E. coli, Enterococci, Klebsiella pneumonia and Bacteroides spp. The human gut is generally regarded to be a reservoir for antibiotic resistance genes,36,39,89 exemplified by the observed prevalence of genes encoding tetracycline resistance in human gut isolates of Bacteriodes species even before the widespread introduction of this antibiotic.36 Furthermore, recent metagenomic analyses have highlighted the potential diversity of resistance determinants that may be encoded by the gut microbiota, and their frequent association with putative MGE.39
Plasmids encoding resistance to a wide range of antibiotics have been identified in members of the human gut microbiota, including those conferring resistance to β-lactams, sulphonamide, tetracycline, chloramphenicol, trimethoprim, kanamycin, erythromycin, streptomycin, clindamycin, and metronidazole.38,79,80,82,84,90,102-115 Such plasmids are also frequently mobile and autonomous transfer between members of the mammalian gut community, as well as between commensal organisms and species considered to be transient colonizers, has been demonstrated both in vitro and in vivo.38,81,82,84,90,102-105,107-109,113-115 Although the majority of studies have been justifiably concerned with the spread of plasmid-encoded antibiotic resistance between common and pathogenic species associated with the gut microbiota, there is also increasing interest in understanding if less well characterized members of the gut microbiota also serve as reservoirs of mobilizable antibiotic resistance determinants, and in particular gut-associated species considered beneficial to human health.
The carriage and transfer of plasmids between bacteria in the GIT, such as Bifidobacteria and lactobacilli spp, is of considerable interest since members of these groups are not only part of the normal gut microbiota, but also important industrial organisms used in the manufacture of many fermented dairy products, as well as being formulated as probiotics.40,41,64,102,103 Plasmids are considered to be common among some groups of Lactic acid bacteria (LAB) and have been shown to encode a wide range of traits that facilitate survival and adaptation to the gut environment.40,41,64,74,75,85,106,114,116-119 Numerous studies have also reported antibiotic resistance among isolates of these organisms, including those obtained from food products, the human GIT, and used as probiotics, and there is increasing interest in the mobility of the relevant genes.102,106,114,115,120-125
Several studies have demonstrated the presence of antibiotic resistance genes encoded by plasmids and other MGE in species of LAB, and highlighted the transfer of both recombinant and natural plasmids to other members of the gut microbiota.102,103,110,114,115,126–128Further research is still required, however, to enhance our understanding of the potential for this important group of gut related bacterial species to harbor antibiotic resistance genes, and transmit these to other members of the gut microbiota, as well as transiently colonizing pathogens. The latter scenario is of particular concern both in terms of patient welfare and escalating levels of resistance among prominent opportunistic pathogens (which include many members of the normal commensal gut microbiota), and the increasing long-term usage of probiotics by the general public.
Adaptation, survival and persistence in the gut environment
While the role of plasmids in the dissemination of antibiotic resistance among clinically relevant gut-associated bacterial species has received much attention, numerous other accessory functions may be encoded by plasmids, and there is clear potential for plasmid-encoded traits to contribute to the development and functioning of the gut microbiota.30-32,50,89 Activities relevant to aspects of community function and host health, including utilization of diverse nutrient sources and degradation of xenobiotic compounds,31,40,41,64,116,118 virulence factors,78,129-139 bacteriocins41,43,117,119,140,142 and adhesion to host epithelial cells,43,78,90,129,130,136,139 have all been identified on plasmids from gut-associated species. In a number of instances, plasmids encoding multiple functions that increase fitness in the GIT have been isolated and characterized.31,40-44,64,78,131,133,137 Many of these traits are of potential pharmaceutical, or biotechnological value, for example in the discovery of novel bioactive compounds or the rational design of more effective probiotics, including probiotic-based platforms for the delivery of drugs, vaccines, or other therapeutic interventions.
Lactobacilli also provide excellent examples of such plasmid-mediated adaptation to the GIT, and biotechnologically relevant plasmid-encoded traits. In particular, recent studies have highlighted the frequent carriage of large megaplasmids by species of lactobacilli isolated from human and animal sources, including numerous strains obtained from faeces or other intestinal samples.40-42 These include the probiotic strain L. salivaris UCC118, which has a genome composed of multiple replicons, including a large 242 Kb megaplasmid designated pMP118.42 The UCC118 megaplasmid encodes multiple traits relevant to the survival of this species in the GIT, including tolerance to bile, bacteriocin production, and contingency pathways for cell wall biosynthesis and redox balance.42
Collectively these functions are proposed to endow L. salivarius UCC118 with a repertoire of accessory traits or pathways that permit a high degree of genetic and phenotypic flexibility, which facilitates survival in the gut environment by enhancing ability to adapt to transient ecological niches that arise in this community, tolerate fluctuations in diet and community structure, and overcome intrinsic barriers to colonization of the GIT.42 Functions encoded by the pMP118 megaplasmid are also involved in the “probiotic effect” of UCC118, and the pMP118 encoded bacteriocin system has been demonstrated to underlie the ability of UCC118 to protect mice against infection by the foodborne pathogen Listeria monocytogenes.42,141 Overall, the lactobacilli megaplasmids serve as an important example of the functions that may be encoded by the gut mobile metagenome and how these can influence community development and impact on host health. Such traits are also of considerable interest in the rational design of more effective probiotics, and probiotic-mediated platforms for therapeutic or prophylactic disease intervention.
Bacteriocins with activity against prominent foodborne pathogens have also been found to be encoded by plasmids harbored by a range of other gut-associated microorganisms, such as E. coli, Pediococcus acidilactici and Bifidobacterium infantis.117,140,142 In light of the rising levels of antibiotic resistance and the relative dearth of programs aimed at developing new antibiotics, alternative strategies to control or prevent infections are of significant interest, and therefore bacteriocins have potential biotechnological value.42,43,140,143-145 This highlights the pharmaceutical potential of the gut mobile metagenome, and illustrates how this sphere of the gut microbiome may also be a source of novel bioactive agents with potential commercial and clinical applications.145-148
Functions relevant to colonization of the gut and survival in this environment have been attributed to plasmid-encoded genes in a number of species.31,42,43,64,78,118,129,130,136,140 These include mechanisms which promote adherence to host tissues, as described for the human gut commensal E. coli SE11 which harbors six plasmids encoding genes involved in bacteriocin production, tetracycline resistance as well as adherence to host cells.43 In the case of attachment to host tissues it has been proposed that this may be a feature of conjugative plasmids and transposons in general.89,130 Pili and associated conjugation machinery assembled by these MGE, in order to transfer plasmids, may also directly facilitate bacterial adhesion to human cells and tissues.90,130,149 In this scenario, species harboring conjugative elements may be better able to colonise the GIT which would serve not only to enhance the fitness of the host organism, but also offset the fitness cost of plasmid carriage by promoting maintenance of plasmid harboring cells, and by default maintaining the plasmids they carry within the gut community.
Other plasmid-encoded colonization factors may include those that mitigate the toxic effects of bile acids. Thus, the observation that bile tolerance or resistance mechanisms are encoded by plasmids in some gut-associated species is also of considerable interest.31,42,63 Conjugated bile acids are key components of bile, and have diverse functions in the host ranging from absorption of dietary lipids to roles as key signaling molecules regulating a variety of systemic metabolic processes.25-27,62,150 These compounds are also toxic to bacteria and constitute a significant barrier to colonization of the GIT.62-64,150 Genes encoding bile salt hydrolase (BSH) activity, which catalyzes the de-conjugation of bile acids, and has been shown to facilitate tolerance to these compounds in a range of gut-associated bacterial species.62-64,151 This activity is widely distributed in the gut community and present in bacterial species from all major phylogenetic divisions, as well as gut associated archaeal species.62
Such wide dissemination provides a high level of functional redundancy for this activity within the community and it seems likely that HGT has contributed significantly to its broad distribution, with evidence for the transfer of relevant genes between distinct domains of life (Bacteria and Archaea) also documented.62-64 However, the role of bile acids as signaling molecules also opens the possibility for microbial BSH activity to influence wider aspects of host physiology through perturbations in bile acid signaling via production of modified bile acid derivatives that display altered binding characteristics for relevant receptors.62 The observation that this function is also encoded in the mobile metagenome of the gut microbiota suggests further exploration of plasmids and other MGE associated with this community will yield important fundamental insights into its development and overall function, as well as mechanisms by which gut bacteria interact with the human host.
Virulence, commensalism and lifestyle diversification
Plasmids and other MGE comprising the gut mobile metagenome may also encode traits involved in virulence and pathogenesis, and a number of members of the normal microbiota in healthy individuals are also prominent opportunistic pathogens.78,129-139,152 A wide range of gut-associated pathogenic species have been reported to harbor plasmids which confer the ability to produce key virulence factors or other traits contributing to disease processes.72129–139,152 Plasmids coding for the production of toxins and virulence factors,78,136,139,152,153 cell invasion and survival within macrophages,31,78,131,137,153-155 attachment to host tissues,78,90,129,130,136,139 immune evasion,153 type III secretion systems and associated effectors,137,153 as well as host-microbe signaling processes relevant to pathogenesis,78,131,135,139,153 have all been described in gut-associated pathogens.
E. coli has been particularly well studied in this regard, and in the case of gut-associated E. coli strains, plasmids encoding a wide range of traits appear to have played a major role in the diversification of lifestyles adopted by this organism, which range from beneficial gut commensal to highly virulent primary pathogen.43,44,78,129,130,134,136,139 Of the six intestinal E. coli pathotypes, all possess virulence plasmids and HGT in general has been pivotal to the emergence of a pathogenic lifestyle in these strains.78,152 A key example is O157:H7 in which several of the major virulence factors have been acquired by HGT, such as the shiga toxin via bacteriophage,152 while plasmids encoding hemolysins, toxins and factors mediating attachment to host cells are also common.78,136,139,152 In particular, plasmid pO157 is reported to be present in around 99–100% of human O157 clinical isolates, and highly conserved, differing only by single nucleotide polymorphisms between many isolates.78,139,152,156 Conversely, beneficial or harmless strains of gut-associated E. coli also appear to frequently harbor plasmids, including those encoding functions which facilitate their commensal lifestyle, many of which are also functions associated with virulence plasmids, such as attachment to host tissues.43,44
Although the presence of plasmids or other MGE encoding potential virulence attributes among members of the normal gut microbiota in healthy individuals may initially appear counterintuitive, a wide variety of mechanisms for host-microbe interaction may be of utility to both commensals and pathogens. In particular, mechanisms that contribute to colonization of and persistence within the human GIT, as well as host-microbe signaling and immune modulation, may be put to use by both beneficial species and pathogens alike, albeit with radically different outcomes for the host. The deployment of the same basic machinery for host cell adhesion by both commensal and pathogenic E. coli species in this ecosystem is a prime example,157 and mechanisms for adherence to host cells and colonization of the gut epithelium, have been shown to be plasmid-encoded in both pathogenic and commensal strains.44,78,129,130,136,139,156 This highlights the potential for the patho-biotechnological exploitation of the gut mobile metagenome in the rational design of probiotics and associated delivery systems.146-150
Overall, the involvement of plasmids as well as other MGE in diversification of lifestyles among distinct strains of the same species colonising the gut environment, highlights the potential for MGE to provide bacteria with a flexible and diverse genetic resource that permits adaptation to a wide range of ecological niches within a complex microbial community, and ultimately enhances the fitness and success of the species as a whole.42,44,78,152 There is also the possibility to extend this basic concept to the gut microbiome, with the mobile metagenome facilitating lifestyle diversification and adaptation of constituent members, generating functional redundancy, and promoting stability. This would enhance the overall fitness not only of the entire community, but also of the associated metazoan host by virtue of generating a stable and productive gut microbiota with associated benefits. Such hypotheses fit well with the proposed hologenome theory of evolution,4,5,30 but at present there is little evidence to confirm or refute their accuracy.
The mobility of virulence factors among gut pathogens also has important implications for the evolution of deleterious host-microbe interactions in this ecosystem. It is currently unclear if plasmids resident in the gut mobile metagenome are an important reservoir of potential virulence attributes which may be accessed by emerging gut pathogens or transiently colonizing species, but the clear contribution of HGT and plasmid-encoded factors to the evolution of virulence in a wide range of prominent gut pathogens,78,129-139,152 argues for further investigation of the human gut mobile metagenome from this perspective.
Plasmid Transfer in the GIT
In light of the increasing burden of antibiotic resistance in bacteria, and the growing appreciation of the gut microbiota and other human associated microbiomes as reservoirs for antibiotic resistance genes, understanding the factors influencing HGT in environments such as the GIT are of considerable importance from this perspective alone. However, such information is also required to complete our fundamental understanding of how the gut microbiota develops and functions, and to access the full biotechnological and clinical potential of this microbial ecosystem. The mammalian GIT is considered a hotspot for HGT, likely due to a high population density coupled with the presence of large numbers of closely related species derived from relatively few phylogenetic divisions, factors known to be beneficial to conjugal transfer of plasmids.20,32,50,90,159,160 In addition, the growth of bacteria as biofilm-like communities associated with the intestinal mucus layers may also promote the transfer of plasmids between constituent species, as well as between indigenous members and transiently colonizing species.161
The in vivo transfer of plasmids in the GITs of mammalian or avian model organisms has been demonstrated between both closely related species, as well as between more distantly related phyla.80,90,102,103,105,107-109,113-115,119,138,158,161-169 These include both Gram-negative and Gram-positive species derived from all major phylogenetic divisions (Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria) of the human microbiota, but most studies have traditionally focused on the transfer of antibiotic resistance genes among potentially pathogenic species that frequently form part of the normal human gut bacterial community. Examples include the transfer of plasmids encoding resistance to ampicillin between distinct strains of E. coli in the infant GIT,108 the acquisition of plasmids encoding carbapenem resistance by commensal E. coli from K. pneumonia strains;105,170 and the transfer of plasmids encoding erythromycin resistance from probiotic lactobacilli species to E. faecalis.102,103
Of particular concern are recent reports detailing the transfer of plasmids harboring extended spectrum β-lactamases among gut-associated species, which not only confer resistance to a wide range of commonly used β-lactam antibiotics including those designed to resist β-lactamase activity, but are also associated with multiple antibiotic resistance phenotypes.105,168 The broader range transfer of plasmids between Gram-negative and Gram-positive members of the gut microbiota has also been demonstrated,169 and recent studies have highlighted how some plasmids found in gut-associated species may function as novel broad range shuttle vectors, and the potential for these to develop naturally in the mobile metagenome.32
Factors influencing plasmid transfer rates
Numerous factors have been found to contribute to the relative rates of transmission for different plasmids. These include attributes of a particular plasmid and recipient/donor species studied, such as the conjugation machinery employed, the physiological status of donor and recipient cells, compatibility of plasmids with host cell replication machinery and potentially other plasmids already harbored by the cell.56,90,159 Collectively these factors will dictate the host range of a particular plasmid and therefore how widely it may be disseminated within a microbial community with a given population structure.
Furthermore, intrinsic features of the plasmid, such as its encoded functions and the level of selection for these, will also influence transfer rates and stability in the GIT. For example, plasmids producing short rigid pili have been observed to transfer more efficiently when donor and recipient cells are associated with a solid surface, while those that utilize long flexible pili can transfer equally well in liquid culture.56,90 In terms of selection and the influence of plasmid-encoded traits, it is well documented that plasmid transfer is generally higher when there is strong positive pressure for traits encoded, as exemplified by studies of plasmids harboring mercury resistance in soil bacteria.170,171 This phenomenon has also been observed for plasmid transfer in the GIT, with selection for plasmid-encoded traits enhancing the rate of transfer and spread,102,172 and indicating some plasmids may have an optimal level of selection at which transfer rates are highest, as opposed to a simple cumulative effect of increasing selective pressure.172
In addition, ability to transfer will also be impacted by aspects of donor and recipient cell abundance, their physiological status, and distribution in this environment. In terms of host cell status, the availability of nutrients, growth phase and general cell health are all implicated in the kinetics of plasmid transfer,52,56,90,159,161 and therefore the relative fitness of donors and recipients within the gut environment will also play a role in the levels of plasmid transfer observed in vivo. However, much accepted wisdom regarding factors affecting plasmid transfer, and the degree to which these influence rates of conjugal transfer, has been derived from laboratory experiments using batch cultures.52,160
Although in vitro systems have provided much insight into the kinetics of plasmid transfer, these effectively represent artificial and simplistic model systems which do not incorporate, or permit assessment of, key parameters that are likely to significantly influence plasmid transfer in vivo.160 As well as factors such as population density of donor and recipient cells, and their relative status within a population (e.g., well adapted allochthonous members of the community vs. transiently colonizing autochthonous species); the complexity of the associated microbial ecosystem, the motility of host and donor species within the environment, the presence of homogenizing mechanical forces or flow, and physicochemical characteristics of the environment can also impact greatly on the observed rate of transfer.52,90,159,161
Factors influencing plasmid transfer in the GI tract
While some studies of plasmid transfer in natural environments, including the gut, have shown a lower rate in vivo than in vitro,173-175 a growing number of studies investigating transfer in the GIT using animal models have described a generally higher rate of transfer for certain plasmids in vivo, compared with in vitro laboratory conditions, and indicate existence of indigenous transfer promoting factors in the GIT.38,89,102,103,138,158,161,162 Furthermore, in vitro assays have been shown to underestimate the true rates of transfer in vivo when factors other than donor and recipient cell densities are accounted for;158,161,162 however, it should also be noted that many studies investigating plasmid transfer in vivo have used gnotobiotic animals which lack a gut microbiota, or models in which the microbiota has been disrupted using antibiotics to permit colonization by donor or recipients (reviewed in ref. 90). As the presence of a complex microbiota may itself be a factor limiting the rate of conjugal transfer in the GIT, experiments in germ free animals likely do not reflect the true rate of plasmid transfer in this environment. Nevertheless, a growing number of studies support the GIT as a hotspot for HGT, and suggest that certain features of this environment may enhance plasmid transfer.
A particular feature of the GIT that is thought to provide favorable conditions for plasmid transfer is the intestinal mucous layer, and the formation of biofilm-like communities within this matrix.90,161 Plasmid transfer is thought to be enhanced by the close cell-cell contacts generated when bacterial biofilms are formed, and rates of transfer for some plasmids are observed to be higher in biofilm communities compared with planktonic cell populations, including studies with gut-associated species.161,176-178 Licht et al. (1999)161 indicated that in biofilms the initial spread of plasmids after introduction of donor cells is rapid, but subsequently reduces to very low levels or ceases completely. This is believed to be due to a combination of fixed spatial positions of donor and recipient cells within the biofilm, and the general limitation of plasmid spread to cells immediately surrounding donors during initial transfer events, rather than continuous rounds of subsequent transfer initiated by newly formed transconjugants.161
The observed lack of subsequent transfer from new trans-conjugants is most likely a result of the physiological status of cells within the biofilm, as well as variation in nutrient availability in different sections of the biofilm.161 The authors also noted that the transfer kinetics of plasmids in vivo were more similar to those observed in biofilm communities than in mixed liquid cultures, which was attributed to growth and transfer within the mucus layer. The observation that conjugative plasmids enhance biofilm formation in vitro on abiotic surfaces by virtue of their transfer machinery, which may also promote adhesion to mucosal surfaces, reinforces the mucosa associated plasmid transfer hypothesis.129,130,149
Importantly, this model makes certain predictions regarding the ability of plasmids to transfer after initial colonization of the mucus layer, and suggests that this feature of the GIT will enhance the initial transfer of plasmids from donor cells to recipients, but subsequent transfer will be limited by the nature and physiological status of surrounding cells.161 If so, then transfer of plasmids to donor cells would be expected to not only occur rapidly after initial colonization of the intestinal tract, but depend greatly on the nature of surrounding cells and the numbers of suitable recipients in the gut community.161 In contrast, the proposed rapid nature of initial transfer events increases the likelihood of plasmid transfer from transiently colonizing species to normal members of the gut microbiota.161 Aside from the acquisition of undesirable traits such as antibiotic resistance, this would also afford the gut microbiota greater opportunity to sample the wider flexible prokaryotic gene pool and aquire new functional capacity of benefit to both host and microbe. The recent observation that the Japanese gut microbiota has been subject to functional enhancement through plasmid-mediated HGT fits well with this scenario.50 In this example, genes required for utilization of seaweed glycans by gut commensals were shown to have been acquired from marine bacteria that naturally colonise dietary seaweeds, and frequently come into contact with gut microbes through the consumption of uncooked seaweed.50
Given the ancient nature of the proposed transfer events underpinning this functional adaptation, the exact mechanisms involved cannot be determined with certainty, however, genetic evidence indicates this gene exchange was mediated by plasmid transfer, and it is tempting to speculate that the kinetics of plasmid transfer observed in other studies of the GIT, particularly the potential for rapid initial transfer from incoming foreign organisms, were important to this event. Regardless of the factors dictating the original transfer event, such studies highlight the potential impact on the host of plasmid transfer in the gut microbiota, in this case mediating access to a new dietary energy source.50
Can host diet influence rates of plasmid transfer in the GI tract?
Other features of the GIT are also thought to be favorable for conjugal transfer. In other habitats, such events have been shown to occur at highest rates when cells are in a nutrient rich environment, with nutrient limitation observed to reduce transfer rates.160,179-182 In general the GIT should provide indigenous microbes with an ample and steady supply of nutrients, and this may also be true for some transiently colonizing organisms. However, in the case of allochthonous species the high level of competition extant in the gut microbiota and the resulting occupation and exploitation of available ecological niches by autochthonous species is likely to effectively exclude many such organisms and limit their ability to proliferate and utilize available nutrient sources.2,8,183 Therefore, the proposed advantages of mucosal associated communities mediating rapid transfer of plasmids may be offset by effects of competition and nutrient limitation in transiently colonizing species, reducing transfer between allochthonous and autochthonous species.
In this context it is of note that for certain plasmids, changes in host diet have been found to have a significant impact on rates of plasmid transfer.103,184 While the GIT may provide a relatively steady source of nutrients overall, natural variation in diet means this is unlikely to be completely consistent, and when considered in terms of the complexity and niche specialization that exists in this community, it seems unlikely that all members will receive a steady and constant supply of readily utilizable nutrients. Furthermore, although in industrialized countries access to food supplies on demand are taken for granted, a significant portion of the extant human population is subject to periods of starvation.
The influence of diet on plasmid transfer could also occur via alternative mechanisms, not least of which is the impact of diet on the physicochemical properties of the intestinal milieu. In addition to factors such as nutrient availability, the fluidity of intestinal contents, the production of metabolites by indigenous community members, effects on community structure, and gut motility, are all factors that may be potentially influenced by diet, and are believed to influence plasmid transfer.89,155,157 There is also potential for diet to influence rates of plasmid transfer by modulating the level of host digestive secretions in the intestinal lumen. A prime example of this is the increased levels of bile secreted in response to diets high in fat. Exposure to bile has been found to increase the rates of curing and plasmid loss in Salmonella enterica, for otherwise highly stable plasmids,185 and the bile salt deoxycholate was subsequently shown to reduce the transfer rates of the virulence plasmid pLST in vitro.138
For E. coli, formation of the conjugation pilus associated with transfer of the F plasmid, has been shown to increase sensitivity of this organism to bile salts commonly encountered in the human GIT, particularly conjugated bile acids.186 In this case, the increased sensitivity of pili expressing cells would likely serve to destabilize plasmids in gut dwelling populations in compartments where levels of bile acids are high (such as the duodenum, jejunum and proximal ileum), and restrict transfer to regions of the GIT where bile levels are low, such as the distal ileum and colon. Of particular interest in this context are the observations by Tuohy et al. (2002), who documented a decreased level of trans-conjugant formation when rats were fed a high fat human diet.103 Although the mechanisms involved in this dietary induced change in plasmid transfer were not elucidated in this study, given that increased fat intake increases levels of bile in the intestine it is possible that this impact of diet on intestinal physiology was involved in the observed reduction in plasmid transfer. Overall, the effects of diet on plasmid transfer merit further investigation, and may provide options for suppressing in vivo transfer among certain groups of bacteria in the human GIT.
This also highlights the potential variation in plasmid transfer rates across different compartments of the GIT, which vary in terms of parameters known to influence plasmid transfer.138 Few studies account for the distinct compartmentalization of the GI tract and the variation in environmental parameters along the length of the intestinal tract such as pH, osmolarity, oxygen tension, and metabolic diversity, which are encountered by microbes from entry to exit. It is likely that the changing environmental factors encountered along the length of the GIT add an additional level of complexity to plasmid transfer in vivo. Plasmid transfer rates at different locations in the GIT have yet to be explored in detail, with the majority of in vivo studies concerned only with the levels of trans-conjugants that appear in faeces. However, evidence for variable plasmid transfer rates among different compartments of the GIT may be exemplified by the impact of bile on the transfer of Salmonella virulence plasmids,138 as well as the potential role of this endogenous host secretion in plasmid curing in this prominent gut pathogen.185,186
Phage-mediated transfer of plasmids
There also exists the potential for plasmid transfer in the gut to occur through mechanisms other than conjugal transfer. In particular, the transfer of plasmids between Lactococcus lactis and Streptococcus themophilus through phage-mediated transduction has recently been demonstrated.187 This adds a further possible mechanism of plasmid transfer relevant to the GIT, the significance of which is yet to be explored in terms of HGT in the gut microbiota and microbial ecosystems in general. Because bacteriophages are abundant in the gut microbiota, and this community is essentially comprised of high numbers of closely related strains and species, it is conceivable that this mechanism plays an important role in the transfer of some plasmids in this community. However, it seems unlikely that a wide range of plasmids will be compatible with this mechanism of transfer, and given the narrow host range of most phage it is probable that the contribution of this form of plasmid transfer in the GIT will be restricted to a relatively low number of plasmids, and their transduction among closely related strains or species.
Future considerations for the study of plasmid transfer in the GI tract
Overall our understanding of the relative rates of plasmid transfer in the gut are limited by available methods to study these events, which typically utilize artificially introduced selectable recipient strains (often through colonization of either germ free animals or post antibiotic disruption of the extant microbiota) and subsequent monitoring of trans-conjugant numbers in faeces. In most cases the motivation for such studies has been to develop an understanding of the transfer of antibiotic resistance genes in the gut microbiota, and the ability of this community to acquire the relevant MGE and act as a reservoir of these traits. As such donor and/or recipient species as well as the plasmids tested may not represent species, elements, or functions prevalent in this community.
The functions encoded by a plasmid are likely to be of much significance to the relative in vivo transfer rate, and many studies have demonstrated that transfer occurs more frequently when selective pressure is introduced for plasmid-encoded traits.102,170-172 As such, utilizing plasmids derived from the gut microbiota itself, and encoding functions relevant to environmental stresses normally encountered by well adapted gut inhabitants should provide a more accurate assessment of the rates of plasmid transfer among members of this community under normal conditions. Furthermore, the importance not only of the prevalence of the donor and recipient cells within a community, but also their physiological status upon rates of plasmid transfer, also suggests that utilizing species normally present in this community and well adapted to life in the gut, in conjunction with naturally occurring plasmids infecting these species, will greatly enhance our understanding of the rates of gene transfer in the gut community.
Thus, our knowledge regarding transfer of plasmids and other MGE that naturally form part of the gut mobile metagenome, as well as the functions they encode and how they contribute to host health is generally limited. Recent metagenomic studies providing evidence that HGT is not only prevalent but important in the evolution of this community and its impact on host health and development, highlight the need for a greater understanding of this aspect of the gut microbiota (Table 1).8,20,32,50-53,62-65 However, a range of methods for plasmid isolation are available and new methodologies have been developed which should permit greater access to the gut mobile metagenome and allow wider study of this important sphere of host-associated microbial ecosystems, including greater access to the biotechnological and pharmaceutical potential of the gut mobile metagenome.
Strategies to Access Plasmids Resident in the Gut Mobile Metagenome
Despite the potential contribution of the gut mobile metagenome to the evolution and functioning of the gut microbiota, including its impact on host health, this flexible gene pool remains largely unexplored. Such studies are technically challenging and investigation of MGE associated with microbial communities has traditionally been impeded by the large number of uncultivated species comprising these ecosystems, including the gut microbiota where ~70–80% of species remain uncultured,2,18,30,31,56,58 as well as the enormous diversity of MGE that may be associated with complex microbial ecosystems.30,31,48,51,58 Furthermore, the high levels of variability in terms of gene content and architecture inherent to MGE such as plasmids, has precluded the development of universally applicable methods for survey and acquisition of certain MGE types comprising the mobile metagenome of a particular microbial ecosystem,
In contrast to studies of the population structure of microbial communities, in which the species present can be identified through a range of universally applicable culture-independent approaches (most commonly 16S rRNA gene amplification and sequencing-based strategies), no such global census can presently be generated for plasmids or other MGE, and given the malleable nature and mosaic structure of many elements characterized in detail so far, coupled with the lack of regions universally conserved in all plasmids (analogous to phylogenetic anchors such as the 16S rRNA genes in bacteria), it is difficult to envision the development of such strategies with current knowledge and available technologies. As such, studies of plasmids associated with bacterial communities are impeded by a fundamental lack of knowledge regarding the composition of the resident plasmid population in terms of numbers and types of elements present. Concordantly, the development of methods to access these elements is challenging, and evaluation of their ability to acquire a representative cross-section of elements present is virtually impossible.
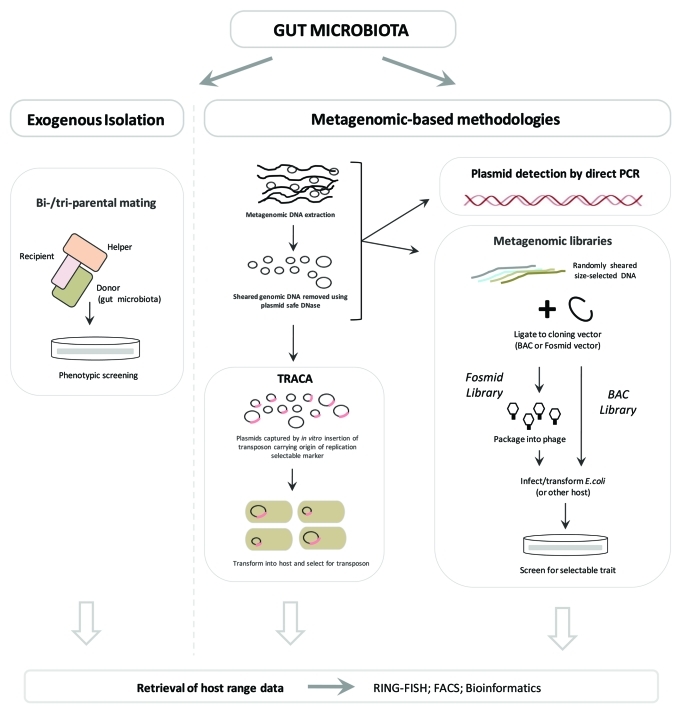
Although technically challenging, a range of methods has been developed for accessing plasmids resident in microbial communities, which have all produced valuable insights into functions of MGE and the bacterial mobile metagenome.20,30-37,42,47,48,50,58,188-193Table 2 summarizes current strategies available for accessing plasmids resident in the gut microbiota and other microbial communities as well as their relative strengths and weaknesses when applied to the study of complex microbial ecosystems such as the gut microbiota. These range from the classical endogenous and exogenous plasmid isolation methods,56,58,189 to more recent metagenomic approaches such as the Transposon Aided Capture System (TRACA)33 system and the direct detection of plasmid sequences in standard metagenomic libraries.33,46,188
Table 2. Relative merits of currently available plasmid isolation strategies for investigation of whole communities (modified from references 31 and 58).
| Plasmid isolation strategy | Advantages | Disadvantages | Potential augmentation/future utility |
|---|---|---|---|
| Endogenous isolation |
• Original bacterial host is known • May be used for all cultivatable bacteria • Applicable to all plasmid types |
• Requires host cultivation restricting utility for study of natural communities • Reliance on plasmid-encoded traits if surrogate host species required for plasmid characterization |
• As new organisms are isolated and culture requirements defined, endogenous isolation will continue to be of utility • Compatible with large-scale whole genome sequencing projects |
| Exogenous isolation |
• Culture-independent • Selective isolation of self-transmissible or mobilizable elements • Potentially capable of isolating all plasmid types (circular and linear), and sizes. • Can isolate plasmids irrespective of abundance in community |
• Relies on plasmid-encoded traits for plasmid transfer, selection, and maintenance in surrogate host • Original bacterial host unknown • Range of plasmids isolated dependant on mating conditions used and dictated by numerous “unknown” environmental variables influencing host cell physiology and plasmid transfer kinetics. |
• Scope for retrieval of plasmid host range information using RING-FISH, or bioinformatic approaches |
| PCR-Based detection |
• Culture-independent • High-throughput • Sensitive • Scope for accurate quantitation of plasmids |
• Original bacterial host unknown • Complete characterization of plasmid detected generally impossible. • Limited to detection of known and characterized plasmid lineages used for primer design. |
• Utility will increase as number of available plasmid sequences grows. • Much scope for combination with other strategies for plasmid isolation to facilitate subsequent analysis of plasmid distribution and abundance. |
| TRACA |
• Culture-independent • Suitable for development of high-throughput strategies • Can isolate plasmids irrespective of abundance in a community • Fully independent of plasmid-encoded traits • Sequence-based characterization of plasmids facilitated by known Tn sequence in plasmids • Potentially applicable to all circular plasmids and bacterial communities • May permit capture of MGE other than plasmids when present as circular DNA molecules205 |
• Original bacterial host unknown • Transposon may inactivate genes of interest, impeding phenotypic characterization • Currently available Tn elements and surrogate host may limit range of plasmids isolated. • Linear plasmids not captured • Transformation step may introduce size bias • Plasmids belonging to same incompatibility group as Tn origin may not be captured due to stability issues in surrogate host. |
• Scope for retrieval of plasmid host range information using RING-FISH, or bioinformatic approaches • Amplification of plasmids and size fractionation prior to capture may further increase range of plasmids isolated and reduce size bias. • Use of a mixture of Tn elements encoding a range of origins of replication, along with alternative surrogate host species (particularly Gram +ve species) will enhance range of plasmids captured and permit functional characterization.205 |
| Standard Metagenomic libraries (BAC/Fosmid) | • Culture-independent • Suitable for development of high-throughput strategies • Initial capture independent of plasmid-encoded traits • Sequence-based characterization facilitated |
• Original bacterial host unknown • Likely bias toward numerically dominant plasmids • Screening relies on plasmid-encoded traits expressed in surrogate host species • Not specifically designed for plasmid capture, and non-plasmid sequences dominate libraries. • Generally only incomplete, partial plasmids identified • General compatibility of library construction methods with plasmid capture unknown. • Plasmids belonging to same incompatibility group as vector (BAC/Fosmid) may not be represented due to instability of clones in surrogate host • Plasmids belonging to same incompatibility group as vector may not be captured due to stability issues in surrogate host. |
• Scope for retrieval of plasmid host range information using RING-FISH, or bioinformatic approaches • Scope for generation of plasmid specific libraries based on cloning endogenously isolated plasmids extracted from whole community. |
Endogenous isolation of plasmids
The most straightforward and well-established method for plasmid acquisition is the direct extraction of plasmids from the native host species.31,56,58 This typically requires cultivation of the plasmid host species, or a mixed microbial population with or without selective enrichment for traits of interest.31,56,58 Subsequently, isolated plasmids are often transformed into a surrogate host, if possible of the same species, and plasmid-encoded traits assessed through predominantly phenotypic assays.31,56,58 The complete nucleotide sequences of isolated plasmids may also be obtained, and plasmid-encoded functions characterized in silico. The generation of plasmid sequences during whole genome sequencing projects may therefore also be considered as examples of the endogenous approach, for instance the characterization of the L. salivarius UCC118 plasmid complement.42,43
A major advantage of this method over other plasmid isolation systems is the unambiguous identification of the natural host species of plasmids isolated.31,58 However, since plasmids are often promiscuous and capable of infecting multiple host species, endogenous isolation does not necessarily provide a complete picture of the range of host species in which a particular plasmid may reside.58 In addition, the culture-dependent nature of this approach severely compromises its utility when applied to study the plasmid populations in complex bacterial ecosystems, such as the gut microbiota.31,33,58 In these cases, the high proportion of uncultivated species comprising such ecosystems greatly restricts the range of plasmids that may be accessed with this approach, rendering it of limited value in this context.31,33
A variation of this approach that may be employed to improve the range of plasmids captured is the direct isolation of plasmid DNA from the mixed microbial population without any prior cultivation, and their subsequent transformation into a surrogate host species for characterization.33 However, this negates a key advantage of the endogenous approach, as information regarding the original host species is lost, and introduces an additional series of potential biases by relying on plasmid-encoded traits, such as the ability to replicate in the surrogate host and the presence of selectable markers, which collectively maintain the restricted utility of this approach for studying the plasmid pool in complex microbial ecosystems.33
Exogenous isolation of plasmids
Due to the inherent limitations of endogenous approaches, a range of culture-independent plasmid isolation methods have been developed, which permit access to a greater range of plasmids resident in bacterial communities (Fig. 1; see refs. 33, 58, 188, 189 and 192). The best established of these methods are the exogenous isolation approaches, which rely on the natural ability of plasmids to transfer between species.189 In this approach, plasmids are acquired by utilizing a selectable surrogate host species in bi-parental or tri-parental mating with the donor population under study, during which plasmids resident in the donor species/community may be transferred via conjugation into the selectable recipient.58,189-193 Subsequently, cells are transferred to media selective for the surrogate recipient strain, as well as plasmid-encoded traits, in order to specifically recover recipient strains that have acquired plasmids.58,189-193 Bi-parental matings can be used to recover self-transmissible conjugative plasmids, while tri-parental matings in which additional donor species carrying a mobilizing helper plasmid are employed, can be used to recover plasmids that are mobilizable but not capable of autonomous transfer.58,189-193
Figure 1. Overview of culture-independent strategies to access plasmids resident in the human gut mobile metagenome.
This exogenous approach permits access to plasmids from the total community regardless of ability to cultivate host species, and has successfully been applied to study a wide range of bacterial ecosystems, including the human gut microbiota.58,189-193 The culture-independent nature of this method is a significant advantage over endogenous approaches, and the acquisition of conjugative or mobilizable plasmids is also of much benefit in identifying elements capable of transfer between host species and important in HGT events within a particular community.31,58,189 However, as with modifications to the endogenous approach described above, information regarding the original host species of plasmids is lost and both methods rely heavily on plasmid-encoded traits such as the presence of selectable markers, or ability to replicate in surrogate host species, which again restricts the range of plasmids that may be isolated.31,58 For example, in this approach recipient cells harboring captured plasmids must be selected for based on the ability of plasmids to confer a selectable phenotype on host cells, most typically resistance to antimicrobial agents such as heavy metals, antibiotics, and biocides is used.31,58,189 While this selectivity may be an advantage in some studies, such as those concerned with specifically understanding the plasmid-mediated spread of antibiotic resistance genes, plasmids not encoding such traits or not expressing these in the surrogate host will not be isolated, limiting the utility of this method for studying the mobile metagenome in general, and its role in fundamental microbial ecology and community evolution.31,33
A variety of other factors also influence the range of plasmids that may be captured using this method, including the mating conditions used.31,56 The duration, temperature, and use of solid or liquid media can affect the range of plasmid types that are acquired, and for some plasmids the physiological status of the host cell, and the presence of environmental cues can influence the initiation of transfer events.31,52,56,90,159,161 Since many organisms in complex communities such as the gut microbiota remain uncultured and almost completely uncharacterized, it seems inevitable that the mating conditions used will be sub-optimal or inhibitory for the transfer of many plasmids, further restricting the fraction of the mobile metagenome that may be sampled and studied with this method.31
Detection of plasmids in standard metagenomic libraries
More recently a range of metagenomic approaches which permit culture-independent access to members of the gut microbiota have been established, and applied to the study of microbial ecosystems, including the mammalian gut microbiota. These include strategies that permit access to plasmids and other MGE either through the construction and screening of metagenomic libraries,46,188 or via the specialized culture-independent TRACA.33 Recently, Kazimierczak et al. (2009) described the isolation of plasmids or plasmid fragments, from standard metagenomic libraries of the organic pig gut microbiota, including those with the ability to replicate autonomously when liberated from the BAC vector and reconstructed by self-ligation.188
While this method successfully identified novel plasmids, drawbacks inherent in exogenous methods remain, including reliance on plasmid-encoded selectable traits, and replication in surrogate hosts for demonstration of autonomous replication.188 Furthermore, this approach is not designed to specifically capture plasmids, but is focused on the acquisition of any metagenomic DNA fragments encoding genes that confer a phenotype of interest on surrogate host cells, in this case tetracycline resistance. As such, sequences captured are not limited to those originating from complete or partial plasmid fragments that have been cloned, and at present this method is not suitable for the specific exploration of the plasmid population associated with the human gut mobile metagenome. Nevertheless, this approach represents a useful strategy for investigating the potential mobility of genes involved in traits of interest within a bacterial community, and is clearly capable of isolating novel, autonomously replicating plasmids.
Transposon aided capture (TRACA) of plasmids
In contrast to the use of standard metagenomic libraries, the TRACA system has been specifically designed to capture plasmids associated with the mobile metagenome of a microbial community.33 This system overcomes some of the key limitations of other plasmid isolation systems, and offers several major advantages including an independence from plasmid-encoded traits for plasmid isolation.31,33 In the TRACA system, plasmids are captured directly from metagenomic DNA extracts, treated with plasmid-safe DNase, by supplying all plasmids with a defined selectable marker and an origin of replication suitable for the chosen surrogate host species (Fig. 1). This is accomplished by delivering the required genes to plasmids extracted from community members via a transposon, in an in vitro reaction.33 Following in vitro transposon tagging, plasmids are transformed into surrogate host cells and recovered on media selective for the transposable element.33 In this way, the transposon effectively retrofits all plasmids in the population with a known selectable marker and suitable origin of replication, permitting subsequent selection and stable maintenance in the surrogate host species independently of plasmid-encoded traits.33 This greatly enhances the range of plasmids that may be captured and application of TRACA to study plasmids resident in the human gut mobile metagenome has demonstrated its ability to isolate novel plasmids devoid of any conventional selectable markers.33
Despite the major advantages offered by the TRACA system, this method does not address all drawbacks of other culture-independent systems, and may also be subject to an additional limitation unique to this approach.30-33 At present all plasmids isolated with the TRACA system have been in the smaller size range (~14 kb or smaller). As this system is yet to be exploited on a large scale and across a wide range of bacterial ecosystems it remains to be seen if this represents a true size bias of the TRACA system, or is a result of a predominance of smaller plasmids in the gut ecosystem.31,33 While there is currently no means to provide a definitive survey of plasmid size in a given bacterial ecosystem, there is evidence that physical features of plasmids including size, are responsive to environmental and ecological factors in the same way as bacterial chromosomes.52
Plasmids appear to follow the same trend as chromosomes, of decreasing size and gene content as complexity and stability of the external environment reduces and increases, respectively.52 Surveys of plasmid size in relation to host habitat have indicated that fewer large plasmids (> 100 kb) are associated with microbial species colonizing higher host organisms (animals and plants), in contrast to terrestrial and aquatic environments.52 It was proposed that this may be due to selection for large plasmids in highly complex and heterogeneous environments, such as soil, which encode a wide range of accessory traits allowing the host to adapt to multiple and diverse ecological niches and environmental stresses encountered.52 However, there may also be mechanisms or selective pressures extant in certain microbial communities which actively limit plasmid size, but at present any such processes remain obscure. As such, there exists the potential for the gut microbiota to be populated by predominantly smaller plasmids, but such hypotheses are not presently supported by any firm evidence and larger plasmids clearly do infect important members of this community and form part of the gut mobile metagenome (exemplified by the lactobacilli and enterococci megaplasmids40-42). In general, elucidation of the average size and dominant plasmid types of elements resident in the human gut is unknown and awaits the implementation of an in-depth census of the gut-associated plasmid pool.
Overall, it is most likely that both the composition of the natural plasmid population and features of the TRACA methodology will contribute to the characteristics of captured plasmids, and the transformation step in the TRACA system represents a key point at which size bias may be introduced, since smaller elements are transformed with much greater efficiency than larger plasmids.31,33 In this regard, size fractionation and amplification of plasmid DNA prior to transposon tagging and transformation may permit larger plasmids to be acquired more readily with this system. In addition, the use of surrogate host species other than E. coli, and transposons with a wider range of replication origins should also increase the plasmid range that can be acquired, further enhancing this method.
As with exogenous isolation and the use of standard metagenomic libraries, plasmids isolated by TRACA are also divorced from the original host species and this phylogenetic host-range information is lost.31,33 Although not required for subsequent characterization of plasmid-encoded functions, knowledge of the original host range of such plasmids is highly desirable and would greatly facilitate our understanding of the role of the gut mobile metagenome in dissemination and distribution of key community functions.30,32 There is, however, significant scope to compensate for loss of host range data in the TRACA system, as well as other culture-independent plasmid capture approaches, by implementing downstream plasmid characterization aimed at retrieving such information.
Retrieval of host-range data following culture-independent plasmid isolation
Several strategies currently exist which theoretically permit the retrieval of host range data following the culture-independent isolation of plasmids; a summary of how these may be integrated into existing culture-independent workflows is presented in Table 2. These include the utilization of fluorescent in situ hybridization (FISH) coupled with fluorescence associated cell sorting (FACS), in which plasmid specific probes may be used in conjunction with FACS to retrospectively acquire intact bacterial cells harboring plasmids of interest.194 Recovered cell populations may then be analyzed to determine phylogenetic origin using 16S rRNA gene sequences.31,195
Alternatively a number of purely bioinformatic approaches to determine the phylogenetic origin or affiliation of DNA fragments have been developed, including several specifically designed to recover plasmid host range data.196,197 In particular, differences in the relative frequencies of di- or tri- nucleotide sequences in plasmids and chromosomes of bacteria have been used to determine evolutionarily distant host ranges of a variety of plasmids.196-200 The absolute average relative abundance of di-nucleotide repeats (or delta-distance) has previously been exploited to identify recently acquired or “foreign” genes in bacterial genomes based on differences in this genomic signature, which remains relatively constant throughout the genome but is noticeably altered in recently acquired DNA.197-200
Since plasmids and other MGE have been demonstrated to acquire the genomic signatures of their long-term hosts over time, and these genomic signatures are discriminatory in terms of bacterial species relatedness,197,198,200 this concept has also been applied to identify host-plasmid associations based on similarities in genomic signatures.197,198,200 The Mahalanobis distance measurement has also been introduced in order to compensate for variance and covariance effects of di- or tri-nucleotide repeats throughout the genome, and shown to be useful for exploring the long-term hosts of plasmids.197
As more complete bacterial genome sequences become available, and in light of the growing number of genome sequences from various human microbiomes including the gut microbiota, the use of powerful metagenomic based methods for plasmid isolation, combined with bioinformatic approaches to recover information on long-term bacterial hosts in a community, is likely to provide much insight into the gut mobile metagenome and its role in community development. The use of the TRACA system in combination with a subsequent in silico metagenomic analysis of plasmid-encoded functions has already been implemented and revealed not only the potential broad geographic distribution of some plasmids or plasmid families in the gut microbiomes of individuals from across the globe, but also highlighted the unexpected enrichment of some plasmid-encoded functions in this community.32
Direct plasmid detection by PCR
Finally, there also exists the option for the direct detection of plasmids in metagenomic DNA extracts through a PCR-based approach, using primers specific to conserved backbone regions of certain plasmid families.56,58 A range of primer sets have been developed to distinguish between plasmids of various incompatibility groups, and these may also be exploited to determine if plasmids of a particular group are present in a microbial population.56,58,192 However, such an approach is extremely limited, and not only is the subsequent characterization of plasmids virtually impossible, the plasmids that may be detected are limited to those for which well-characterized relatives already exist which can be used for primer design.31
Despite this, such approaches will likely be of value in augmenting other culture-independent metagenomic strategies, facilitating elucidation of plasmid host ranges, geographic distribution, and inter-individual variation between gut microbiomes (Table 2). PCR-based approaches are also well suited to determine the general prevalence and overall abundance of particular plasmids of interest within a community through quantitative PCR, and would permit fluctuations in the population size of plasmids of interest isolated by methods such as TRACA, to be monitored over time within an individual or in response to changes in environmental parameters.
Summary and Future Perspectives: What Can the Mobile Metagenome Tell Us?
Although MGE have been extensively studied from the perspective of gene flow among bacteria, the evolution of bacterial species, and the spread of antibiotic resistance genes, few studies have considered the role of MGE at the community level and the contribution of the mobile metagenome to the development of community functions. In particular, the study of the mobile metagenome in microbial ecosystems that have co-evolved with higher host organisms, such as the gut microbiota, are likely to reveal important insights into the development of this community, its functional output, and interaction with the metazoan host.30-33,89
There is increasing evidence that the gut mobile metagenome reflects the co-evolution of host and microbe (Table 1),20,30,32,48-50,201 and as such contains MGE and encodes functions important to bacteria inhabiting this environment. Recently, the integration of the mobile metagenome into the hologenome model of evolution has been proposed and described.30 The hologenome theory builds on the increasing appreciation that humans and other complex metazoans are in essence amalgams of both prokaryotic and eukaryotic cells, resulting from the co-evolution of host and microbe,4,5,8,30 and proposes that the basic unit of selection in evolution consists of the total genetic content of the eukaryotic host plus that of all associated microbial partners.4,5 However, it should be noted that alternative and popular evolutionary theories place the emphasis on selection at the level of the gene, but this view is not necessarily incompatible with the hologenome theory (in which success of the holobiont would also equate to success of genes constituting the hologenome) and it is likely that selection acts at multiple levels.
One way in which the hologenome arrangement is proposed to greatly benefit the human host is by increasing adaptability to new environmental conditions and exploitation of food sources.4,5,30 This stems from the plasticity of prokaryotic genomes, which by virtue of their short generation times and ability to acquire new functions through HGT, are infinitely more flexible than the relatively static “human” genome.4,5,30 In the proposed integration of the gut mobile metagenome into this model, the genetic information encoded by the prokaryotic complement of metazoans constitutes a secondary accessory genome, and the mobile metagenome a highly flexible tertiary gene pool which provides access to the wider prokaryotic gene pool, and serves to further enhance the adaptability of the holobiont as a whole through HGT.30
Furthermore, MGE have also been proposed to be important in the maintenance of cooperation between bacteria, and therefore the mobile metagenome may illuminate mechanisms through which the community maintains cooperation and social stability, and curtails the ingress of cheaters.202,203 Many genes involved in the production of beneficial traits that may be exploited by a population as a whole (public goods) such as secreted enzymes, are encoded by MGE.202,203 There is growing evidence that genes required for the production of such public goods may be stratified between MGE and core bacterial genomes, with the mobile metagenome encoding a greater proportion of activities involved in the production of these public goods.202,203
The maintenance of such genes on MGE, rather than their fixation into core genomes of relevant bacterial species, may be due to a number of selective pressures. This includes temporal and spatial variation in selective pressure for the traits encoded, but also the possibility that mobility of these genes permits the population to rehabilitate individuals that loose genes required for generation of public goods, and subsequently exploit a public good without contributing to its production (referred to as cheaters).8,203 These cheaters generally exhibit a fitness advantage over other members of the population and in the case of the human gut microbiota, it is predicted that the emergence of cheaters will destabilize community functions adversely affecting the human host, and so have deleterious effects on the community and the holobiont as a whole.8 It has been proposed that the infection or re-infection of these cheaters with MGE encoding genes required for production of the public good in question, serves as a mechanism by which their emergence in a population may be controlled and sociality maintained.202,203
The role of the gut mobile metagenome in regulating this aspect of community function has not yet been subject to significant study, but the contribution of this flexible gene pool to policing or rehabilitation of cheaters, and maintenance of cooperation in the gut microbiota presents an intriguing possibility that should be investigated in more detail. Of particular interest in this context is the potential prevalence of RelBE TA modules in gut-associated plasmids, which enhance the stability of MGE in a bacterial host through a post segregation killing mechanism (in which loss of MGE encoding the TA module results in growth arrest or cell death), and so could ensure contribution to the production of a public good is maintained by enforcing carriage of MGE encoding the necessary genetic information.32,202 Recently, Noguiera et al. (2009) highlighted not only the association of genes in the E. coli secretome (producing public goods) with MGE, but also the association of these with “mafia strategy” addictive systems such as TA modules, which were proposed to promote their maintenance.202
Overall, the gut mobile metagenome constitutes a vast and untapped reservoir of novel genetic information, which has the potential to greatly enhance our understanding of this complex community, as well as facilitating the development of strategies to modulate the functions of this ecosystem for the benefit of human health. Therefore, accessing the wealth of novel genetic data that is likely to reside in the human gut mobile metagenome will not only illuminate important, fundamental aspects of community function and development, but will also facilitate the discovery and exploitation of novel MGE or MGE-encoded activities of biotechnological or clinical value. For example, MGE enriched or specific to this ecosystem have already been exploited as novel tools for microbial source tracking to determine the origin of faecal pollution in surface waters.47
The study of host-associated microbial ecosystems such as the gut microbiota, and in particular the community level analysis of associated mobile metagenomes, constitutes a new frontier in microbiology, the opening of which will to lead great intellectual reward. Although significant technological development is still required to facilitate the full exploration of this new territory, strategies that permit the first forays into this promised land are now in place, and initial expeditions have been mounted. However, few studies have so far examined the mobile metagenomes of such communities in detail, and future projects aimed at characterizing host associated microbiomes should incorporate the study of associated mobile metagenomes as a key component.
Acknowledgments
The authors are extremely grateful to Ms. Cinzia Dedi for expert technical assistance and general laboratory support, and Dr. Caroline Jones for insightful discussion and constructive comments during manuscript preparation. Dr. Lesley Ogilvie is supported by funding from the Medical Research Council (Grant ID number G0901553 awarded to Dr. B.V. Jones). Research in the laboratory of Dr. B.V. Jones is also supported by funding from the Hospital Infection Society, The Royal Society, The Society for Applied Microbiology, The University of Brighton, and the European Union (FP7 Marie Curie IAPP scheme).
Glossary
Abbreviations:
- BSH
bile salt hydrolase
- FACS
fluorescence associated cell sorting
- FISH
fluorescent in situ hybridization
- GIT
gastrointestinal tract
- HGT
horizontal gene transfer
- LAB
lactic acid bacteria
- MGE
mobile genetic elements
- RING-FISH
recognition of individual genes-FISH
- TRACA
transposon aided capture
Footnotes
Previously published online: www.landesbioscience.com/journals/biobugs/article/17883
References
- 1.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–83. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 3.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–93. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zilber-Rosenberg I, Rosenberg E. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev. 2008;32:723–35. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 5.Rosenberg E, Sharon G, Zilber-Rosenberg I. The hologenome theory of evolution contains Lamarckian aspects within a Darwinian framework. Environ Microbiol. 2009;11:2959–62. doi: 10.1111/j.1462-2920.2009.01995.x. [DOI] [PubMed] [Google Scholar]
- 6.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–8. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilks M. Bacteria and early human development. Early Hum Dev. 2007;83:165–70. doi: 10.1016/j.earlhumdev.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 9.Qin J, Li R, Raes J, Arumugaum M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bäckhed F, Ding H, Wang T, Hooper LV, Koh Y, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–23. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–33. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Natl Acad Sci USA. 1999;96:9833–8. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stappenbeck TS, Hooper LV, Gordon JI. Development regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. 2002;99:15451–5. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zoetendal EG, Vaughan EE, de Vos WM. A microbial world within us. Mol Microbiol. 2006;59:1639–50. doi: 10.1111/j.1365-2958.2006.05056.x. [DOI] [PubMed] [Google Scholar]
- 15.Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci USA. 2010;107:7503–8. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet J-P, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–84. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- 17.Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799–807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–9. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–81. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNeil NI. The contribution of the large intestine to energy supplies in man. Am J Clin Nutr. 1984;39:338–42. doi: 10.1093/ajcn/39.2.338. [DOI] [PubMed] [Google Scholar]
- 22.Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361:512–9. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 23.Cummings JH, Antoine J-M, Azpiroz F, Bourdet-Sicard R, Brandtzaeg P, Calder PC, et al. PASSCLAIM--gut health and immunity. Eur J Nutr. 2004;43:118–73. doi: 10.1007/s00394-004-1205-4. [DOI] [PubMed] [Google Scholar]
- 24.Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol. 2007;9:1101–11. doi: 10.1111/j.1462-2920.2007.01281.x. [DOI] [PubMed] [Google Scholar]
- 25.Houten SM, Watanabe M, Auwerx J. Endocrine functions of bile acids. EMBO J. 2006;25:1419–25. doi: 10.1038/sj.emboj.7601049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–9. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 27.Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678–93. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 28.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–6. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Jones BV. The human gut mobile metagenome: A metazoan perspective. Gut Microbes. 2010;1:415–31. doi: 10.4161/gmic.1.6.14087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones BV, Marchesi JR. Accessing the mobile metagenome of the human gut microbiota. Mol Biosyst. 2007;3:749–58. doi: 10.1039/b705657e. [DOI] [PubMed] [Google Scholar]
- 32.Jones BV, Sun F, Marchesi JR. Comparative metagenomic analysis of plasmid encoded functions in the human gut microbiome. BMC Genomics. 2010;11:46. doi: 10.1186/1471-2164-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones BV, Marchesi JR. Transposon-aided capture (TRACA) of plasmids resident in the human gut mobile metagenome. Nat Methods. 2007;4:55–61. doi: 10.1038/nmeth964. [DOI] [PubMed] [Google Scholar]
- 34.Shoemaker NB, Vlamakis H, Hayes K, Salyers AA. Evidence for extensive of resistance gene transfer among Bacteroides spp. And among Bacteroides and among other genera in the human colon. Appl Environ Microbiol. 2001;67:561–8. doi: 10.1128/AEM.67.2.561-568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta A, Vlamakis H, Shoemaker N, Salyers AA. A new Bacteroides conjugative transposon that carries an ermB gene. Appl Environ Microbiol. 2003;69:6455–63. doi: 10.1128/AEM.69.11.6455-6463.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salyers AA, Gupta A, Wang YP. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004;12:412–6. doi: 10.1016/j.tim.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 37.Melville CM, Brunel R, Flint HJ, Scott KP. The Butyrivibrio fibrisolvens tet(W) gene is carried on the novel conjugative transposon TnB1230, which contains duplicated nitroreductase coding sequences. J Bacteriol. 2004;186:3656–9. doi: 10.1128/JB.186.11.3656-3659.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trobos M, Lester CH, Olsen JE, Frimondt-Møller N, Hmmerum AM. Natural transfer of sulphonamide and amplicllin resistance between Escherichia coli residing in the human intestine. J Antimicrob Chemother. 2009;63:80–6. doi: 10.1093/jac/dkn437. [DOI] [PubMed] [Google Scholar]
- 39.Sommer MOA, Dantas G, Church GM. Functional characterisation of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–31. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Canchaya C, Fang F, Raftis E, Ryan KA, van Pijkeren J-P, et al. Distribution of megaplasmids in Lactobacillus salivarius and other Lactobacilli. J Bacteriol. 2007;189:6128–39. doi: 10.1128/JB.00447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Claesson MJ, Li Y, Leahy S, Canchaya C, van Pijkeren J-P, Cerdeño-Tárraga C, et al. Multireplicon genome architecture of Lactobacillus salivarius. Proc Natl Acad Sci USA. 2006;103:6718–23. doi: 10.1073/pnas.0511060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, Vrijenhoek JE, Bonten MJ, Willems RJ, van Schaik W. A genetic element on megaplsmids allows Enterococcus faecium to use raffinose as carbon source. Environ Microbiol. 2011;13:518–28. doi: 10.1111/j.1462-2920.2010.02355.x. [DOI] [PubMed] [Google Scholar]
- 43.Oshima K, Toh H, Ogura Y, Sasamoto H, Morita H, Park S-H, et al. Complete genome sequence and comparative analysis of the wild-type commensal Escherichia coli strain SE11 isolated from a healthy adult. DNA Res. 2008;15:375–86. doi: 10.1093/dnares/dsn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blum-Oehler G, Oswald S, Eitejörge K, Sonnenborn U, Schulze J, Kruis W, et al. Development of strain-specific PCR reactions for the detection of the probiotic Escherichia coli strain Nissle 1917 in faecal samples. Res Microbiol. 2003;154:59–66. doi: 10.1016/S0923-2508(02)00007-4. [DOI] [PubMed] [Google Scholar]
- 45.Shankar N, Baghdayan AS, Gilmore MS. Modulation of virulence within an pathogenicity island in a vancomycin resistant Enterococcus faecalis. Nature. 2002;417:746–50. doi: 10.1038/nature00802. [DOI] [PubMed] [Google Scholar]
- 46.Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–3. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ebdon J, Muniesa M, Taylor H. The application of a recently isolated strain of Bacteroides (GB-124) to identify human sources of faecal pollution in a temperate river catchment. Water Res. 2007;41:3683–90. doi: 10.1016/j.watres.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 48.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F. Viruses in the faeca microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–8. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zaneveld JR, Lozupone C, Gordon JI, Knight R. Ribosomal RNA diversity predicts genome diversity in gut bacteria and their relatives. Nucleic Acids Res. 2010;38:3869–79. doi: 10.1093/nar/gkq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hehemann J-H, Correc G, Barbeyon T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–12. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 51.Rohwer F, Prangishvili D, Lindell D. Roles of viruses in the environment. Environ Microbiol. 2009;11:2771–4. doi: 10.1111/j.1462-2920.2009.02101.x. [DOI] [PubMed] [Google Scholar]
- 52.Slater FR, Bailey MJ, Tett AJ, Turner SL. Progress towards understanding the fate of plasmids in bacterial communities. FEMS Microbiol Ecol. 2008;66:3–13. doi: 10.1111/j.1574-6941.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- 53.Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64:69–114. doi: 10.1128/MMBR.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 55.Wilson BA, Salyers AA. Is the evolution of bacterial pathogens an out-of-body experience? Trends Microbiol. 2003;11:347–50. doi: 10.1016/S0966-842X(03)00179-3. [DOI] [PubMed] [Google Scholar]
- 56.Smalla K, Osburne AM, Wellington EMH. In: ed. Thomas CM, The Horizontal Gene Pool, Bacterial Plasmids and Gene Spread. Amsterdam: Harwood Academic Publishers, 2000: 207-248. [Google Scholar]
- 57.Springael D, Top EM. Horizontal gene transfer and microbial adaptation to xenobiotics: new types of mobile genetic elements and lessons from ecological studies. Trends Microbiol. 2004;12:53–8. doi: 10.1016/j.tim.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 58.Smalla K, Sobecky PA. The prevalence and diversity of mobile genetic elements in bacterial communities of different environmental habitats: insights gained from different methodological approaches. FEMS Microbiol Ecol. 2002;42:165–75. doi: 10.1111/j.1574-6941.2002.tb01006.x. [DOI] [PubMed] [Google Scholar]
- 59.Lozupone CA, Hamady M, Cantral BL, Coutinho PM, Henrissat B, Gordon JI, et al. The convergence of carbohydrate active gene repertoires in human gut microbes. Proc Natl Acad Sci USA. 2008;105:15076–81. doi: 10.1073/pnas.0807339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu J, Mahowals MA, Ley RE, Lozupone CA, Hamady M, Martens EC, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Louis P, McCrae SI, Charrier C, Flint HJ. Organization of butyrate synthetic genes in human colonic bacteria: phylogenetic conservation and horizontal gene transfer. FEMS Microbiol Lett. 2007;269:240–7. doi: 10.1111/j.1574-6968.2006.00629.x. [DOI] [PubMed] [Google Scholar]
- 62.Jones BV, Begley M, Hill C, Gahan CGM, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci USA. 2008;105:13580–5. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fang F, Li Y, Bumann M, Raftis EJ, Casey PG, Cooney JC, et al. Allelic variation of bile salt hydrolase genes in Lactobacillus salivarius does not determine bile resistance levels. J Bacteriol. 2009;191:5743–57. doi: 10.1128/JB.00506-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fang F, Flynn S, Li Y, Claesson M, van Pijkeren J-P, Collins JK, et al. Characterisation of endogenous plasmids from Lactobacillus salivarius UCC118. Appl Environ Microbiol. 2008;74:3216–28. doi: 10.1128/AEM.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Begley M, Hill C, Gahan CG. Bile salt hydrolase activity in probiotics. Appl Environ Microbiol. 2006;72:1729–38. doi: 10.1128/AEM.72.3.1729-1738.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gelvin SB. Agrobacterium and plant genes involved in t-DNA transfer and integration. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:223–56. doi: 10.1146/annurev.arplant.51.1.223. [DOI] [PubMed] [Google Scholar]
- 67.Zambryski P. Basic processes underlying Agrobacterium-mediated DNA transfer to plant cells. Annu Rev Genet. 1988;22:1–30. doi: 10.1146/annurev.ge.22.120188.000245. [DOI] [PubMed] [Google Scholar]
- 68.Heinemann JA, Sprague GF., Jr Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989;340:205–9. doi: 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- 69.Waters VL. Conjugation between bacterial and mammalian cells. Nat Genet. 2001;29:375–6. doi: 10.1038/ng779. [DOI] [PubMed] [Google Scholar]
- 70.Wachino J, Shibayama K, Kurokawa H, Kimura K, Yamane K, Suzuki S, et al. Novel plasmid-mediated 16S rRNA M1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob Agents Chemother. 2007;51:4401–9. doi: 10.1128/AAC.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schlüter A, Szczepanowski R, Pühler A, Top EM. Genomics of IncP-1 anitbiotic resistance plasmids isolated from wastewater plants provides evidence for a widely accessible resistance gene pool. FEMS Microbiol Rev. 2007;31:449–77. doi: 10.1111/j.1574-6976.2007.00074.x. [DOI] [PubMed] [Google Scholar]
- 72.Yin S, Hao Y, Zhai Z, Li R, Huang Y, Tian H, et al. Characterisation of a cryptic plasmid from Lactobacillus plantarum M4. FEMS Microbiol Lett. 2008;285:183–7. doi: 10.1111/j.1574-6968.2008.01229.x. [DOI] [PubMed] [Google Scholar]
- 73.Chen D-Q, Zheng X-c, Lu Y-j. Identification and characterisation of novel ColE1-type high-copy number plasmid mutants in Legionella pneumophila. Plasmid. 2006;56:167–78. doi: 10.1016/j.plasmid.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 74.Lee J-H, Halerson JS, Kim J-H, O’Sullivan DJ. Comparative sequence analysis of plasmids from Lactobacillus delbrueckii and construction of a shuttle cloning vector. Appl Environ Microbiol. 2007;73:4417–24. doi: 10.1128/AEM.00099-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee J-H, O’Sullivan DJ. Sequence analysis of two cryptic plasmids from Bifidobacteriaum longum DJO10A and construction of a shuttle cloning vector. Appl Environ Microbiol. 2006;72:527–35. doi: 10.1128/AEM.72.1.527-535.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao J-Y, Zhong L, Shen M-j, Xia Z-j, Cheng Q-x, Sun X, et al. Discovery of the autonomously replicating plasmid pMF1 from Myxococcus fulvus and development of a gene cloning system in Myxococcus xanthus. Appl Environ Microbiol. 2008;74:1980–7. doi: 10.1128/AEM.02143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wexler HM. 2007, Bacteriodes: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Johnson TJ, Nolan LK. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol Mol Biol Rev. 2009;73:750–74. doi: 10.1128/MMBR.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tannock GW, Fuller R, Smith SL, Hall MA. Plasmid profiling of members of the family Enterobacteriaceae Lactobacilli, and Bifidobacteria to study the transmission of bacteria from mother to infant. J Clin Microbiol. 1990;28:1225–8. doi: 10.1128/jcm.28.6.1225-1228.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sóki J, Gal M, Brazier JS, Rotimi VO, Urbán E, Nagy E, et al. Molecular investigation of genetic elements contributing to metronidazole resistance in Bacteriodes strains. J Antimicrob Chemother. 2006;57:212–20. doi: 10.1093/jac/dki443. [DOI] [PubMed] [Google Scholar]
- 81.Trinh S, Reysset G. Detection by PCR of the nim Genes encoding 5-nitroimidazole resistance in Bacteriodes spp. J Clin Microbiol. 1996;34:2078–84. doi: 10.1128/jcm.34.9.2078-2084.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reysset G, Haggoud A, Sebald M. Genetics of resistance of Bacteriodes species to 5-nitroimidazole. Clin Infect Dis. 1993;16:S401–3. doi: 10.1093/clinids/16.Supplement_4.S401. [DOI] [PubMed] [Google Scholar]
- 83.Ruiz-Barba JL, Floriano B, Maldonado-Barragán A, Jiménez-Díaz R. Molecular analysis of the 21kb bacteriocin-encoding plasmid pEF1 from Enterococcus faecium 6T1a. Plasmid. 2007;57:175–81. doi: 10.1016/j.plasmid.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 84.Shears P, Suliman G, Hart CA. Occurrence of multiple antibiotic resistant and R plasmids in Enterobacteriaceae isolated from children in the Sudan. Epidemiol Infect. 1988;100:73–81. doi: 10.1017/S0950268800065572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alvarez-Martín P, Flórez AB, Mayo B. Screening for plasmids among human Bifidobacteria species: Sequencing and analysis of pBC1 from Bifidobacterium catenulatum L48. Plasmid. 2007;57:165–74. doi: 10.1016/j.plasmid.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 86.Iwata M, Morishita T. The presence of plasmids in Bifidobacterium breve. Lett Appl Microbiol. 1989;9:165–8. doi: 10.1111/j.1472-765X.1989.tb00315.x. [DOI] [Google Scholar]
- 87.Park MS, Lee KH, Ji GE. Isolation and characterisation of two plasmids from Bifidobacterium longum. Lett Appl Microbiol. 1997;25:5–7. doi: 10.1046/j.1472-765X.1997.00059.x. [DOI] [PubMed] [Google Scholar]
- 88.Sgorbati B, Scardovi V, Leblanc DJ. Plasmids in the genus Bifidobacterium. J Gen Microbiol. 1982;128:2121–31. doi: 10.1099/00221287-128-9-2121. [DOI] [PubMed] [Google Scholar]
- 89.Smith CJ, Tribble GD, Bayley DP. Genetic elements of Bacteriodes species: A moving story. Plasmid. 1998;40:12–29. doi: 10.1006/plas.1998.1347. [DOI] [PubMed] [Google Scholar]
- 90.Licht TR, Wilcks A. Conjugative gene transfer in the gastrointestinal environment. Adv Appl Microbiol. 2005;58:77–95. doi: 10.1016/S0065-2164(05)58002-X. [DOI] [PubMed] [Google Scholar]
- 91.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–76. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Van Melderen L, De Bast MS. Bacterial toxin-antitoxinsystems: More than selfish entities? PLoS Genet. 2009;5:e1000437. doi: 10.1371/journal.pgen.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Christensen SK, Pedersen K, Hansen FG, Gerdes K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol. 2003;332:809–19. doi: 10.1016/S0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- 94.Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. ReIE, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci USA. 2001;98:14328–33. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Anantharaman V, Aravind L. New connections in the prokaryotic toxin-antitoxin network: Relationship with the eukaryotic nonsense-mediated RNA decay System. Genome Biol. 2003;4:R81. doi: 10.1186/gb-2003-4-12-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vázquez-Laslop N, Lee H, Neyfakh AA. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J Bacteriol. 2006;188:3494–7. doi: 10.1128/JB.188.10.3494-3497.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yamamoto TA, Gerdes K, Tunnacliffe A. Bacterial toxin RelE induces apoptosis in human cells. FEBS Lett. 2002;519:191–4. doi: 10.1016/S0014-5793(02)02764-3. [DOI] [PubMed] [Google Scholar]
- 98.Kristoffersen P, Jensen GB, Gerdes K, Piskur J. Bacterial toxin-antitoxin gene system as containment control in yeast cells. Appl Environ Microbiol. 2000;66:5524–6. doi: 10.1128/AEM.66.12.5524-5526.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Andreev D, Hauryliuk V, Terenin I, Dmitriev S, Ehrenberg M, Shatsky I. The bacterial toxin ReIE induces specific mRNA cleavage in the A site of the eukaryote ribosome. RNA. 2008;14:233–9. doi: 10.1261/rna.693208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pecota DC, Wood TK. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J Bacteriol. 1996;178:2044–50. doi: 10.1128/jb.178.7.2044-2050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nieto C, Sadowy E, de la Campa AG, Hryniewicz W, Espinosa M. The relBE2Spn toxin-antitoxin system of Streptocococcus pneumonia: Role in antibiotic tolerance and functional conservation in clinical isolates. PLoS ONE. 2010;5:e11289. doi: 10.1371/journal.pone.0011289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Feld L, Schjørring S, Hammer K, Licht RT, Danielsen M, Krogfelt K, et al. Selective pressure affects transfer and establishment of a Lactobacillus plantarum plasmid in the gastrointestinal environment. J Antimicrob Chemother. 2008;61:845–52. doi: 10.1093/jac/dkn033. [DOI] [PubMed] [Google Scholar]
- 103.Tuohy K, Davies M, Rumsby P, Rumney C, Adams MR, Rowland IR. Monitoring transfer of recombinant and non-recombinant plasmids between Lactococcus lactis strains and members of the human gastrointestinal microbiota in vivo -impact of donor cell number and diet. J Appl Microbiol. 2002;93:954–64. doi: 10.1046/j.1365-2672.2002.01770.x. [DOI] [PubMed] [Google Scholar]
- 104.Clewell DB, Yagi Y, Dunny GM, Schultz SK. Characterisation of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: Identification of a plasmid determining erythromycin resistance. J Bacteriol. 1974;117:283–9. doi: 10.1128/jb.117.1.283-289.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goren MG, Carmeli Y, Schwaber MJ, Chmelnitsky I, Schechner V, Navon-Venezia S. Transfer of carbapenem-resistant plasmid from Klebsiella pneumonia ST258 to Escherichia coli in patient. Emerg Infect Dis. 2010;16:1014–7. doi: 10.3201/eid1606.091671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lin CF, Fung ZF, Wu CL, Chung TC. Molecular characterisation of a plasmid-borne (pTC82) chloramphenicol resistance determinant (cat-TC) from Lactobacillus reuteri plasmid G4. Plasmid. 1996;36:116–24. doi: 10.1006/plas.1996.0039. [DOI] [PubMed] [Google Scholar]
- 107.Obaseiki-Ebor EE, Abiodun PO, Emina PA. Faecal Escherichia coli mediating transferable multi-antibiotic resistance and undesirable extra-chromosomal genes. Ann Trop Paediatr. 1986;6:283–6. doi: 10.1080/02724936.1986.11748457. [DOI] [PubMed] [Google Scholar]
- 108.Karami N, Martner A, Enne VI, Swerkersson S, Alderberth I, Wold AE. Transfer of an ampicillin resistance gene between two Escherichia coli strains in the bowel microbiota of an infant treated with antibiotics. J Antimicrob Chemother. 2007;60:1142–5. doi: 10.1093/jac/dkm327. [DOI] [PubMed] [Google Scholar]
- 109.Martínez-súrez JV, Baquero F, Reig M, Pérez-Díaz JC. Transferable plasmid-linked chloramphenicol acetyletransferase conferring high-level resistance in Bacteriodes uniformis. Antimicrob Agents Chemother. 1995;28:113–7. doi: 10.1128/aac.28.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aquilanti L, Garofalo C, Osimani A, Silvestri G, Vignaroli C, Clementi F. Isolation and molecular characterisation of antibiotic-resistant lactic acid bacteria from poultry and swine meat products. J Food Prot. 2007;70:557–65. doi: 10.4315/0362-028x-70.3.557. [DOI] [PubMed] [Google Scholar]
- 111.Nakano V, Padilla G. Marques M do V, Avila-Campos MJ. Plasmid-related β-lactamase production in Bacteriodes fragilis strains. Res Microbiol. 2004;155:843–6. doi: 10.1016/j.resmic.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 112.Wallace BL, Bradley JE, Rogolsky M. Plasmid analyses in clinical isolates of Bacteriodes fragilis and other Bacteriodes species. J Clin Microbiol. 1981;14:383–8. doi: 10.1128/jcm.14.4.383-388.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Licht TR, Laugesen D, Jensen LB, Jacobsen BL. Transfer of the pheromone-inducible plasmid pCF10 among Enterococcus faecalis microorganisms colonizing the intestine of mini-pigs. Appl Environ Microbiol. 2002;68:187–93. doi: 10.1128/AEM.68.1.187-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gevers D, Huys G, Swings J. In vitro conjugal transfer of tetracycline resistance from Lactobacillus isolates to other Gram-positive bacteria. FEMS Microbiol Lett. 2003;225:125–30. doi: 10.1016/S0378-1097(03)00505-6. [DOI] [PubMed] [Google Scholar]
- 115.Jacobsen L, Wilcks A, Hammer K, Huys G, Gevers D, Andersen SR. Horizontal transfer of tet(M) and erm(B) resistance plasmids from food strains of Lactobacillus plantarum to Enterococcus faecalis JH2-2 in the gastrointestinal tract of gnotobiotic rats. FEMS Microbiol Ecol. 2007;59:158–66. doi: 10.1111/j.1574-6941.2006.00212.x. [DOI] [PubMed] [Google Scholar]
- 116.Wang T-T, Lee BH. Plasmids in Lactobacillus. Crit Rev Biotechnol. 1997;17:227–72. doi: 10.3109/07388559709146615. [DOI] [PubMed] [Google Scholar]
- 117.Cheikhyoussef A, Cheikhyoussef N, Chen H, Zhao J, Tang J, Zhang H, et al. Bifidin I. A new bacteriocin produced by Bifidobacterium infantis BCRC 14602: Purification and partial amino acid sequence. Food Contr. 2010;21:746–53. doi: 10.1016/j.foodcont.2009.11.003. [DOI] [Google Scholar]
- 118.Chassy B, Gibson EM, Giuffrida A. Evidence for plasmid associated lactose metabolism in Lactobacillus casei ssp. casei. Curr Microbiol. 1978;1:141–4. doi: 10.1007/BF02601666. [DOI] [PubMed] [Google Scholar]
- 119.Muriana PM, Klaenhammer RT. Conjugal transfer of plasmid-encoded determinants for bacteriocin production and immunity in Lactobacillus acidophilus 88. Appl Environ Microbiol. 1987;53:553–60. doi: 10.1128/aem.53.3.553-560.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hummel AS, Hertel C, Holzapfel WH, Franz CM. Antibiotic resistances of starter and probiotic strains of lactic acid bacteria. Appl Environ Microbiol. 2007;73:730–9. doi: 10.1128/AEM.02105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Klare I, Konstabel C, Werner G, Huys G, Vankerckhoven V, Kahlmeter G, et al. Antimicrobial susceptibilities of Lactococcus, Pediococcus and Lactococcus human isolates and cultures intended for probiotic or nutritional use. J Antimicrob Chemother. 2007;59:900–12. doi: 10.1093/jac/dkm035. [DOI] [PubMed] [Google Scholar]
- 122.Flórez AB, Delgado S, Mayo B. Antimicrobial susceptibility of lactic acid bacteria from a cheese environment. Can J Microbiol. 2005;51:51–8. doi: 10.1139/w04-114. [DOI] [PubMed] [Google Scholar]
- 123.Çataloluk O, Gogebakan B. Presence of drug resistance in intestinal lactobacilli of dairy and human origin in Turkey. FEMS Microbiol Lett. 2004;236:7–12. doi: 10.1016/j.femsle.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 124.D’Aimmo MR, Modesto M, Biavati B. Antibiotic resistance of lactic acid bacteria and Bifidobacterium spp. Isolated from dairy and pharmaceutical products. Int J Food Microbiol. 2007;115:35–42. doi: 10.1016/j.ijfoodmicro.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 125.Gevers D, Huys G, Swings J. Molecular characterisation of tet(M) genes in Lactobacillus isolates from different types of fermented dry sausage. Appl Environ Microbiol. 2003;69:1270–5. doi: 10.1128/AEM.69.2.1270-1275.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Egervärn M, Lindmark H, Olsson J, Roos S. Transferability of a tetracycline resistance gene from Lactobacillus reuteri to bacteria in the gastrointestinal tract of humans. Antonie van Leeuwenhoek. 2010;97:189–200. doi: 10.1007/s10482-009-9401-0. [DOI] [PubMed] [Google Scholar]
- 127.Dahl KH, Mater DD, Flores MJ, Johnsen PJ, Midtvedt T, Corthier G, et al. Transfer of plasmid and chromosomal glycopeptides resistance determinants occurs more readily in the digestive tract of mice than in vitro and exconjugants can persist stably in vivo in the absence of glycopeptides selection. J Antimicrob Chemother. 2007;59:478–86. doi: 10.1093/jac/dkl530. [DOI] [PubMed] [Google Scholar]
- 128.Lester CH, Frimodt-Moller N, Hammerum AM. Conjugal transfer of aminoglycoside and macrolide resistance between Enterococcus faecium isolates in the intestine of streptomycin-treated mice. FEMS Microbiol Lett. 2004;235:385–91. doi: 10.1111/j.1574-6968.2004.tb09614.x. [DOI] [PubMed] [Google Scholar]
- 129.Tzipori S, Gibson R, Montanaro J. Nature and distribution of mucosal lesions associated with enteropathogenic and enterohemorrhagic Escherichia coli in piglets and the role of plasmid-mediated factors. Infect Immun. 1989;57:1142–50. doi: 10.1128/iai.57.4.1142-1150.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dudley EG, Abe C, Ghigo J-M, Latour-Lambert P, Hormazabal JC, Nataro JP. An IncI1 plasmid contributes to the adherence of the atypical enteroaggregative Escherichia coli train C1096 to cultured cells and abiotic surfaces. Infect Immun. 2006;74:2102–14. doi: 10.1128/IAI.74.4.2102-2114.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rotger R, Casadesús J. The virulence plasmids of Salmonella. Int Microbiol. 1999;2:177–84. [PubMed] [Google Scholar]
- 132.Boyd EF, Hartl D. Salmonella virulence plasmid: Modular acquisition of the spv virulence region by an F-plasmid in Salmonella enterica subspecies I and insertion into the chromosome of subspecies II, IIa, IV and Vii isolates. Genetics. 1998;149:1183–90. doi: 10.1093/genetics/149.3.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Venkatesan MM, Goldberg MB, Rose DJ, Grotebeck EJ, Burland V, Blattner FR. Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect Immun. 2001;69:3271–85. doi: 10.1128/IAI.69.5.3271-3285.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zagaglia C, Casalino M, Colonna B, Conti C, Calconi A, Nicoletti M. Virulence plasmids of enteroinvasive Escherichia coli and Shigella flexneri integrate into a specific site on the host chromosome: Integration greatly reduces expression of plasmid-carried virulence genes. Infect Immun. 1991;59:729–31. doi: 10.1128/iai.59.3.792-799.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.McCormick BA, Siber AM, Maurelli AT. Requirement of the Shigella flexneri virulence plasmid in the ability to induce trafficking of neutrophils across polarized monolayers of the intestinal epithelium. Infect Immun. 1998;66:4237–43. doi: 10.1128/iai.66.9.4237-4243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yamamoto T, Honda T, Miwatani T, Yokota T. A virulence plasmid in Escherichia coli enterotoxigenic for humans: intergenic transfer and expression. J Infect Dis. 1984;150:688–98. doi: 10.1093/infdis/150.5.688. [DOI] [PubMed] [Google Scholar]
- 137.Buchrieser C, Glaser P, Rusniok C, Nedjari H, d’Hauteville H, Kunst F, et al. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol. 2000;38:760–71. doi: 10.1046/j.1365-2958.2000.02179.x. [DOI] [PubMed] [Google Scholar]
- 138.García-Quintanilla M, Ramos-Morales F, Casadesús J. Conjugal transfer of the Salmonella enterica virulence plasmid in the mouse intestine. J Bacteriol. 2008;190:1922–7. doi: 10.1128/JB.01626-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Burland V, Shao Y, Perna NT, Plunkett G, Sofia HJ, Blattner FR. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 1998;26:4196–204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Millette M, Dupont C, Sharek F, Ruiz MT, Archambault D, Lacroix M. Purification and identification of the pediocine produced by Pediococcus acidilactici MM33, a new human intestinal strain. J Appl Microbiol. 2008;104:269–75. doi: 10.1111/j.1365-2672.2007.03583.x. [DOI] [PubMed] [Google Scholar]
- 141.Corr SC, Li Y, Riedel CU, O’Toole PW, Hill C, Gahan CGM. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci USA. 2007;104:7617–21. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Šmajs D, Strouhal M, Matĕjková P, Čejková D, Cursino L, Charton-Souza E, et al. Complete sequence of low-copy-number plasmid MccC7-H22 of probiotic Escherichia oli H22 and the prevalence of mcc genes among human E. coli. Plasmid. 2008;59:1–10. doi: 10.1016/j.plasmid.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 143.Cotter PD, Hill C, Ross P. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005;3:777–88. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 144.Deegan LH, Cotter PD, Hill C, Ross P. Bacteriocins: Biological tools for bio-preservation and shelf-life extension. Int Dairy J. 2006;16:1058–71. doi: 10.1016/j.idairyj.2005.10.026. [DOI] [Google Scholar]
- 145.Riley MA, Wertz JE. Bacteriocins: evolution, ecology and application. Annu Rev Microbiol. 2002;56:117–37. doi: 10.1146/annurev.micro.56.012302.161024. [DOI] [PubMed] [Google Scholar]
- 146.Sleator RD. Probiotic therapy – recruiting old friends to fight new foes. Gut Pathog. 2010;2:5. doi: 10.1186/1757-4749-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sleator RD, Hill C. Designer probiotics: a potential therapeutic for Clostridium difficile? J Med Microbiol. 2008;57:793–4. doi: 10.1099/jmm.0.47697-0. [DOI] [PubMed] [Google Scholar]
- 148.Culligan EP, Hill C, Sleator RD. Probiotics and gastrointestinal disease: successes, problems and future prospects. Gut Pathog. 2009;1:19. doi: 10.1186/1757-4749-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gigho J-M. Natural conjugative plasmids induce bacterial biofilm development. Nature. 2001;412:4442–5. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- 150.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–59. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 151.Dussurget O, Cabanes D, Dehoux P, Lecuit M, Buchrieser C, Glaser P, et al. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol Microbiol. 2002;45:1095–106. doi: 10.1046/j.1365-2958.2002.03080.x. [DOI] [PubMed] [Google Scholar]
- 152.Croxen MA, Finlay BB. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol. 2010;8:26–38. doi: 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- 153.Cornelis GR, Boland A, Boyd AP, Geuijen C, Iriarte M, Neyt C, et al. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–52. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gulig PA, Danbara H, Guiney DG, Lax AJ, Norel F, Rhen M. Molecular analysis of spv virulence genes of the Salmonella virulence plasmids. Mol Microbiol. 1993;7:825–30. doi: 10.1111/j.1365-2958.1993.tb01172.x. [DOI] [PubMed] [Google Scholar]
- 155.Parsot C. Shigella spp. and enteroinvasive Escherichia coli pathogenicity factors. FEMS Microbiol Lett. 2005;252:11–8. doi: 10.1016/j.femsle.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 156.Zhang W, Qi W, Albert TJ, Motiwala AS, Alland D, Hyytia-Trees EK. Probing genomic diversity and evolution of Escherichia coli O157 by single nucleotide polymorphisms. Genome Res. 2006;16:757–67. doi: 10.1101/gr.4759706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rendón MA, Saldaña Z, Erdem AL, Monteiro-Neto V, Vázquez A, Kaper JB. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc Natl Acad Sci USA. 2007;104:10637–42. doi: 10.1073/pnas.0704104104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Netherwood T, Bowden R, Harrison P, O'Donnell AG, Parker DS, Gilbert HJ. Gene transfer in the gastrointestinal tract. Appl Environ Microbiol. 1999;65:5139–41. doi: 10.1128/aem.65.11.5139-5141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Corpet DE. Ecological factors influencing the transfer of plasmids in vitro and in vivo. J Antimicrob Chemother. 1986;18:127–32. doi: 10.1093/jac/18.supplement_c.127. [DOI] [PubMed] [Google Scholar]
- 160.van Elsas JD, Fry J, Hirsch P, Molin S. In: Thomas CM, ed. The Horizontal Gene Pool, Bacterial Plasmids and Gene Spread. Amsterdam: Harwood Academic Publishers 2000: 175–206. [Google Scholar]
- 161.Licht TR, Christensen BB, Krogfelt KA, Molin S. Plasmid transfer in the animal intestine and other dynamic bacterial populations: the role of community structure and environment. Microbiology. 1999;145:2615–22. doi: 10.1099/00221287-145-9-2615. [DOI] [PubMed] [Google Scholar]
- 162.Dahl KH, Mater DDG, Flores MJ, Johnsen PJ, Midtvedt T, Corthier G, et al. Transfer of plasmid and chromosomal glycopeptides resistance determinants occurs more readily in the digestive tract of mice than in vitro and exconjugants can persist in vivo in the absence of glycopeptides selection. J Antimicrob Chemother. 2007;59:478–86. doi: 10.1093/jac/dkl530. [DOI] [PubMed] [Google Scholar]
- 163.McConnell MA, Mercer AA, Tannock GW. Transfer of plasmid pAMβ1 between members of the normal microflora inhabiting the murine digestive tract and modification of plasmids in Lactobacillus reuteri host. Microb Ecol Health Dis. 1991;4:343–55. doi: 10.3109/08910609109140149. [DOI] [Google Scholar]
- 164.Morelli L, Sarra PG, Bottazzi V. In vivo transfer of pAM beta 1 from Lactobacillus reuteri to Enterococcus faecalis. J Appl Bacteriol. 1988;65:371–5. doi: 10.1111/j.1365-2672.1988.tb01905.x. [DOI] [PubMed] [Google Scholar]
- 165.Vescovo M, Morelli L, Bottazzi V, Gasson MJ. Conjugal transfer of broad-host range plasmid pAMβ1 into enteric species of lactic acid bacteria. Appl Environ Microbiol. 1983;46:753–5. doi: 10.1128/aem.46.3.753-755.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Flint HJ, Thomson AM, Bisset J. Plasmid-associated transfer of tetracycline resistacne in Bacteriodes ruminicola. Appl Environ Microbiol. 1988;54:855–60. doi: 10.1128/aem.54.4.855-860.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Igimi S, Ryu CH, Park SH, Sasaki Y, Sasaki T, Kumagai S. Transfer of conjugative plasmid pAM beta 1 from Lactococcus lactis to mouse intestinal bacteria. Lett Appl Microbiol. 1996;23:31–5. doi: 10.1111/j.1472-765X.1996.tb00023.x. [DOI] [PubMed] [Google Scholar]
- 168.Bidet P, Burghoffer B, Gautier V, Brahimi N, Mariani-Kurkdjian P, El-Ghoneimi A, et al. In vivo transfer of plasmid-encoded ACC-1 AmpC from Klebsiella pneumoniae to Escherichia coli in an infant and selection of impermeability to imipenem in K. pneumoniae. Antimicrob Agents Chemother. 2005;49:3562–5. doi: 10.1128/AAC.49.8.3562-3565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Charpentier E, Gerbaud G, Courvalin P. Conjugative mobilization of the rolling-circle plasmid pIP823 from Listeria monocytogenes BM4293 among Gram-positive and Gram-negative bacteria. J Bacteriol. 1999;181:3368–74. doi: 10.1128/jb.181.11.3368-3374.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Dørnen AK, Torsvik V, Goksoyr J, Top EM. Effect of mercury addition on plasmid incidence and gene mobilizing capacity in bulk soil. FEMS Microbiol Ecol. 1998;27:381–94. doi: 10.1016/S0168-6496(98)00085-3. [DOI] [Google Scholar]
- 171.Smit E, Wolters A, van Elsas JD. Self-transmissible mercury resistance plasmids with gene-mobilizing capacity in soil bacterial populations: influence of wheat roots and mercury addition. Appl Environ Microbiol. 1998;64:1210–9. doi: 10.1128/aem.64.4.1210-1219.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Licht TR, Struve C, Christensen BB, Poulsen RL, Molin S, Krogfelt KA. Evidence of increased spread and establishment of plasmid RP4 in the intestine under sub-inhibitory tetracycline concentrations. FEMS Microbiol Ecol. 2003;44:217–23. doi: 10.1016/S0168-6496(03)00016-3. [DOI] [PubMed] [Google Scholar]
- 173.Mach PA, Grimes DJ. R-plasmid transfer in a wastewater treatment plant. Appl Environ Microbiol. 1982;44:1395–403. doi: 10.1128/aem.44.6.1395-1403.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Marcinek H, Wirth R, Muscholl-Silberhorn A, Gauer M. Enterococcus faecalis gene transfer under natural conditions in municipal sewage water treatment plants. Appl Environ Microbiol. 1998;64:626–32. doi: 10.1128/aem.64.2.626-632.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Schlundt J, Saadbye P, Lohmann B, Jacobsen BL, Nielsen EM. Conjugal transfer of plasmid DNA between Lactococcus lactis strains and distribution of transconjugants in the digestive tract of gnotobiotic rats. Microb Ecol Health Dis. 1994;7:59–69. doi: 10.3109/08910609409141574. [DOI] [Google Scholar]
- 176.Molin S, Tolker-nielsen T. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol. 2003;14:255–61. doi: 10.1016/S0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- 177.Kajiura T, Wada H, Ito K, Anzai Y, Kato F. Conjugative plasmid transfer in the biofilm formed by Enterococcus faecalis. J Health Sci. 2006;52:358–67. doi: 10.1248/jhs.52.358. [DOI] [Google Scholar]
- 178.Król J, Nguyen HD, Rogers LM, Beyanal H, Krone SM, Top EM. Increased transfer of a multi-drug resistance plasmid in E. coli biofilms at the air-liquid interface. Appl Environ Microbiol. 2011 doi: 10.1128/AEM.00090-11. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Van Elsas JD, Trevors JT, Starodub M-E. Bacterial conjugation between pseudomonas in the rhizoshpere of wheat. FEMS Microbiol Ecol. 1988;53:299–306. [Google Scholar]
- 180.Top E, Mergeay M, Sprigael D, Versraete W. Gene escape model: transfer of heavy metal resistance from Escherichia coli to Alcaligenes eutrophus on agar plates and in soil samples. Appl Environ Microbiol. 1990;56:2471–9. doi: 10.1128/aem.56.8.2471-2479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wellington EMH, Cresswell N, Saunders VA. Growth and survival of Streptomycete inoculants and extent of plasmid transfer in sterile and non-sterile soil. Appl Environ Microbiol. 1990;56:1413–9. doi: 10.1128/aem.56.5.1413-1419.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pukall R, Tschape H, Smalla K. Monitoring the spread of broad-host range and narrow host-range plasmids in soil microcosms. FEMS Microbiol Ecol. 1996;20:53–66. doi: 10.1111/j.1574-6941.1996.tb00304.x. [DOI] [Google Scholar]
- 183.Guarner F, Malagelada J-R. Gut flora in health and disease. Lancet. 2003;361:512–9. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 184.Duval-Iflah Y, Maisonneuve S, Ouriet MF. Effect of fermented milk intake on plasmid transfer and on the persistence of transconjugants in the digestive tract of gnotobiotic mice. Antonie van Leeuwenhoek. 1998;73:95–102. doi: 10.1023/A:1000603828184. [DOI] [PubMed] [Google Scholar]
- 185.García-Quintanilla M, Prieto AI, Barnes L, Ramos-Morales F, Casadesús J. Bile-induced curing of the virulence plasmid in Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:7963–5. doi: 10.1128/JB.00995-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bidlack JE, Silverman PM. An active type IV secretion system encoded by the F plasmid sensitizes Escherichia coli to bile salts. J Bacteriol. 2004;186:5202–9. doi: 10.1128/JB.186.16.5202-5209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ammann A, Neve H, Geis A, Heller KJ. Plasmid transfer via transduction from Streptococcus thermophilus to Lactococcus lactis. J Bacteriol. 2008;190:3083–7. doi: 10.1128/JB.01448-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kazimierczak KA, Scott KP, Kelly D, Aminov RI. Tetracycline resistome of the organic pig gut. Appl Environ Microbiol. 2009;75:1717–22. doi: 10.1128/AEM.02206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bale MJ, Day MJ, Fry JC. Novel method for studying plasmid transfer in undisturbed river epilithon. Appl Environ Microbiol. 1988;54:2756–8. doi: 10.1128/aem.54.11.2756-2758.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lilley AK, Fry JC, Bailey MJ, Day MJ. Comparison of aerobic heterotrophic taxa isolated from four root domains of mature sugar beet (Beta vulgaris) FEMS Microbiol Ecol. 1996;21:231–42. doi: 10.1111/j.1574-6941.1996.tb00350.x. [DOI] [Google Scholar]
- 191.Lilley AK, Fry JC, Day MJ, Bailey MJ. In situ transfer of an exogenously isolated plasmid between Pseudomonas spp. in sugar-beet rhizosphere. Microbiology. 1994;140:27–33. doi: 10.1099/13500872-140-1-27. [DOI] [Google Scholar]
- 192.Smalla K, Heuer H, Götz A, Niemeyer D, Krögerrecklenfort E, Tietze E. Exogenous isolation of antibiotic resistance plasmids from piggery manure slurries reveals a high prevalence and diversity of IncQ-like plasmids. Appl Environ Microbiol. 2000;66:4854–62. doi: 10.1128/AEM.66.11.4854-4862.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Top E, Desmet I, Verstraete W, Dijkmans R, Mergeay M. Exogenous Isolation of mobilizing plasmids from polluted soils and sludges. Appl Environ Microbiol. 1994;60:831–9. doi: 10.1128/aem.60.3.831-839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zwirglmaier K, Ludwig W, Schleifer KH. Recognition of individual genes in a single bacterial cell by fluorescence in situ hybridization–RING-FISH. Mol Microbiol. 2004;51:89–96. doi: 10.1046/j.1365-2958.2003.03834.x. [DOI] [PubMed] [Google Scholar]
- 195.Pratscher J, Stichternoth C, Fichtl K, Schleifer KH, Braker G. Application of recognition of individual genes-fluorescence in situ hybridization (RING-FISH) to detect nitrite reductase genes (nirK) of denitrifiers in pure cultures and environmental samples. Appl Environ Microbiol. 2009;75:802–10. doi: 10.1128/AEM.01992-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Suzuki H, Yano H, Brown CJ, Top EM. Predicting plasmid promiscuity based on genomic signature. J Bacteriol. 2010;192:6045–55. doi: 10.1128/JB.00277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Suzuki H, Sota M, Brown CJ, Top EM. Using Mahalanobis distance to compare genomic signatures between bacterial plasmids and chromosomes. Nucleic Acids Res. 2008;36:e147. doi: 10.1093/nar/gkn753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Karlin S, Burge C. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet. 1995;11:283–90. doi: 10.1016/S0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- 199.Karlin S. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 2001;9:335–43. doi: 10.1016/S0966-842X(01)02079-0. [DOI] [PubMed] [Google Scholar]
- 200.Campbell A, Mrazek J, Karlin S. Genome signature comparisons among prokaryote, plasmid, and mitochondrial DNA. Proc Natl Acad Sci USA. 1999;96:9184–9. doi: 10.1073/pnas.96.16.9184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Qu A, Brulc JM, Wilson MK, Law BF, Theoret JR, Joens LA, et al. Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome. PLoS ONE. 2008;3:e2945. doi: 10.1371/journal.pone.0002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Nogueira T, Rankin DJ, Touchon M, Taddei F, Brown SP, Rocha EP. Horizontal gene transfer of the secretome drives the evolution of bacterial cooperation and virulence. Curr Biol. 2009;19:1683–91. doi: 10.1016/j.cub.2009.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Rankin DJ, Rocha EP, Brown SP. What traits are carried on mobile genetic elements, and why? Heredity. 2011;106:1–10. doi: 10.1038/hdy.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lepage P, Colombet J, Marteau P, Sime-Ngando T, Doré J, Leclerc M. Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut. 2008;57:424–5. doi: 10.1136/gut.2007.134668. [DOI] [PubMed] [Google Scholar]
- 205.Warburton PJ, Allan E, Hunter S, Ward J, Booth V, Wade G, et al. Isolation of bacterial extra-chromosomal DNA from human dental plaque associated with periodontal disease, using transposon-aided capture (TRACA) FEMS Microbiol Ecol. 2011 doi: 10.1111/j.1574-6941.2011.01166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]