Abstract
Transcription factors of the basic Helix-Loop-Helix (bHLH) family play a central role in cell proliferation, determination, and differentiation. In plants, the regulatory functions of bHLHs in phenylpropanoid biosynthesis have been well established with regard to other interacting-proteins; i.e., MYB and WD40 repeat proteins. On the other hand, those in alkaloid biosynthesis are greatly limited due to the limited distribution of alkaloids in plant species. Recently, several groups have reported the regulatory functions of bHLH in alkaloid biosynthesis: novel CjbHLH1 in isoquinoline alkaloid biosynthesis in Coptis japonica, and Jasmonate-inducible MYC2-type bHLHs in nicotine-alkaloid biosynthesis in Nicotiana plants and indole alkaloid biosynthesis in Catharanthus roseus. We report here the JA-inducibility of CjbHLH1 and discuss the similarity and differences of non-MYC2-resemblant CjbHLH1 and MYC2-type bHLHs in nicotine and indole alkaloid biosynthesis.
Keywords: alkaloid biosynthesis, bHLH, indole alkaloid, isoquinoline alkaloid, jasmonate signaling, MYC2, nicotine alkaloid
The bHLH family of proteins is a wide group of functionally diverse transcription factors that are distributed in both animals and plants. In plants, bHLHs are key components of transcriptional networks that regulate several biological processes, such as light signaling, hormone signaling, development, and secondary metabolism.1,2 In secondary metabolism, bHLHs involved in phenylpropanoid biosynthesis, which is found in all plant species, have been well characterized in association with other interacting-proteins; i.e., MYB and WD40 repeat proteins.3 On the other hand, those in alkaloid biosynthesis are greatly limited due to the limited distribution of alkaloids in specific plant species and the scarce characterization of the molecular mechanism of biosynthesis.4 The first bHLH to be identified in alkaloid biosynthesis was CrMYC1, in terpenoid indole alkaloid biosynthesis in Catharanthus roseus, although its function remained unclear.5 Recently, some MYC2-type bHLHs, such as NbbHLH1/NbbHLH2/NtMYC2 and CrMYC2, have been reported to be directly involved in nicotine biosynthesis in Nicotiana plants and terpenoid indole alkaloid biosynthesis in C. roseus, respectively.6-8 These bHLHs function commonly through a signaling cascade of jasmonate (JA), a well-known phytoalexin inducer.
On the other hand, we recently identified a unique bHLH-type transcription factor, CjbHLH1, which regulates isoquinoline alkaloid biosynthesis in C. japonica.9 Since CjbHLH1 showed low sequence similarity to MYC2-type bHLHs and formed a distinct isoquinoline alkaloid-specific clade, we have characterized the responsiveness of CjbHLH1 to JA and discuss the similarity and differences in the signaling cascade among alkaloid biosynthesis.
MeJA rapidly induces the expression of the CjbHLH1 gene
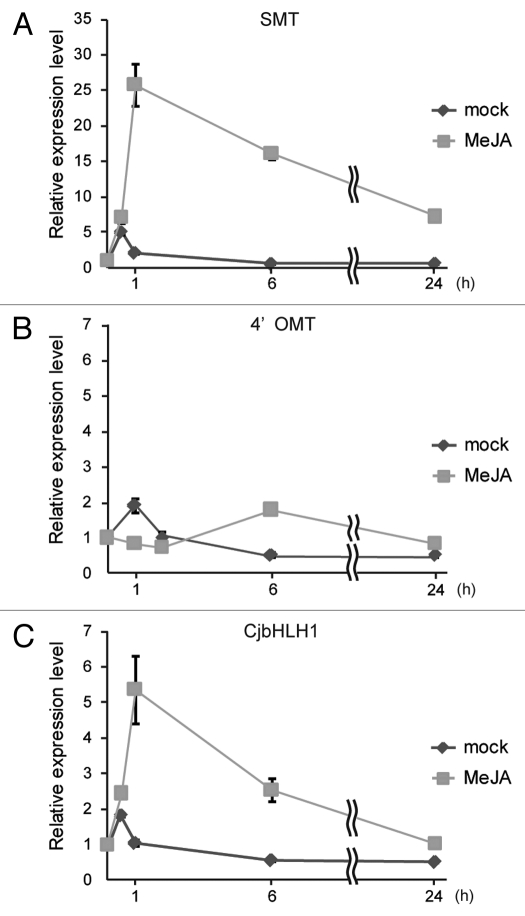
JA, an efficient inducer of secondary metabolism in the defense response, has also been reported to influence isoquinoline alkaloid biosynthesis.10,11 The involvement of CjbHLH1 in MeJA induced-isoquinoline alkaloid biosynthesis was examined with low berberine-producing suspension-cultured C. japonica cells (CjY line), since high-secondary metabolite-producing lines are often insensitive to treatment with JA. Treatment of CjY cells with MeJA clearly induced the expression of (S)-scoulerine-9-O-methyltransferase (SMT), which converts (S)-scoulerine to (S)-tetrahydrocolumbamine, while the expression of 3′-hydroxy-N-methylcoclaurine-4’-O-methyltransferase (4’OMT), which converts (S)-4’-hydroxy-N-methylcoclaurine to (S)-reticuline in berberine biosynthesis, was scarcely induced (Fig. 1). The transcript rapidly increased at 1 h after treatment with MeJA and remained at a high level for 24 h compared with treatment with dimethyl sulfoxide (DMSO mock). Treatment with MeJA also significantly induced the expression of CjbHLH1. The expression level was about 5 times higher than that with mock treatment at 0.5–1 h after treatment with MeJA, but decreased to the same level as with mock treatment after 24 h. This result and previous results9 on the transcriptional activity of CjbHLH1 clearly indicated that CjbHLH1 was involved in the response to MeJA in berberine biosynthesis.
Figure 1.
MeJA induced the expression of CjbHLH1 gene in low alkaloid-producing cultured C. japonica cells. Total RNA, prepared from 100 μM methyl-jasmonate (MeJA)- or 0.1% DMSO (mock)-treated cells, was used for reverse transcription after treatment with DNase I. The expression levels of SMT (A), 4’OMT (B) and CjbHLH1 (C) were determined using quantitative RT-PCR. The relative expression levels show the values standardized by that for the 0 h sample as 1. The results shown are mean values ± SD of three measurements.
Diversified transcriptional regulation network in alkaloid biosynthesis
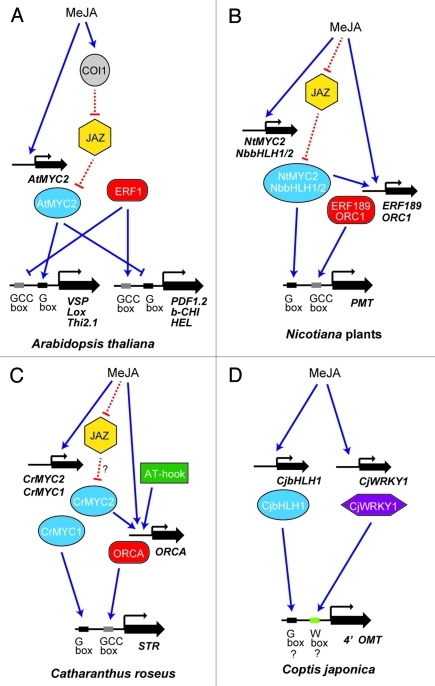
CjbHLH1 is a bHLH protein that is distinct from the MYC2 clade,9 but its expression was induced by MeJA, similar to Arabidopsis MYC2 (AtMYC2). AtMYC2 is a key component of the JA signaling pathway, and binds to the G-box in the promoter of JA-induced genes (Fig. 2A).12,13 In addition to JA-responsiveness, the transcriptional activity of AtMYC2 is also regulated by JAZ repressors in the JA signal pathway. Thus, JA promotes the interaction of JAZ repressors with the SCFCOI1 complex, and degrades JAZ by 26S proteasome to activate the expression of JA-responsive genes.14-16
Figure 2.
Simplified model of the MeJA-mediated signaling cascade in Arabidopsis thaliana (A), nicotine biosynthesis in Nicotiana plants (B), terpenoid indole alkaloid biosynthesis in C. roseus (C) and isoquinoline alkaloid biosynthesis in C. japonica (D). Solid (blue) lines show transcriptional regulation and dotted (red) lines show post-translational regulation via protein-protein interaction. Arrows indicate activation and T-shaped lines indicate inhibition. The detailed regulation of gene expression is discussed in the text.
NbbHLH1/2 and NtMYC2, which show high sequence similarity to AtMYC2, were also JA-responsive and interacted with tobacco JAZ repressor in yeast (Fig. 2B).7,17 Nicotiana MYC2-type bHLHs also directly bind to G-box in the target promoter of alkaloid biosynthetic genes, i.e., putrescine N-methyltransferase (PMT). Thus, MYC2-clade bHLH proteins directly regulated tobacco nicotine alkaloid biosynthesis in the JA-response, whereas tobacco MYC2-type bHLHs also regulated AP2/ERF-domain transcription factors ERF189/ORC1, and both transcription factors coordinately regulated the biosynthetic enzyme genes.7,17,18
On the other hand, CrMYC2, the closest homolog of AtMYC2 in C. roseus, did not directly bind to the promoter of biosynthetic enzyme genes, i.e., strictosidine synthase (STR), but was MeJA-responsive and regulated biosynthetic genes through the activation of Catharanthus AP2-domain protein (ORCA) transcription factor (Fig. 2C).8,19,20 This amplification of signal of JA via bHLH resembles the regulation of ERF expression via tobacco MYC2 in nicotine biosynthesis. Also, the high sequence similarity between CrMYC2 and AtMYC2 suggests that these activities may be regulated by JAZ repressors.8
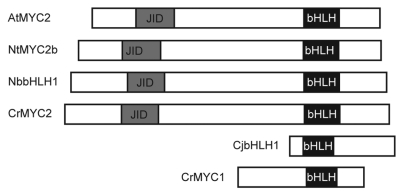
In the case of isoquinoline alkaloid biosynthesis, CjbHLH1 was MeJA-responsive but showed low homology to AtMYC2, especially at the N-terminal half of the JAZ interaction domain (JID) in MYC2,21 which suggests that transcriptional regulation by CjbHLH1 might be JAZ-independent (Fig. 3). It would be interesting to characterize the JAZ-interacting proteins in isoquinoline alkaloid biosynthesis.
Figure 3.
Domain organization in bHLH transcription factors. Gray boxes show the conserved JAZ interaction domain (JID) and black boxes show the bHLH domain.
As mentioned above, the cascade of signal transduction in the biosynthesis of different alkaloids would be diverse. Whereas the transcriptional regulation network is not yet sufficiently characterized, there may be differences in a subset of transcription factors among the pathways, as well as in the cascade (Figs. 2A-D). In nicotine biosynthesis, NbARF1 (auxin responsive factor) and NbHB1 (homeobox gene) have also been reported to play a role in transcriptional regulation.6 In indole alkaloid biosynthesis, AT-hook protein has been reported to regulate the expression of ORCA, although the details of this regulation are not yet clear.22
In isoquinoline alkaloid biosynthesis, CjWRKY1 is another direct activator of biosynthetic enzyme genes.23 In addition to our work, Apuya et al. reported that WRKY, AP2/ERF domain and other transcription factors are involved in isoquinoline alkaloid biosynthesis based on their results with an artificial reporter assay system.24 Whereas we have not yet detected the transcriptional regulation activity of ERF genes in isoquinoline alkaloid biosynthesis, several ERF-like genes were found in high berberine-producing C. japonica expression sequence tag (EST). Characterization of ERF in isoquinoline alkaloid biosynthesis, WRKY in nicotine or indole alkaloid biosynthesis and non-MYC2 type CrMYC1 in indole alkaloid biosynthesis would clarify the similarities and differences in the transcriptional regulation networks in alkaloid biosynthesis.
While the biosynthesis of alkaloids, which are powerful components of the plant chemical defense mechanism, is commonly regulated by a key signal regulator, jasmonate, it is also quite plant family-specific4 since the biosynthetic enzyme genes, such as CYP719A1 in isoquinoline alkaloid biosynthesis, only exist in specific plant families.25 Different types of alkaloids and secondary metabolites may have different transcriptional networks. Apuya et al. did not identify the transcriptional activity of Arabidopsis bHLH in isoquinoline alkaloid biosynthesis, whereas bHLH is a common transcription factor and AtMYC2 is an ortholog of NbbHLH1 and CrMYC2. It would be interesting to elucidate whether AtMYC2 can substitute for NbbHLH1 or CrMYC2. Our preferred hypothesis is that bHLHs may have diverse functionality in alkaloid biosynthesis. Also, in Arabidopsis, ERF and MYC2 antagonistically regulate the downstream genes, whereas they act synergistically in nicotine and indole alkaloid biosynthesis. Elucidation of the distinct mechanisms by which alkaloid biosynthesis is regulated by transcription factors including bHLHs should help to clarify how specific plant families evolved systems to produce their own alkaloids.
Acknowledgment
This research was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan [Grant-in-Aid (No. 21248013 to F. S.)]; the Japan Society for the Promotion of Science [fellowship to Y.Y.].
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/17599
References
- 1.Ledent V, Vervoort M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 2001;11:754–70. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martínez-García JF, Bilbao-Castro JR, Robertson DL. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010;153:1398–412. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 4.Liscombe DK, Facchini PJ. Evolutionary and cellular webs in benzylisoquinoline alkaloid biosynthesis. Curr Opin Biotechnol. 2008;19:173–80. doi: 10.1016/j.copbio.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 5.Chatel G, Montiel G, Pré M, Memelink J, Thiersault M, Saint-Pierre B, et al. CrMYC1, a Catharanthus roseus elicitor- and jasmonate-responsive bHLH transcription factor that binds the G-box element of the strictosidine synthase gene promoter. J Exp Bot. 2003;54:2587–8. doi: 10.1093/jxb/erg275. [DOI] [PubMed] [Google Scholar]
- 6.Todd AT, Liu E, Polvi SL, Pammett RT, Page JE. A functional genomics screen identifies diverse transcription factors that regulate alkaloid biosynthesis in Nicotiana benthamiana. Plant J. 2010;62:589–600. doi: 10.1111/j.1365-313X.2010.04186.x. [DOI] [PubMed] [Google Scholar]
- 7.Shoji T, Hashimoto T. Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant Cell Physiol. 2011;52:1117–30. doi: 10.1093/pcp/pcr063. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Hedhili S, Montiel G, Zhang Y, Chatel G, Pré M, et al. The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 2011;67:61–71. doi: 10.1111/j.1365-313X.2011.04575.x. [DOI] [PubMed] [Google Scholar]
- 9.Yamada Y, Kokabu Y, Chaki K, Yoshimoto T, Ohgaki M, Yoshida S, et al. Isoquinoline alkaloid biosynthesis is regulated by a unique bHLH-type transcription factor in Coptis japonica. Plant Cell Physiol. 2011;52:1131–41. doi: 10.1093/pcp/pcr062. [DOI] [PubMed] [Google Scholar]
- 10.Pauwels L, Inzé D, Goossens A. Jasmonate-inducible gene: What does it mean? Trends Plant Sci. 2009;14:87–91. doi: 10.1016/j.tplants.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Ikezawa N, Iwasa K, Sato F. Molecular cloning and characterization of methylenedioxy bridge-forming enzymes involved in stylopine biosynthesis in Eschscholzia californica. FEBS J. 2007;274:1019–35. doi: 10.1111/j.1742-4658.2007.05652.x. [DOI] [PubMed] [Google Scholar]
- 12.Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16:1938–50. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell. 2007;19:2225–45. doi: 10.1105/tpc.106.048017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signaling. Nature. 2007;448:661–5. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 15.Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, et al. The JAZ family of repressors is the missing link in jasmonate signaling. Nature. 2007;448:666–71. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 16.Chico JM, Chini A, Fonseca S, Solano R. JAZ repressors set the rhythm in jasmonate signaling. Curr Opin Plant Biol. 2008;11:486–94. doi: 10.1016/j.pbi.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 17.De Boer K, Tilleman S, Pauwels L, Vanden Bossche R, De Sutter V, Vanderhaeghen R, et al. APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J. 2011;66:1053–65. doi: 10.1111/j.1365-313X.2011.04566.x. [DOI] [PubMed] [Google Scholar]
- 18.Shoji T, Kajikawa M, Hashimoto T. Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell. 2010;22:3390–409. doi: 10.1105/tpc.110.078543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menke FL, Champion A, Kijne JW, Memelink J. A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J. 1999;18:4455–63. doi: 10.1093/emboj/18.16.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Fits L, Memelink J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science. 2000;289:295–7. doi: 10.1126/science.289.5477.295. [DOI] [PubMed] [Google Scholar]
- 21.Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23:701–15. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vom Endt D, Soares e Silva M, Kijne JW, Pasquali G, Memelink J. Identification of a bipartite jasmonate-responsive promoter element in the Catharanthus roseus ORCA3 transcription factor gene that interacts specifically with AT-Hook DNA-binding proteins. Plant Physiol. 2007;144:1680–9. doi: 10.1104/pp.107.096115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato N, Dubouzet E, Kokabu Y, Yoshida S, Taniguchi Y, Dubouzet JG, et al. Identification of a WRKY protein as a transcriptional regulator of benzylisoquinoline alkaloid biosynthesis in Coptis japonica. Plant Cell Physiol. 2007;48:8–18. doi: 10.1093/pcp/pcl041. [DOI] [PubMed] [Google Scholar]
- 24.Apuya NR, Park JH, Zhang L, Ahyow M, Davidow P, Van Fleet J, et al. Enhancement of alkaloid production in opium and California poppy by transactivation using heterologous regulatory factors. Plant Biotechnol J. 2008;6:160–75. doi: 10.1111/j.1467-7652.2007.00302.x. [DOI] [PubMed] [Google Scholar]
- 25.Ikezawa N, Tanaka M, Nagayoshi M, Shinkyo R, Sakaki T, Inouye K, et al. Molecular cloning and characterization of CYP719, a methylenedioxy bridge-forming enzyme that belongs to a novel P450 family, from cultured Coptis japonica cells. J Biol Chem. 2003;278:38557–65. doi: 10.1074/jbc.M302470200. [DOI] [PubMed] [Google Scholar]