Abstract
We carried out a genome-wide association study (GWAS) of LDL-c response to statin using data from participants in the Collaborative Atorvastatin Diabetes Study (CARDS; n = 1,156), the Anglo-Scandinavian Cardiac Outcomes Trial (ASCOT; n = 895), and the observational phase of ASCOT (n = 651), all of whom were prescribed atorvastatin 10 mg. Following genome-wide imputation, we combined data from the three studies in a meta-analysis. We found associations of LDL-c response to atorvastatin that reached genome-wide significance at rs10455872 (P = 6.13 × 10−9) within the LPA gene and at two single nucleotide polymorphisms (SNP) within the APOE region (rs445925; P = 2.22 × 10−16 and rs4420638; P = 1.01 × 10−11) that are proxies for the ϵ2 and ϵ4 variants, respectively, in APOE. The novel association with the LPA SNP was replicated in the PROspective Study of Pravastatin in the Elderly at Risk (PROSPER) trial (P = 0.009). Using CARDS data, we further showed that atorvastatin therapy did not alter lipoprotein(a) [Lp(a)] and that Lp(a) levels accounted for all of the associations of SNPs in the LPA gene and the apparent LDL-c response levels. However, statin therapy had a similar effect in reducing cardiovascular disease (CVD) in patients in the top quartile for serum Lp(a) levels (HR = 0.60) compared with those in the lower three quartiles (HR = 0.66; P = 0.8 for interaction). The data emphasize that high Lp(a) levels affect the measurement of LDL-c and the clinical estimation of LDL-c response. Therefore, an apparently lower LDL-c response to statin therapy may indicate a need for measurement of Lp(a). However, statin therapy seems beneficial even in those with high Lp(a).
Keywords: genetics, low density lipoprotein, LDL/metabolism, lipoprotein(a), statins
Statin therapy is now widely accepted for the primary and secondary prevention of cardiovascular disease (CVD) in certain patient groups. However, there is considerable variation in response to statin therapy that remains poorly understood. For example, in the Collaborative Atorvastatin Diabetes Study (CARDS) trial (1), among self-reported and pill count-validated compliant recipients of atorvastatin 10 mg daily, the absolute change in LDL-c at one month post-randomization varied from −2 to −0.6 mmol/l, (5th and 95th centiles of the range), and the percentage lowering from baseline varied from 67% to 22%. Understanding the pathways and determinants involved in this variation in response to therapy could lead to improved treatments. Even without understanding the pathways, identifying predictors of poorer response could identify those most in need of additional or alternative therapeutic strategies.
Two genome-wide association studies (GWAS) of statin response and several candidate gene association studies have been reported (2–5). From these, the only consistent finding is that variants in the APOE gene region are associated with variation in LDL response. Here, we report a genome-wide analysis of LDL-c response from two randomized clinical trials of atorvastatin, CARDS and the Anglo-Scandinavian Outcomes Trial (ASCOT) (6), to investigate genetic effects on LDL-c response to atorvastatin. We chose to model genetic determinants of LDL-c response to atorvastatin among those assigned to atorvastatin in these trials. An alternative approach would be to model the interaction of genotype on the effect of atorvastatin on LDL-c using data from both placebo and active treatment groups. However, we did not consider this latter approach as optimal as testing for interactions is much less powerful than direct tests of association and as, in any case, we did not consider genetic effects on change LDL-c in the placebo groups to be plausible.
MATERIALS AND METHODS
Study populations and phenotype definition
Both trials were conducted with Ethics Committee/IRB approval, under good clinical practice guidelines and in accordance with the Declaration of Helsinki principles. Patients gave consent for genetic studies.
CARDS
Methods in CARDS have been described previously. In brief, 2,838 patients with type 2 diabetes and no previous CVD were randomized to receive either placebo or atorvastatin 10 mg once daily and followed for a median of 3.7 years. Allocation was double blinded. Mean serum LDL-c concentration during baseline visits prior to randomization had to be ≤ 4.14 mmol/l (160 mg/dl) and serum triglycerides ≤ 6.78 mmol/l (600 mg/dl). After randomization, total cholesterol (TC), HDL-C, and triglycerides were measured at one, two, and three months, and then every six months. Patients attended after an overnight fast. LDL-c was calculated with the Friedewald formula (7), or if serum triglycerides exceeded 4.0 mmol/l, by removing VLDL by ultracentrifugation and then measuring the change in infranatant cholesterol content when LDL was removed by precipitation of apolipoprotein B-containing lipoproteins. For this genome-wide study, the analyses were restricted to those randomized to atorvastatin, and the mean of two pretreatment LDL-c measurements was used as the baseline LDL-c and a weighted average of five post-randomization values within the first year post-randomization was the outcome measure or “on treatment LDL-c,” with weights (0.6 for month 1 and then 0.1 for measurements at 2, 3, 6, and 12 months). Lipoprotein(a) concentrations were determined by an immunoturbidimetric assay with Immuno LEIA® reagents from Technoclone Ltd., Dorking, UK (now www.PathwayDiagnostics.com), which is calibrated against the IFCC standard preparation PRM02.
ASCOT
Of 19,342 hypertensive patients (40–79 years of age with at least three other cardiovascular risk factors) who were randomized to one of two antihypertensive regimens in ASCOT, 10,305 with nonfasting TC concentrations of 6.5 mmol/l or less (measured at the nonfasting screening visit) had been randomly assigned additional atorvastatin 10 mg or placebo. These patients formed the lipid-lowering arm of the study. For this genome-wide study, two subpopulations from ASCOT were included. The first subpopulation included individuals randomized to 10 mg atorvastatin in whom pretreatment LDL-c was measured at the (fasting) randomization visit and on-treatment LDL-c was calculated as the simple average of measures at the (fasting) visits 6 months and 12 months post-randomization. LDL-c was estimated using the Friedewald equation as in CARDS. Following the end of the randomization phase, there was an observational period. The second subpopulation included all individuals not originally randomized to 10 mg atorvastatin (i.e., those randomized to placebo and those not eligible for the LLA) who were subsequently prescribed atorvastatin 10 mg. For these individuals, pretreatment LDL-c was defined as the measurement on the last visit before or equal to date of starting atorvastatin, and on-treatment LDL-c was defined as the measurement taken from the first visit after date of starting atorvastatin.
PROSPER (replication cohort)
All data were from the PROspective Study of Pravastatin in the Elderly at Risk (PROSPER) (8). PROSPER was a prospective multicenter randomized placebo-controlled trial to assess whether treatment with pravastatin diminishes the risk of major vascular events in elderly. Between December 1997 and May 1999, we screened and enrolled subjects in Scotland (Glasgow), Ireland (Cork), and the Netherlands (Leiden). Men and women 70–82 years of age were recruited if they had preexisting vascular disease or increased risk of such disease because of smoking, hypertension, or diabetes. A total number of 5,804 subjects were randomly assigned to pravastatin or placebo, of which 2,550 subjects assigned to the Pravastatin arm of the trial were included in the present study. TC, HDL-C, and triglycerides were assessed after an overnight fast, at baseline, and at 3, 6, 12, 24, and 36 months post-randomization. LDL-C was calculated by the Friedewald formula. The pretreatment measurement was at baseline before randomization, and the posttreatment was the mean of the lipid measurements after randomization.
Phenotype transformation
To maximize power to detect associations and to improve test statistic behavior under the null for low minor allele frequency (MAF) single nucleotide polymorphisms (SNP), we transformed measured LDL-c levels to conform to the distributional assumptions made by our association analysis model using the same transformation for off- and on-treatment measures to preserve the relationship between the two. We maximized the fit of the residuals in a regression of on-treatment on the pretreatment value to a Gaussian distribution. We used a 2-parameter Box-Cox transform of the form applied to baseline and on-treatment LDL-c values. The parameter values α and β were chosen by maximizing the likelihood of a model with linear regressions of the transformed pretreatment and response (transformed pretreatment minus transformed pretreatment) values on the covariates (age and sex), with the joint distribution of the residuals from the two regression models being bivariate Gaussian.
The parameter values obtained were α = 0.156, β = −0.505 mmol/l in CARDS. In ASCOT, the parameters were α = 0.6807, β = 0.8850 mmol/l in the randomized dataset, and α = 0.4805, β = 0.5813 mmol/l in the observational dataset. This transformation has the same motivation as the inverse normal transform used in some GWAS applications (9, 10), but the use of a parametric transform preserves the relationship between pre and on-treatment measures, thereby allowing the difference between the two, adjusted for pretreatment value, to be used as a response variable as was done in ASCOT or as simply the on-treatment adjusted for pretreatment value as in CARDS (these are equivalent). The effect sizes in discovery cohorts (CARDS and ASCOT) and the replication cohort (PROSPER) were scaled so that the residuals had unit variance, thereby allowing studies using different transforms to be combined.
DNA extraction and genotyping
CARDS.
DNA was extracted from whole-blood EDTA samples. DNA was isolated from 10 ml of frozen blood using the Gentra Puregene DNA Isolation Kit from Qiagen (Cat. no. 158389). Briefly, RBC was lysed with an anionic detergent in the presence of a DNA stabilizer that limits the activity of intracellular DNases. White blood cells were collected by centrifugation at 2,000 g for 2 min. RNA was removed by treatment with RNase A. Protein was removed by salt precipitation (centrifugation at 2000 g for 5 min). Genomic DNA was recovered by precipitation with isopropanol and centrifugation at 2,000 g for 5 min, the DNA pellet was washed in 70% ethanol, air dried, and dissolved in hydration solution (1 mM EDTA, 10 mM Tris·CI, pH 7.5). Purified DNA was stored at −20°C. DNA aliquots were genotyped at Perlegen Sciences using a proprietary SNP set comprising 599,164 SNPs. Of these, 243 SNPs that had discrepant map positions between HapMap and Perlegen were dropped. We set a minimum SNP call rate threshold of 80% for including SNPs in the analysis, and we required that the P-value for a test of deviation from Hardy-Weinberg equilibrium (HWE) was not < 10−5. This gave 517,746 SNPs for analysis. The average call rate was 98%, with 86.25% SNPs with a call rate of greater than 90%. SNP annotation was based on build 36 of the Human Genome Sequence. All SNPs were used in the analysis regardless of allele frequency, but the allele frequency was considered when evaluating putative associations. Allele frequency was below 1% at 6% of SNPs. We selected samples from those people who had been allocated atorvastatin 10 mg daily, had given consent for genotyping, and had a sample SNP call rate > 80%. After applying the exclusions of HWE, we estimated relatedness with PLINK, and those individuals with Pi_HAT > 0.25(excluding first- and second-degree relatives) were removed (n = 0). Only LDL-c values from time points at which the person was compliant with atorvastatin (based on pill count > 80%) were used.
ASCOT genotyping.
Genotyping was carried out on HumanCNV370 (Illumina) array on 3,868 individuals at Centre National de Génotypage (CNG) in two batches. Samples were excluded if they had ≥ 5% missing data (two samples). SNPs were excluded based on the following criteria: i) they had been mapped to different chromosomes or positions in the different releases (two SNPs), or ii) they were polymorphic A/T or C/G in either release or in the combined dataset, or iii) they had call rate ≤ 97% in either release or in the combined dataset (47,744 SNPs), or iv) they had HWE P-value ≤ 10−7 in either release or in the combined dataset (8,502 SNPs). After applying the above exclusions, samples were excluded if they had estimated relatedness > 0.1875 (halfway cut point between second- and third-degree relatives), estimated using a using a subset of 101,954 SNPs obtained by linkage disequalibrium (LD)-based pruning (87 duplicates, 15 first-degree relatives and 4 presumed second-degree relatives removed. Then SNPs were excluded if they showed significant differences in allele frequency between the different batches at P < 10−7 (20 SNPs), if they were monomorphic in the combined dataset (3,838 SNPs), if they were not in HapMap r22 (12,817 SNPs) or had different alleles to HapMap r22 (6 SNPs), or if they showed significant differences (P < 10−7 using Fisher's exact test) in allele frequency between the combined dataset and HapMap r22 (308 SNPs). After applying all the above exclusions, ancestry outliers were excluded (n = 143) by using ancestry principal component analysis (11) on a subset of 100,905 SNPs selected by LD-based pruning, and ancestry principal components (PCs) were calculated for the remaining 3,804 individuals.
PROSPER genotyping.
A whole genome-wide screening was performed in the sequential PHASE project. DNA was available for genotyping 5,763 subjects. Genotyping was performed with the Illumina 660K beadchip. After QC (call rate < 95%), 5,244 subjects and 557,192 SNPs were left for analysis (12).
Statistical analysis
Imputation of genotypes.
The CARDS genotype data were combined with phased haplotypes from HapMap phase II CEU r22 to compute posterior probability distribution of genotype at all HapMap loci using the IMPUTE program (13). For ASCOT and PROSPER, genotypes at unmeasured SNPs were imputed using MACH (14) and phased haplotypes from HapMap CEU r22. For ASCOT, a randomly chosen subset of 400 individuals was used to estimate transition and emission probabilities (i.e., to estimate recombination rates between SNPs and per-SNP genotyping error rates) using MACH options “-greedy -r 100” for each (entire) chromosome in turn. Using these estimated rates (the .rec and .erate files), genotypes were imputed for the whole sample of 3,804 individuals using MACH options “-greedy-mle-mldetails” for each (entire) chromosome in turn.
CARDS data analysis.
The EIGENSTRAT program (15) was used to adjust for population structure. Using PLINK (16), we generated a pruned subset of 152,587 SNPs that are in approximate linkage equilibrium with each other in the CARDS dataset. Principal components analysis was undertaken using this subset of SNPs. Thirty-seven individuals identified as outliers in the initial principal components analysis were excluded from the subsequent computation of principal components, leaving 1174 persons evaluable for statin response. The first three principal components were retained and included as covariates in all tests of association.
On-treatment values for LDL-c for each individual at 1, 2, 3, 6, and 12 months post-randomization were available. We initially used the first available post-randomization LDL-c and established that the previously reported APOE genotype at rs445925 was the strongest association in a genome-wide analysis of response at P = 1.1 × 10−13. To maximize the power to detect any further new associations, we trained the weighting of post-randomization LDL-c time points to maximize the strength of the association of LDL response with APOE genotype at rs445925. Based on this, the nonmissing values for each individual were combined in a weighted average, with the one-month value allocated a weight of 0.6 and the four subsequent values, weights of 0.1 each (P-value for rs445925 with these weights = 2.2 × 10−16). SNPTEST (13) was used to test for association of LDL response with genotype in a linear regression with the weighted average post-randomization LDL value as dependent variable and with covariates, including transformed pretreatment LDL-c, age, sex, and scores on the first three principal components of population stratification. The missing-data likelihood option was used to allow for uncertainty of genotypes at each imputed locus. In practice, the use of several weighted post-randomization LDL-c values rather than a single first value made very little difference to the results (see supplementary table II).
We used the conditional analysis test in PLINK (16) to test for independence of SNP associations over short regions within the same gene; a null model based on equating the effects of haplotypes that differed only at the SNP under test was compared with a more general model in which the effects of these haplotypes were unconstrained. The null hypothesis is that the SNP under test accounts for all associations of haplotypes with response. Other analyses included those carried out to explore initial associations, including a test of whether LPA genotype modifies the effect of atorvastatin on CVD. This was carried out by estimating the hazard ratio associated with allocation to atorvastatin in a Cox regression model of time to first CVD event and using a likelihood ratio test comparing a model with this main treatment effect and one including a term for interaction of genotype × treatment effect.
ASCOT data analysis.
We regressed the response variable (transformed on-treatment minus transformed pretreatment LDL-c) onto imputed expected genotype dosage as implemented in ProbABEL (14, 17). This is asymptotically equivalent to score test for taking into account uncertainty in imputed genotypes (as in SNPTEST) but with improved finite sample size operating characteristics (18). Age, sex, age*sex, and transformed pretreatment LDL were used as covariates, plus 10 ancestry principal components.
PROSPER data analysis.
The response variable was regressed (natural log of transformed on-treatment minus natural log of pretreatment LDL-c) onto imputed expected genotype dosage as implemented in SNPTEST. Age, sex, transformed pretreatment LDL, and top three principal components were used as covariates.
Meta-analysis.
The score and observed information for the effect parameter were summed over studies to obtain a summary score test. This is algebraically equivalent (based on the quadratic approximation of the log-likelihood) to obtaining a weighted average of the maximum likelihood estimates with weights inversely proportional to the squared standard errors, with the useful feature that the ratio of observed to complete information (calculated by summing numerators and denominators over the three studies) is obtained as a summary measure of the efficiency of genotype imputation. For concise presentation, we focus here on showing the results of the meta-analysis rather than each study separately and provide study-specific estimates of effect only at the most extreme significance levels. In the data presentation, those loci at which the overall proportion of information extracted was less than 30% across the studies have been excluded. We have used the P-value threshold of <5 × 10−8 as the threshold for declaring a genome-wide significant association.
Distinguishing indirect and direct effects of genotype on on-treatment LDL.
Effects of genetic variation on treatment response as measured by on-treatment LDL-c could be mediated through effects on the pretreatment LDL-c. To evaluate whether genetic on-treatment LDL-c likely reflects residual effect on pretreatment LDL-c, it is necessary to adjust for the pretreatment LDL-c levels and to correct the maximum likelihood estimate of the adjusted effect of genotype on on-treatment value for the noise in pretreatment values (the noise is both random measurement error and intra-individual variation in usual LDL-c). From the rules of path analysis, we calculated the direct effect γ of genotype on an on-treatment trait value as β − αδ (1 − ρ) / ρ, where β is the coefficient of regression for on-treatment trait value on genotype adjusted for measured pretreatment value, ρ is the intraclass correlation between replicate measurements of pretreatment values, and δ is the coefficient of regression for on-treatment value on observed pretreatment value. For these calculations, we used ρ = 0.8 as a plausible value for the intraclass correlation based on the within-person correlation in LDL-c values taken over two pretreatment visits in CARDS.
RESULTS
Table 1 compares baseline characteristics of participants in the three studies. Fig. 1 shows a quantile-quantile plot of the −log10 P-values for association of each SNP with LDL-c response to treatment, obtained by meta-analyzing effect size estimates across the CARDS and ASCOT datasets. This plot shows that the cumulative distribution of test statistics approximates the null distribution over most of its range but that there is a tail of extreme results. Fig. 2 shows a Manhattan plot of the −log10 P-values by map position. Table 2 shows all loci at which the summary test for association yielded a nominal P-value < 10−6. The estimates of effect (β) are for the transformed response variable (see Materials and Methods). In CARDS, the response variable was transformed on-treatment LDL-c with transformed pretreatment LDL-c entered as a covariate in the model. This is mathematically equivalent to modeling change in LDL-c with pretreatment LDL-c as a covariate (i.e., the difference in transformed on-treatment and adjustment for pretreatment LDL) as was done in ASCOT. A negative β for an allele means that the modeled allele is associated with a bigger reduction in posttreatment LDL-c and a better response to statins.
TABLE 1.
Characteristics of patients and studies included in the meta-analysis
| CARDS | ASCOT-R | ASCOT-Obs | |
| n = 1194 | n = 895 | n = 691 | |
| Age (mean years ± SD) | 61.6 ± 8.2 | 64.1 ± 8.0 | 64.2 ± 8.6 |
| Ethnicity | Caucasian (UK and Ireland) | Caucasian (UK and Ireland) | Caucasian (UK and Ireland) |
| Women (%) | 47 | 11.0 | 13.1 |
| Diabetes (%) | 100 | 21 | 21 |
| Follow-up years (median IQR) | 3.9 years (3.0–4.7) | First year was used | First year was used |
| Hypertension (%) | 87 | 100 | 100 |
| LDL-c level at baseline (mean mmol/l ± SD) | 3.04 ± 0.71 | 3.47 ± 0.70 | 3.75 ± 0.85a |
| Lipid entry criterion | Fasting LDL-c ≤ 4.14 mmol/l | Non-fasting TC ≤ 6.5 mmol/l | None |
| Fasting status for lipidsb | Overnight fast | Fasting | Fasting |
| Statin dose | Atorvastatin 10 mg daily | Atorvastatin 10 mg daily | Atorvastatin 10 mg daily |
| Platform | Perlegen 6 | Illumina HumanCNV370 | Illumina HumanCNV370 |
| pHWEc exclusion | 10 | 10 | 10 |
| Imputation software | IMPUTE 2 | MACH | MACH |
| NCBI build for imputation | HapMap CEU r22 | HapMap CEU r22 | HapMap CEU r22 |
ASCOT-Obs, observational arm of ASCOT; ASCOT-R, randomized arm of ASCOT.
In N = 656 with nonmissing LDL-c at baseline; the missingness is nonrandom because these are individuals with baseline triglycerides too high for Friedewald formula.
Fasting status for LDL-c at baseline (see previous row) and for response to statin measure.
cthinspP: -value threshold for exclusion of SNPs not in HWE.
Fig. 1.
Quantile-quantile plot of meta-analysis P-values for statin response. A plot of the quantiles of observed and expected distribution of P-values against each other.
Fig. 2.
Manhattan plot of P-values from meta-analysis of all SNPs that passed stringent quality control. The Manhattan plots [also known as −log10 (P) association plots[ show the chromosomal position of SNPs exceeding the genome-wide significance threshold (P < 5 × 10−8) as indicated by the solid red line.
TABLE 2.
Combined analysis (CARDS, ASCOT randomized, and ASCOT observational)
| CHR | POS (cM) | SNP | Modeled Allele | Minor Allele (Frequency) | CARDS | Ascot-R | Ascot-Obs | Meta-analysis | P | Genec | |||||
| β | SE | β | SE | β | SE | Rsqa | βb | SE | |||||||
| 6 | 195.419 | rs10455872 | A | G (0.07) | −0.35 | 0.08 | −0.36 | 0.11 | −0.1 | 0.18 | 0.54 | −0.35 | 0.06 | 6.13E-09 | LPA |
| 12 | 55.598 | rs1627770 | G | T (0.2) | 0.18 | 0.05 | 0.13 | 0.05 | 0.17 | 0.06 | 1 | 0.17 | 0.03 | 1.81E-07 | LOC390301-ALG10 |
| 12 | 55.598 | rs863626 | C | T (0.2) | 0.18 | 0.05 | 0.13 | 0.05 | 0.17 | 0.06 | 1 | 0.18 | 0.03 | 1.39E-07 | LOC390301-ALG10 |
| 12 | 55.598 | rs11053045 | A | T (0.2) | 0.18 | 0.05 | 0.13 | 0.05 | 0.17 | 0.06 | 1 | 0.18 | 0.03 | 1.34E-07 | LOC390301-ALG10 |
| 12 | 55.598 | rs1619785 | A | A (0.2) | −0.18 | 0.05 | −0.13 | 0.05 | −0.17 | 0.06 | 1 | −0.18 | 0.03 | 1.28E-07 | LOC390301-ALG10 |
| 12 | 55.599 | rs10844779 | A | A (0.2) | −0.18 | 0.05 | −0.14 | 0.05 | −0.17 | 0.06 | 1 | −0.18 | 0.03 | 1.44E-07 | ALG10-LOC260338 |
| 12 | 55.599 | rs11053068 | C | C (0.2) | −0.18 | 0.05 | −0.14 | 0.05 | −0.17 | 0.06 | 1 | −0.18 | 0.03 | 1.44E-07 | ALG10-LOC260338 |
| 12 | 55.599 | rs5004272 | A | G (0.21) | 0.18 | 0.05 | 0.12 | 0.05 | 0.17 | 0.06 | 1 | 0.17 | 0.03 | 2.81E-07 | ALG10-LOC260338 |
| 12 | 55.599 | rs10844823 | C | C (0.21) | −0.18 | 0.05 | −0.12 | 0.05 | −0.17 | 0.06 | 0.99 | −0.17 | 0.03 | 2.86E-07 | ALG10-LOC260338 |
| 16 | 27.656 | rs721843 | C | G (0.46) | 0.13 | 0.04 | 0.12 | 0.05 | 0.14 | 0.05 | 0.97 | 0.14 | 0.03 | 6.05E-07 | LOC653737-GRIN2A |
| 19 | 80.713 | rs4803760 | C | T (0.2) | 0.24 | 0.06 | 0.08 | 0.05 | 0.19 | 0.07 | 0.97 | 0.18 | 0.04 | 4.23E-07 | BCAM-PVRL2 |
| 19 | 80.766 | rs1985096 | A | A (0.16) | −0.33 | 0.07 | −0.16 | 0.06 | −0.28 | 0.08 | 0.8 | −0.27 | 0.04 | 9.49E-11 | BCAM-PVRL2 |
| 19 | 80.877 | rs395908 | A | A (0.16) | −0.21 | 0.06 | −0.1 | 0.06 | −0.23 | 0.07 | 0.92 | −0.19 | 0.04 | 3.46E-07 | PVRL2-BCAM-TOMM40 |
| 19 | 80.954 | rs6857 | C | T (0.14) | −0.32 | 0.07 | −0.06 | 0.07 | −0.23 | 0.08 | 0.93 | −0.23 | 0.04 | 7.43E-08 | PVRL2-BCAM-TOMM40 |
| 19 | 81.023 | rs405509 | G | T (0.48) | −0.17 | 0.05 | −0.1 | 0.04 | −0.21 | 0.05 | 0.99 | −0.17 | 0.03 | 3.46E-09 | APOE-TOMM40-APOE |
| 19 | 81.051 | rs445925 | A | A (0.11) | −0.44 | 0.08 | −0.36 | 0.07 | −0.34 | 0.09 | 0.77 | −0.42 | 0.05 | 1.59E-17 | LOC100129500-APOE APOC1 |
| 19 | 81.081 | rs4420638 | A | G (0.16) | −0.44 | 0.08 | −0.15 | 0.07 | −0.32 | 0.09 | 0.56 | −0.33 | 0.05 | 1.12E-11 | APOC1 APOC1 APOC4 |
SNPs associated with LDL-c response to statins with meta-analysis values of P < 10−6 and Rsq > 0.30.
Ascot-Obs, ASCOT observational; Ascot-R, ASCOT randomized; CHR, chromosome; POS, position.
Estimate of squared correlation between imputed and true genotypes.
A positive β value means that the modeled allele is associated with a bigger posttreatment LDL-c and, therefore, a lower response to statins. A negative β value means that the modeled allele is associated with lower posttreatment LDL-c and, therefore, a better response to statins.
For SNPs that lie in the intergenic regions, the location of the nearby genes is shown.
The strongest associations were with rs10455872 in the LPA gene on chromosome 6, and with SNPs in the BCAM/PVRL2/APOE/APOC1 gene region on chromosome 19, where genome-wide significant associations were found. The SNPs in the LPA and APOE region explained 4% of the variance in LDL-c response in CARDS. The next most significant P-value was that for the ALG10 region on chromosome 12, but this did not reach genome-wide significance. There was no evidence of gene-gender interaction for all the top SNPs reported in the study. The effect sizes for all top SNPs were similar in CARDS, where all the participants had type 2 diabetes, and in ASCOT, where 21% of the participants had type 2 diabetes (Table 2), suggesting that diabetes per se was not a strong determinant of the genetic effect of the top SNPs.
LPA
In the LPA gene SNP, rs10455872 showed a genome-wide significant association with LDL-c response (Fig. 3). The effect at rs10455872 was modest; the β shown in Table 2 is not easily directly interpretable given the transformation used, but in CARDS, for example, the percentage change in LDL-c with statin therapy at one month was approximately −43% in those with at least one “G” allele at rs10455872 (MAF = 8%) compared with −46.5% in homozygotes for the “A” allele. There was no significant effect of this SNP on change in LDL-c post-randomization in those in the placebo group (P = 0.28).
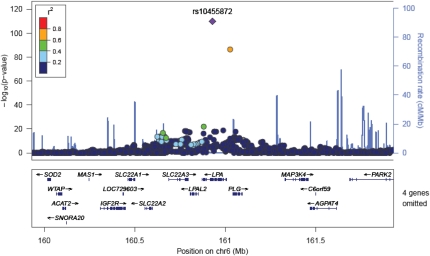
Fig. 3.
Regional association plot of LPA locus with statin response. Correlations between the target SNP (the SNP with the lowest P value, depicted in purple) and nearby SNPs within a 500 kb region. The r2 values were based on the HapMap CEU population.
To investigate the association with LPA genotype further, we first confirmed that LPA genotypes predicted serum lipoprotein(a) [Lp(a)] levels, which had been measured in CARDS but not in ASCOT. Fig. 4 shows the results of the GWAS for serum Lp(a) levels; all significantly associated loci were in the LPA region, consistent with other reports (19). In a linear regression model that included age, sex, and population structure covariates, 12 SNPs in the LPA region had independent effects on serum Lp(a) (rs10455872, rs5014650, rs783147, rs6919346, rs3103349, rs2063347, rs6415084, rs10455782, rs394487, rs6926458, rs316174, rs3127569). Together, these SNPs explained 40% variation in the serum Lp(a) levels in CARDS; however, most of this was attributable to the rs10455872 SNP (30%) with median levels being 7.6 mg/dl [interquartile range (IQR) 4.1–14 mg/dl], 50.5 mg/dl (IQR 37–68 mg/dl) and 55.2 mg/dl (IQR 51–113 mg/dl) in those with AA, AG, and GG genotype, respectively. We then adjusted the association of genotype at SNPs in the LPA gene with LDL-c response to statin for measured serum Lp(a) levels in the CARDS data to test whether the genetic effects seen are likely to be mediated through the effect of LPA on serum Lp(a) levels (Table 3). The estimate of the standardized regression coefficient at the associated SNP (rs10455872) in LPA in CARDS was reduced from −0.35 (±0.08) to −0.09 (±0.08), consistent with the effect of genotype on apparent response to statin being mediated through Lp(a) levels. We noted that Lp(a) levels had an independent association with apparent LDL-c response to statin beyond genotype in these analyses (P = 0.001). Further analysis in CARDS also confirmed that there was no effect of statin on Lp(a) levels; β was −0.23 mg/dl, (95% CI: −2.25 to 1.80) for difference in Lp(a) levels with atorvastatin versus placebo at one year post-randomization, adjusted for baseline Lp(a), age, and sex.
Fig. 7.
Regional association plot of LPA locus with Lp(a) levels in the CARDS dataset. Correlations between the target SNP (the SNP with the lowest P value, depicted in purple) and nearby SNPs within a 500 kb region. The r2 values were based on the HapMap CEU population.
TABLE 3.
Effect of adjustment for serum Lp(a) levels in CARDS
| CHR | POS (cM) | SNP | Modeled Allele | Minor Allele (Frequency) | Before Lp(a) Adjustment | After Lp(a) Adjustment | |||||
| β | SE | P | β | SE | P | Genea | |||||
| 6 | 195.419 | rs10455872 | A | G (0.07) | −0.35 | 0.08 | 1.12E-05 | −0.09 | 0.08 | 2.96E-01 | LPA |
| 12 | 55.598 | rs1627770 | G | T (0.2) | 0.18 | 0.05 | 5.08E-04 | 0.19 | 0.05 | 3.21E-04 | LOC390301-ALG10 |
| 12 | 55.598 | rs863626 | C | T (0.2) | 0.18 | 0.05 | 3.83E-04 | 0.19 | 0.05 | 2.48E-04 | LOC390301-ALG10 |
| 12 | 55.598 | rs11053045 | A | T (0.2) | 0.18 | 0.05 | 3.73E-04 | 0.2 | 0.05 | 2.40E-04 | LOC390301-ALG10 |
| 12 | 55.598 | rs1619785 | A | A (0.2) | −0.18 | 0.05 | 3.60E-04 | −0.2 | 0.05 | 2.28E-04 | LOC390301-ALG10 |
| 12 | 55.599 | rs10844779 | A | A (0.2) | −0.18 | 0.05 | 4.22E-04 | −0.19 | 0.05 | 2.79E-04 | ALG10-LOC260338 |
| 12 | 55.599 | rs11053068 | C | C (0.2) | −0.18 | 0.05 | 4.27E-04 | −0.19 | 0.05 | 2.82E-04 | ALG10-LOC260338 |
| 12 | 55.599 | rs5004272 | A | G (0.21) | 0.18 | 0.05 | 5.82E-04 | 0.19 | 0.05 | 4.14E-04 | ALG10-LOC260338 |
| 12 | 55.599 | rs10844823 | C | C (0.21) | −0.18 | 0.05 | 5.88E-04 | −0.19 | 0.05 | 4.19E-04 | ALG10-LOC260338 |
| 16 | 27.656 | rs721843 | C | G (0.46) | 0.13 | 0.05 | 4.67E-01 | 0.13 | 0.04 | 3.69E-01 | LOC653737-GRIN2A |
| 19 | 80.713 | rs4803760 | C | T (0.2) | 0.24 | 0.06 | 3.43E-05 | 0.23 | 0.06 | 7.73E-05 | BCAM-PVRL2 |
| 19 | 80.766 | rs1985096 | A | A (0.16) | −0.33 | 0.07 | 8.31E-07 | −0.33 | 0.07 | 1.39E-06 | BCAM-PVRL2 |
| 19 | 80.877 | rs395908 | A | A (0.16) | −0.21 | 0.06 | 2.10E-04 | −0.21 | 0.06 | 3.65E-04 | PVRL2-BCAM- TOMM40 |
| 19 | 80.954 | rs6857 | C | T (0.14) | −0.32 | 0.07 | 1.85E-06 | −0.3 | 0.07 | 1.75E-05 | PVRL2-BCAM- TOMM40 |
| 19 | 81.023 | rs405509 | G | T (0.48) | −0.17 | 0.05 | 3.36E-04 | −0.14 | 0.05 | 4.31E-03 | APOE-TOMM40- APOE |
| 19 | 81.051 | rs445925 | A | A (0.11) | −0.44 | 0.08 | 1.13E-08 | −0.42 | 0.08 | 1.39E-07 | LOC100129500- APOE APOC1 |
| 19 | 81.081 | rs4420638 | A | G (0.16) | −0.44 | 0.08 | 1.65E-08 | −0.43 | 0.08 | 1.39E-07 | APOC1 APOC1 APOC4 |
Effect of adjustment for serum Lp(a) levels in CARDS for SNPs associated with an LDL-c response to statins with a meta-analysis of P <10−6.
CHR, chromosome; POS, position.
For SNPS that lie in the intergenic regions, the location of the nearby genes is shown.
To assess whether serum Lp(a) levels might alter efficacy of statin therapy on CVD itself, we examined whether there was any evidence of interaction (deviation from a multiplicative model of joint effects on a hazard scale) between high serum Lp(a) levels and atorvastatin on CVD end points in CARDS. The hazard ratio for CVD events associated with statin use was 0.60 (95% CI: 0.32–1.13) among those in the top quartile for serum Lp(a) (>22 mg/dl) compared with 0.66 (95% CI: 0.46–0.93) among those with serum Lp(a) below this level (likelihood ratio test for interaction P = 0.8).
Nor was there any evidence of interaction between rs10455872 genotype at LPA and atorvastatin for effects on CVD end points (P = 0.27 for the interaction of genotype at rs10455872 locus). Here the HR associated with atorvastatin in those homozygous for the A allele was 0.58 (95% CI: 0.41–0.83) and the HR in those with at least one G allele was 1.03 (95% CI: 0.38–2.78). However, the power to detect such an interaction was limited as there were only 16 events among the 294 trial participants with at least one copy of the G allele.
Replication of the LPA SNP
We tested the effect of LPA SNP (rs10455872) in 2,550 participants in the PROSPER trial randomized to 40 mg/day of pravastatin. In this study, “A” allele of rs10455872 was also associated with lower response to statins with a scaled β of –0.18 ± 0.04, P = 0.009. The combined P-value for the three studies was 1.2E-09 (β = −0.28 ± 0.04).
APOE
Several SNPs in the BCAM/PVRL2/APOE/APOC1/APOE gene region reached genome-wide significance for statin response (Fig. 5). The effect on LDL-c response to statin therapy associated with these SNPs at in this region was modest; in CARDS for example the % change in LDL-c with statin therapy at one month was approximately -51% in those with at least one “A” allele at rs445925 compared with -45% in common GG homozygotes and was approximately -37% in those with at least one “G” allele at rs4420638 compared with -47% in common “AA” homozygotes. These effects were independent of the effect of genotype at rs10455872 in LPA and of Lp(a) levels. In the CARDS dataset we confirmed that there was no significant effect of these SNPs on change in LDL-c post randomization in those in the placebo group (P = 0.47).
Fig. 5.
Regional association plot of APOE locus with statin response. Correlations between the target SNP (the SNP with the lowest P value, depicted in purple) and nearby SNPs within a 500 kb region. The r2 values were based on the HapMap CEU population.
We examined whether the effects in this region could be accounted for by the known ϵ2/ϵ3/ϵ4 protein polymorphism of apolipoprotein E, which corresponds to APOE SNP haplotypes T-T, T-C, and C-C, respectively, at rs429358 and rs7412. The presence of the “T” allele at rs7412 contrasts the ϵ2 protein variant with other protein variants, whereas presence of the “C” allele at rs429358 contrasts the ϵ4 protein variant with other protein variants. These two SNPS were not directly typed and could not be imputed as they are not in the HapMap II.
The “A” allele at rs445925, which we found to be associated with a higher statin response (β = −0.44) is in LD with the “T” allele at rs7412 with a reported r2 of 0.76; thus, it is a proxy for the ϵ2 protein variant (20). The “G” allele at rs4420638, which was associated with lower response to statin, is in LD with the “C” allele at rs429358 with reported r2 of 0.62 but with a low r2 of 0.01 for rs7412 (21); thus, it is a proxy for the ϵ4 protein variant.
These two proxy SNPs are in the HapMap and could be imputed in this analysis with percentage information content (i.e., imputation quality) of 77% and 56%, respectively. Thus we tested for residual effects of SNP haplotypes conditioning either on rs445925 (as a proxy for rs7412) or on rs4420638 (as a proxy for rs429358). When conditioned on rs4420638, the ϵ4 proxy, the additional percentage variance explained by residual haplotype effects is 0.7% (F statistic with 8 and 854, df = 3.21, P = 0.001). When conditioned on rs445925, the proxy for ϵ2, the additional percentage variance explained by residual haplotype effects is only 0.2% (F statistic with 8 and 854, df = 1.76, P = 0.08), suggesting that ϵ2 accounts for most of the variance in response at this locus.
ALG10
Beyond these associations of LDL-c response with APOE and LPA, no other genome-wide significant associations were found. The next most significant SNPS were those in the ALG10 gene region (Fig. 6) on chromosome 12 where several SNPs had P < 10−6. ALG10 codes for asparagine-linked glycosylation protein 10 homolog A. Of these SNPs, most map to intergenic regions either side of the ALG10 gene itself with one imputed SNP within ALG10 having a P-value for association with statin response of 6.79 × 10−6.
Fig. 6.
Regional association plot of ALG10 locus with statin response before Lp(a) adjustments. Correlations between the target SNP (the SNP with the lowest P value, depicted in purple) and nearby SNPs within a 500 kb region. The r2 values were based on the HapMap CEU population.
Effect of pretreatment LDL-C
To demonstrate that these findings are unlikely to be confounded by baseline LDL-c, Table 4 shows unadjusted, adjusted, and corrected estimates of the direct effect of genotype on posttreatment LDL at the strongest SNPs for the APOE region LPA and ALG10 in the CARDS dataset. At the APOE ϵ2 proxy SNP (rs445925), without adjusting for baseline LDL-c, the apparent LDL-c response to statins would be more than double that observed in our baseline-adjusted model (β = −1.01 vs. −0.44 per copy of “A” allele), emphasizing the effect of adjusting for baseline LDL-c. However, adjusting our effect size estimate further by modeling measurement noise at baseline reduced the apparent effect just slightly to β = −0.30, suggesting there is little residual effect of baseline due to measurement noise. At the APOE ϵ4 proxy SNP and at the LPA SNP, the estimated effect of baseline LDL-c adjustment is much less, and thus, the adjustment for measurement noise alters the association only slightly.
TABLE 4.
Effect of genotype on posttreatment LDL-c (CARDS only)
| SNP | β unadjusted for baseline LDL | β adjusted for observed baseline LDL but uncorrected for measurement noise | β adjusted for baseline LDL and corrected for measurement noise |
| rs445925 | −1.01 | −0.44 | −0.38 |
| rs4420638 | −0.54 | −0.44 | −0.42 |
| rs10455872 (LPA) | −0.49 | −0.35 | −0.32 |
| rs10844779 (ALG10) | −0.18 | −0.18 | −0.18 |
With and without correction for measurement noise in baseline LDL.
Other genes of interest
Previously reported variants associated with statin response in the PCSK9 (rs11591147), HMGCR (rs1047443, rs17671591, rs6453131), KIF-6 (rs20455), ABCB1 (1236/2677/3435 TTT haplotype), CLMN (rs80141914, associated with TC response to statin), and GCKR (rs1260326 associated with triglyceride level response to statin) genes were not significantly associated at with LDL-c response to statin in this study at an accepted genome-wide association threshold (P ≤ 10−8) or even at thresholds typically expected to declare replication (say, P ≤ 10−2). However, PCSK9 (rs11591147) and GCKR (rs1260326) were significant at a threshold of 0.05 (see supplementary Table I). We have refrained from comparing the directionality and magnitude of these effects in the present study because of the different phenotype characterization and transformations across the studies and, in some studies, lack of information about the modeled alleles.
DISCUSSION
In this genome-wide association study of LDL-c response to atorvastatin therapy, we report that those with genotypes in the LPA gene that lead to higher Lp(a) levels have an apparently lower LDL-c response to statin, and we replicate the previously reported association of a higher response to statin in those with the A allele at the APOE ϵ2 locus. The top three SNPs in the study, rs10455872 in LPA and the APOE ϵ2 and APOE ϵ4 variants, explained only 4% variance in the LDL-c response to statin treatment; however, it is possible that that larger studies might detect more SNPs with smaller effect sizes or that there are larger effects at rarer variants not captured by our imputed genotypes.
LPA
Lipoprotein(a) is a plasma lipoprotein consisting of a cholesterol-rich LDL particle with one molecule of apolipoprotein B100 and an additional protein, apolipoprotein(a), attached via a disulfide bond. Serum levels of Lp(a) have a highly skewed distribution; for example, in CARDS, the median serum Lp(a) was 8.9 mg/dl (IQR 4.5–21.3 mg/dl) and with values as high as 238 mg/dl. Approximately 30% of variance in Lp(a) levels has been reported as determined by the kringle IV type 2 (KIV-2) copy number variant in LPA, which is known to encode variability in the size of apo(a). Some variance in measured Lp(a) attributable to genes is also due to apo(a) size heterogeneity affecting the results of the immunochemical methods used to quantify Lp(a), as is the case with the assay we used (22). That is, genotype can induce some measurement error in Lp(a), although recent data from the Framingham study suggest the measurement error is likely to be of little practical importance (23). The Lp(a)-raising genotype associated with the kringle repeat and high Lp(a) levels themselves have also been reported to be associated with increased cardiovascular risk in several studies (24–28). As such, recent guidelines emphasize the importance of detecting high Lp(a) phenotype and possible intervention with niacin (26).
The rs10455872 SNP that we found associated with LDL-c response is in strong LD with the KIV-2 copy number variant in Lp(a) (29). Consistent with this, variation at rs10455872 accounted for 30% of variance in Lp(a) in the CARDS data. However, the explanation for the apparently lower LDL-c response in those with genotypes associated with high Lp(a) lies in understanding what LDL-c estimation actually captures. The standard Friedewald formula calculates LDL-c levels from TC, HDL-cholesterol, and plasma triglyceride and actually includes the cholesterol that resides in Lp(a). For most patients, this is of little importance as usually only about 5% of what is measured as LDL-cholesterol is estimated to reside in Lp(a). However, it is estimated that about 8% of apparent LDL-c resides in Lp(a) if Lp(a) levels are in the range 30–60 mg/dl and as much as 20% if Lp(a) is > 60 mg/dl (30). As we show definitively here in the CARDS trial, statin therapy did not lower Lp(a) levels. Thus, individuals who had an appreciable fraction of their total plasma cholesterol carried on Lp(a) particles had some cholesterol in statin-responsive LDL particles and some in statin-unresponsive Lp(a) particles. For such patients, true LDL-c response will be underestimated because apparent on-treatment LDL-c will comprise truly falling LDL-c but static Lp(a) levels. This phenomenon has previously been noted in the context of nephrotic syndrome (31) and has been emphasized by Scanu et al. (32). Our estimate that those with at least one copy of the Lp(a)-raising G allele at rs10455872 have about a 5 percentage points lower apparent statin response (45% in “GG” and “AG” genotype vs. 40% in “AA” genotype) and that this association disappears when adjusted for Lp(a) levels is consistent with these observations.
Although the effect of the G allele on statin response is modest, this allele only accounts for about 30% of variance in Lp(a) levels. The data highlight a more general clinical point that individuals with raised Lp(a) levels for any reason have a somewhat lower apparent response to statin therapy and, therefore, that an apparently lower LDL-c response to statin may be an indication for checking Lp(a) levels. However, we also show here that similar relative protection from CVD with atorvastatin therapy was found in those with and without elevated Lp(a). It is increasingly accepted that elevated Lp(a) increases CVD risk (24, 25). Therefore, it is important that, although Lp(a) levels themselves are not changed and statin effects on LDL-c appear erroneously low, statin therapy be continued in individuals with high Lp(a).
We confirmed the association of LPA SNP rs10455872, that is, the association of “A” allele with a lower response to statins in an independent cohort of 2,550 subjects randomized to 40 mg of pravastatin. This is the first report of a successful replication of genetic response to statin treatment beyond the APOE region in a genome-wide association study.
APOE
We replicated the previous finding that genotype at the APOE ϵ2 locus is associated with variation in statin response. Having at least one “A” allele at rs445925, which is in strong LD with the locus determining the ϵ2 protein variant, was associated with both higher baseline LDL-c and greater response to statin, whereas the proxy for ϵ4 protein variant was associated with lower LDL-c response to statin. The conditional haplotype analysis suggests most of the variation is attributable to the number of ϵ2 copies rather than to the number of ϵ4 copies, but this is not definitive given that there is uncertainty in the haplotypes and that the HapMap SNPs are imperfect proxies. As noted previously, individuals in whom a higher proportion of cholesterol is synthesized rather than taken up via diet, such as ϵ2 carriers, are more susceptible to inhibition of cholesterol synthesis (2), and in addition, there may be more remnant and IDL-like particles contributing to apparent LDL-c in those patients with an APO ϵ2 allele. Statins are very good at removing these larger LDL particles through LDL receptor upregulation.
ALG10
We found some suggestion of an association between statin response and variants in the ALG10 gene region, but this did not reach the usual genome-wide significance threshold of P < 10−8; therefore, it requires confirmation in other studies before considering it other than a spurious association. ALG10 codes for asparagine-linked glycosylation protein 10 homolog A, which adds the third glucose residue to the lipid-linked oligosaccharide precursor for N-linked glycosylation. Its relevance to statin response remains to be established, though of course, protein modification by N-linked glycosylation is relevant to diverse aspects of human biology, including functional modification of many enzymes (33).
Previous studies
Two GWAS of LDL-c response to statin therapy have previously been reported (2, 3). In the Treating to New Targets (TNT) trial dataset with 1,984 treated individuals typed with a genome-wide panel, there were no loci at which P-values for association were less than 10−6, but in a superset of 5,745 individuals studied for candidate gene associations only, three SNPs in APOE and one SNP in PCSK9 reached genome-wide significance (2). In a meta-analysis of three trials that included 3,932 treated subjects (3) a SNP in the CLMN gene was significant at P < 10−7 for association with TC response, and there was a weak association with SNP in APOE. In the same study, polymorphism in the GCKR gene was shown to be associated with statin-induced change in triglycerides. Candidate gene analyses have shown that a common LDLR 3-UTR haplotype is associated with attenuated lipid-lowering response to simvastatin treatment (34). In the same study, HMGCR gene polymorphisms were also associated with reduced plasma LDL-c and with reduced LDL-c response to simvastatin. The association of HMGCR gene with statin response was also reported in a population-based cohort of patients with diabetes (35). These effects were more evident in African-Americans than in European-Americans. In a separate study, carried out in acute coronary syndrome patients, carriers of a polymorphism in kinesin-like protein 6 (KIF-6) have been reported to have greater benefit from pravastatin versus placebo with respect to CVD outcome but not with respect to lipid or C-reactive peptide response (36). Additionally, association of the ABCB1 gene with statin response has been reported (4, 37). Apart from the APOE association, none of these other associations were replicated here.
Finally, we note that the effects identified in this study are of modest size: the importance of further studying the genetics of response to statin therapy may be not in predicting who will benefit from statins but in identifying other therapeutic targets.
Acknowledgments
The authors thank the other investigators, the staff, and the participants of the CARDS and ASCOT studies for their important contributions. Generated data for the study: Helen M. Colhoun, D. John Betteridge, John H. Fuller, Shona Livingstone, Valentine Charlton-Menys, Andrew Neil, Neil Poulter, Peter Sever, Denis Shields, Alice Stanton, Alu Chatterjee, Craig Hyde, Roberto A. Calle, David A. DeMicco, Graham A. Hitman, Mark Caulfield, Stella Trompet, Iris Postmus, Ian Ford, and J. Wouter Jukema. Analyzed data: Paul M. McKeigue, Toby Johnson, Harshal Deshmukh, Helen M. Colhoun, Graham Hitman, Mark Caulfield, Stella Trompet, and J. Wouter Jukema. Drafted initial manuscript: Helen M. Colhoun, Paul M. McKeigue, Harshal Deshmukh, Toby Johnson, Graham Hitman, and Mark Caulfield. Edited manuscript: All authors. See Refs. 1, 6, and 8 for a list of CARDS, ASCOT, and PROSPER investigators, respectively.
Footnotes
Abbreviations:
- ASCOT
- Anglo-Scandinavian Cardiac Outcomes Trial
- CARDS
- Collaborative Atorvastatin Diabetes Study
- CVD
- cardiovascular disease
- GWAS
- genome-wide association study
- HWE
- Hardy-Weinberg equilibrium
- IQR
- interquartile range
- LD
- linkage disequilibrium
- Lp(a)
- lipoprotein(a)
- PROSPER
- PROspective Study of Pravastatin in the Elderly at Risk
- SNP
- single nucleotide polymorphism
- TC
- total cholesterol
The CARDS trial was cofunded by Pfizer Ltd., Diabetes UK, and National Health Service R&D. Genotyping was funded by Pfizer Ltd. The Anglo-Scandinavian Cardiac Outcomes Trial and establishment of the associated genetic repository were funded by Pfizer Inc. Genotyping was funded by Barts, the London School of Medicine and Dentistry, and by the Centre Nationale de Genotypage Paris. The PROSPER study was supported by an investigator-initiated grant obtained from Bristol-Myers Squibb. Prof. Dr. J. W. Jukema is an established clinical investigator of the Netherlands Heart Foundation (Grant 2001-D-032). Support for genotyping was provided by the Seventh Framework Program of the European Commission (Grant 223004) and by the Netherlands Genomics Initiative (Netherlands Consortium for Healthy Aging Grant 050-060-810). For the PROSPER/PHASE program, the research leading to results received funding from the European Commission's Seventh Framework Program (FP7/2007–2013) under Grant HEALTH-F2-2009-223004 PHASE. CARDS registration number: NCT00327418. ASCOT registration number: EUDRACT2008-007494-20.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three tables.
REFERENCES
- 1.Colhoun H. M., Betteridge D. J., Durrington P. N., Hitman G. A., Neil H. A., Livingstone S. J., Thomason M. J., Mackness M. I., Charlton-Menys V., Fuller J. H. 2004. Primary prevention of cardiovascular disease with atorvastatin in type 2 diabetes in the Collaborative Atorvastatin Diabetes Study (CARDS): multicentre randomised placebo-controlled trial. Lancet. 364: 685–696 [DOI] [PubMed] [Google Scholar]
- 2.Thompson J. F., Hyde C. L., Wood L. S., Paciga S. A., Hinds D. A., Cox D. R., Hovingh G. K., Kastelein J. J. 2009. Comprehensive whole-genome and candidate gene analysis for response to statin therapy in the Treating to New Targets (TNT) cohort. Circ. Cardiovasc. Genet. 2: 173–181 [DOI] [PubMed] [Google Scholar]
- 3.Barber M. J., Mangravite L. M., Hyde C. L., Chasman D. I., Smith J. D., McCarty C. A., Li X., Wilke R. A., Rieder M. J., Williams P. T., et al. 2010. Genome-wide association of lipid-lowering response to statins in combined study populations. PLoS ONE. 5: e9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson J. F., Man M., Johnson K. J., Wood L. S., Lira M. E., Lloyd D. B., Banerjee P., Milos P. M., Myrand S. P., Paulauskis J., et al. 2005. An association study of 43 SNPs in 16 candidate genes with atorvastatin response. Pharmacogenomics J. 5: 352–358 [DOI] [PubMed] [Google Scholar]
- 5.Donnelly L. A., Palmer C. N., Whitley A. L., Lang C. C., Doney A. S., Morris A. D., Donnan P. T. 2008. Apolipoprotein E genotypes are associated with lipid-lowering responses to statin treatment in diabetes: a Go-DARTS study. Pharmacogenet. Genomics. 18: 279–287 [DOI] [PubMed] [Google Scholar]
- 6.Sever P. S., Dahlof B., Poulter N. R., Wedel H., Beevers G., Caulfield M., Collins R., Kjeldsen S. E., Kristinsson A., McInnes G. T. 2003. Prevention of coronary and stroke events with atorvastatin in hypertensive patients who have average or lower-than-average cholesterol concentrations, in the Anglo-Scandinavian Cardiac Outcomes Trial–Lipid Lowering Arm (ASCOT-LLA): a multicentre randomised controlled trial. Lancet. 361: 1149–1158 [DOI] [PubMed] [Google Scholar]
- 7.Friedewald W. T., Levy R. I., Fredrickson D. S. 1972. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 18: 499–502 [PubMed] [Google Scholar]
- 8.Shepherd J., Blauw G. J., Murphy M. B., Cobbe S. M., Bollen E. L., Buckley B. M., Ford I., Jukema J. W., Hyland M., Gaw A., et al. 1999. The design of a prospective study of Pravastatin in the Elderly at Risk (PROSPER). PROSPER Study Group. PROspective Study of Pravastatin in the Elderly at Risk. Am. J. Cardiol. 84: 1192–1197 [DOI] [PubMed] [Google Scholar]
- 9.Lindgren C. M., Heid I. M., Randall J. C., Lamina C., Steinthorsdottir V., Qi L., Speliotes E. K., Thorleifsson G., Willer C. J., Herrera B. M., et al. 2009. Genome-wide association scan meta-analysis identifies three loci influencing adiposity and fat distribution. PLoS Genet. 5: e1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willer C. J., Speliotes E. K., Loos R. J., Li S., Lindgren C. M., Heid I. M., Berndt S. I., Elliott A. L., Jackson A. U., Lamina C., et al. 2009. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 41: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patterson N., Price A. L., Reich D. 2006. Population structure and eigenanalysis. PLoS Genet. 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trompet S., de Craen A. J., Postmus I., Ford I., Sattar N., Caslake M., Stott D. J., Buckley B. M., Sacks F., Devlin J. J., et al. 2011. Replication of LDL GWAs hits in PROSPER/PHASE as validation for future (pharmaco)genetic analyses. BMC Med. Genet. 12: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchini J., Howie B., Myers S., McVean G., Donnelly P. 2007. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39: 906–913 [DOI] [PubMed] [Google Scholar]
- 14.Li Y., Willer C., Sanna S., Abecasis G. 2009. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 10: 387–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909 [DOI] [PubMed] [Google Scholar]
- 16.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J., et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aulchenko Y. S., Struchalin M. V., van Duijn C. M. 2010. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 11: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutalik Z., Johnson T., Bochud M., Mooser V., Vollenweider P., Waeber G., Waterworth D., Beckmann J. S., Bergmann S. 2011. Methods for testing association between uncertain genotypes and quantitative traits. Biostatistics. 12: 1–17 [DOI] [PubMed] [Google Scholar]
- 19.Zabaneh D., Kumari M., Sandhu M., Wareham N., Wainwright N., Papamarkou T., Hopewell J., Clarke R., Li K., Palmen J., et al. 2011. Meta analysis of candidate gene variants outside the LPA locus with Lp(a) plasma levels in 14,500 participants of six white European cohorts. Atherosclerosis. 217: 447–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith E. N., Chen W., Kahonen M., Kettunen J., Lehtimaki T., Peltonen L., Raitakari O. T., Salem R. M., Schork N. J., Shaw M., et al. 2010. Longitudinal genome-wide association of cardiovascular disease risk factors in the Bogalusa heart study. PLoS Genet. 6: e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ken-Dror G., Talmud P. J., Humphries S. E., Drenos F. 2010. APOE/C1/C4/C2 gene cluster genotypes, haplotypes and lipid levels in prospective coronary heart disease risk among UK healthy men. Mol. Med. 16: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcovina S. M., Koschinsky M. L., Albers J. J., Skarlatos S. 2003. Report of the National Heart, Lung, and Blood Institute Workshop on Lipoprotein(a) and Cardiovascular Disease: recent advances and future directions. Clin. Chem. 49: 1785–1796 [DOI] [PubMed] [Google Scholar]
- 23.Lamon-Fava S., Marcovina S. M., Albers J. J., Kennedy H., Deluca C., White C. C., Cupples L. A., McNamara J. R., Seman L. J., Bongard V., et al. 2011. Lipoprotein(a) levels, apo(a) isoform size, and coronary heart disease risk in the Framingham Offspring Study. J. Lipid Res. 52: 1181–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erqou S., Kaptoge S., Perry P. L., Di Angelantonio E., Thompson A., White I. R., Marcovina S. M., Collins R., Thompson S. G., Danesh J. 2009. Lipoprotein(a) concentration and the risk of coronary heart disease, stroke, and nonvascular mortality. JAMA. 302: 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genser B., Dias K. C., Siekmeier R., Stojakovic T., Grammer T., Maerz W. 2011. Lipoprotein (a) and risk of cardiovascular disease–a systematic review and meta analysis of prospective studies. Clin. Lab. 57: 143–156 [PubMed] [Google Scholar]
- 26.Nordestgaard B. G., Chapman M. J., Ray K., Boren J., Andreotti F., Watts G. F., Ginsberg H., Amarenco P., Catapano A., Descamps O. S., et al. 2010. Lipoprotein(a) as a cardiovascular risk factor: current status. Eur. Heart J. 31: 2844–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraft H. G., Lingenhel A., Kochl S., Hoppichler F., Kronenberg F., Abe A., Muhlberger V., Schonitzer D., Utermann G. 1996. Apolipoprotein(a) kringle IV repeat number predicts risk for coronary heart disease. Arterioscler. Thromb. Vasc. Biol. 16: 713–719 [DOI] [PubMed] [Google Scholar]
- 28.Clarke R., Peden J. F., Hopewell J. C., Kyriakou T., Goel A., Heath S. C., Parish S., Barlera S., Franzosi M. G., Rust S., Bennett D., et al. 2009. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N. Engl. J. Med. 361: 2518–2528 [DOI] [PubMed] [Google Scholar]
- 29.Lanktree M. B., Anand S. S., Yusuf S., Hegele R. A. 2010. Comprehensive analysis of genomic variation in the LPA locus and its relationship to plasma lipoprotein(a) in South Asians, Chinese, and European Caucasians. Circ. Cardiovasc. Genet. 3: 39–46 [DOI] [PubMed] [Google Scholar]
- 30.Li K. M., Wilcken D. E., Dudman N. P. 1994. Effect of serum lipoprotein(a) on estimation of low-density lipoprotein cholesterol by the Friedewald formula. Clin. Chem. 40: 571–573 [PubMed] [Google Scholar]
- 31.Kronenberg F., Lingenhel A., Lhotta K., Rantner B., Kronenberg M. F., Konig P., Thiery J., Koch M., von Eckardstein A., Dieplinger H. 2004. Lipoprotein(a)- and low-density lipoprotein-derived cholesterol in nephrotic syndrome: impact on lipid-lowering therapy? Kidney Int. 66: 348–354 [DOI] [PubMed] [Google Scholar]
- 32.Scanu A. M., Hinman J. 2002. Issues concerning the monitoring of statin therapy in hypercholesterolemic subjects with high plasma lipoprotein(a) levels. Lipids. 37: 439–444 [DOI] [PubMed] [Google Scholar]
- 33.Ben-Zeev O., Stahnke G., Liu G., Davis R. C., Doolittle M. H. 1994. Lipoprotein lipase and hepatic lipase: the role of asparagine-linked glycosylation in the expression of a functional enzyme. J. Lipid Res. 35: 1511–1523 [PubMed] [Google Scholar]
- 34.Mangravite L. M., Medina M. W., Cui J., Pressman S., Smith J. D., Rieder M. J., Guo X., Nickerson D. A., Rotter J. I., Krauss R. M. 2010. Combined influence of LDLR and HMGCR sequence variation on lipid-lowering response to simvastatin. Arterioscler. Thromb. Vasc. Biol. 30: 1485–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donnelly L. A., Doney A. S., Dannfald J., Whitley A. L., Lang C. C., Morris A. D., Donnan P. T., Palmer C. N. 2008. A paucimorphic variant in the HMG-CoA reductase gene is associated with lipid-lowering response to statin treatment in diabetes: a GoDARTS study. Pharmacogenet. Genomics. 18: 1021–1026 [DOI] [PubMed] [Google Scholar]
- 36.Iakoubova O. A., Tong C. H., Rowland C. M., Kirchgessner T. G., Young B. A., Arellano A. R., Shiffman D., Sabatine M. S., Campos H., Packard C. J., et al. 2008. Association of the Trp719Arg polymorphism in kinesin-like protein 6 with myocardial infarction and coronary heart disease in 2 prospective trials: the CARE and WOSCOPS trials. J. Am. Coll. Cardiol. 51: 435–443 [DOI] [PubMed] [Google Scholar]
- 37.Becker M. L., Visser L. E., van Schaik R. H., Hofman A., Uitterlinden A. G., Stricker B. H. 2009. Common genetic variation in the ABCB1 gene is associated with the cholesterol-lowering effect of simvastatin in males. Pharmacogenomics. 10: 1743–1751 [DOI] [PubMed] [Google Scholar]