Abstract
Cytosolic lipid droplets (LDs) are storage organelles for neutral lipids derived from endogenous metabolism. Acyl-CoA synthetase family proteins are essential enzymes in this biosynthetic pathway, contributing activated fatty acids. Fluorescence microscopy showed that ACSL3 is localized to the endoplasmic reticulum (ER) and LDs, with the distribution dependent on the cell type and the supply of fatty acids. The N-terminus of ACSL3 was necessary and sufficient for targeting reporter proteins correctly, as demonstrated by subcellular fractionation and confocal microscopy. The N-terminal region of ACSL3 was also found to be functionally required for the enzyme activity. Selective permeabilization and in silico analysis suggest that ACSL3 assumes a hairpin membrane topology, with the N-terminal hydrophobic amino acids forming an amphipathic helix restricted to the cytosolic leaflet of the ER membrane. ACSL3 was effectively translocated from the ER to nascent LDs when neutral lipid synthesis was stimulated by the external addition of fatty acids. Cellular fatty acid uptake was increased by overexpression and reduced by RNA interference of ACSL3. In conclusion, the structural organization of ACSL3 allows the fast and efficient movement from the ER to emerging LDs. ACSL3 not only esterifies fatty acids with CoA but is also involved in the cellular uptake of fatty acids, presumably indirectly by metabolic trapping. The unique localization of the acyl-CoA synthetase ACSL3 on LDs suggests a function in the local synthesis of lipids.
Keywords: endoplasmic reticulum, lipid metabolism, fluorescence microscopy, membrane anchor, subcellular targeting
Lipid-containing membranes are essential for living cells, and the storage of lipids for times of need presumably developed very early during evolution. Cytosolic lipid droplets (LDs) are the main reservoir of lipids and are common to many if not all eukaryotic cells (1). LDs have gained much recent interest because of their regulatory role in lipid homeostasis and their implication in metabolic diseases such as obesity and type 2 diabetes (2–4). They have a unique structure composed of a hydrophobic core surrounded by a phospholipid monolayer containing a specific protein composition. Perilipin family proteins and lipid metabolizing enzymes are the most abundant proteins (5, 6), and proteomic studies have identified many additional constituents (7). Little is known how proteins are specifically targeted to lipid droplets (8, 9). The biogenesis of LDs likely involves the endoplasmic reticulum, but the mechanism has not been solved and is debated intensely (2, 3, 10).
Triglycerides are the main species of neutral lipids stored within the LDs of most cell types. The predominant biosynthetic pathway requires three activated fatty acids for each triglyceride molecule. The fatty acids are taken up from the extracellular medium or are derived from endogenous metabolism by fatty acid synthase. In fact, addition of fatty acids is a very efficient way to induce the formation of LDs (4). However, fatty acids are chemically quite inert and need to be activated by esterification with CoA (11). This activation is catalyzed by the family of acyl-CoA synthetases (12); physiologically highly relevant are the long chain (ACSL1, -3, -4, -5, -6) and very long chain (ACSVL1, -2, -3, -4, -5, and -6) fatty acyl-CoA synthetase subfamilies (13). Apart from their obvious enzymatic role, additional functions have been suggested for ACS(V)L family proteins: metabolic channeling of fatty acids toward specific metabolic fates [e.g., phospholipid synthesis vs. β-oxidation (13)] and fatty acid uptake driven by vectorial acylation at the plasma membrane (14). Furthermore, the ACSVL proteins have also been put forward as fatty acid transporters (FATPs) at the cell surface (15).
ACSL3 was cloned initially from rat brain (16) but is ubiquitously expressed (17) and has a substrate preference for polyunsaturated fatty acids (16, 18). ACSL3 was found to be an abundant protein on lipid droplets prepared from Huh7 human hepatoma cells (19). Interestingly, the distribution of ACSL3 between LDs and heavier cell membrane fractions correlated with the amount of intracellular lipid droplets (20).
Here, we present evidence that the N-terminus of ACSL3 is necessary and sufficient for the localization to lipid droplets. Our data suggest that N-terminal anchoring of ACSL3 to the cytosolic leaflet of the endoplasmic reticulum (ER) by an amphipathic helix allows efficient translocation to emerging lipid droplets. Depletion of ACSL3 by RNAi caused a significant reduction in fatty acid uptake, suggesting that the activation of fatty acids serves not only to provide reactive molecules for lipid metabolism but is also a driving force for the transport of fatty acids into the cell.
MATERIALS AND METHODS
Antibodies
Antiserum against human ACSL3 was raised in rabbits using recombinant ACSL3 comprising amino acids 504–720. For affinity purification, recombinant protein coupled to a solid support was used. Other antibodies used were: rabbit anti-GFP (21), rabbit anti-rat protein disulfide isomerase A6/CaBP1 (22), mouse anti-HA (sc-7392; Santa Cruz, CA), mouse anti-β-actin (clone AC-15; Sigma, St. Louis, MO), guinea pig anti-TiP47 (gp32; PROGEN Biotechnik, Heidelberg, Germany;). Donkey anti-rabbit coupled to Cy3 and anti-mouse/rabbit-HRP were from Jackson ImmunoResearch, West Grove, PA.
Plasmids
Human adipose differentiation-related protein (ADRP/perilipin 2) followed by mRFP (23) was used as a lipid droplet marker protein. RFP-ER contains mRFP fused to human Sec61β (24), which is a widely used ER marker protein. CaBP1/PDI A6 is a rat liver protein disulfide isomerase (22) and is a soluble lumenal ER protein. ACSL3HA is full-length human ACSL3, tagged at the C terminus with the hemagglutinin epitope. The full-length cDNA for human ACSL3 (clone MGC 48741) was obtained from the RZPD (Deutsches Ressourcenzentrum fuer Genomforschung; now imaGenes, Germany), and the cDNA was verified. The sequence was identical to the reference sequence (gb: NM_004457.3). PCR with primers sA3kpn (5′-ACGTGGTACCACCATGAATAACCACGTGTCTTC-3′) and aA3 (5′-A CGTCTCGAGTCAAGCGTAATCTGGAACATCGTATGGGTATTTTCTTCCATACATTCGCTCAATGTCC-3′) yielded a cDNA coding for C-terminally HA (hemagglutinin) epitope tagged ACSL3. This was digested with KpnI and XhoI and ligated into pcDNA3 (Invitrogen). GFP-ACSL3HA codes for GFP, and HA tagged full-length human ACSL3. The cDNA of ACSL3HA was moved with KpnI and ApaI into pEGFP-C1 (Clontech, Mountain View, CA). GFP-ΔNt-ACSL3HA lacks the cDNA part, which codes for amino acids 1–68 of human ACSL3. GFP-ACSL3HA was digested with BglII, the released fragment was removed by gel electrophoresis, and the remaining vector was religated. ΔNt-ACSL3HA was also cloned but did express only very poorly. A3Nt-GFP contains the amino acids 1–135 of human ACSL3 followed by GFP. The cDNA of Caco-2 cells was used as a template for PCR with primers s-H3-ACSL3 (5′-GAATTCAAGCTTACCATGAATAACCACGTGTCTTC-3′) and a-ACSL3GFP (5′-ACGTACCGGTGGATCCGCAAGCCAATTATACTGTCC-3′). The PCR product was digested with HindIII and AgeI and ligated into pEGFP-N1 (Clontech). A3Nt-RFP is comprised of amino acids 1–135 of human ACSL3 followed by mRFP. For this, the plasmid A3Nt-GFP was digested with HindIII and BamHI and ligated into Murr1-RFP.pcDNA3 (25); this replaced the Murr1 cDNA and yielded a cDNA containing monomeric RFP in frame (26). Transcription of the ACSL3RNAi plasmid generates a small hairpin RNA. The target sequence of ACSL3 comprised nucleotides 1540–1558 of the human cDNA. Oligos sA3J1 (5′-GATCTCCGGTGGATACTTTAATACTGTTCAAGAGACAGTATTAAAGTATCCACCTTTTTTGGAAC-3′) and aA3J1 (5′-TCGAGTTCCAAAAAAGGTGGATACTTTAATACTGTCTCTTGAACAGTATTAAAGTATCCACCGGA-3′) were annealed and ligated into pSuper digested with BglII and XhoI (27). Subcloning was into the retroviral plasmid pRVH1-puro (28). The cDNA of RFP-FATP4 is an N-terminal mRFP followed by full-length mouse FATP4. Plasmid FATP4.pcDNA3 (29) was digested with ApaI and HindIII and ligated into RFP-ER containing N-terminal mRFP, thereby replacing the cDNA coding for Sec61β. The dominant-negative cav1DGI-GFP has a deletion of amino acids 1–81 of canine caveolin-1. This construct was modeled after cav3DGV and has an intact scaffolding domain (30). PCR of canine caveolin1-GFP (31) with primers s-DGI (5′-ACGTAGATCTCGAGACCATGGATGGCATCTGGAAGGCC-3′) and a-CV-GFP (5′-CCATGGTGGCGACCGG-3′) was followed by digestion with XhoI and AgeI and reinsertion into the caveolin-GFP plasmid. All cDNAs derived from PCR products were fully sequenced.
Cell culture, transient and stable expression
All cell lines were cultivated in media supplemented with 10% FCS, glutamate and penicillin/streptomycin (all reagents from Invitrogen, Carlsbad, CA): COS-7 (ATCC CRL-1651; DMEM 4.5 g/l glucose), Vero (CCL-81; DMEM 1.0 g/l glucose), Ptk2 (CCL-56; MEM, 0.1 mM nonessential amino acids), A431 (CRL-1555; DMEM 4.5 g/l glucose), and HepG2 (HB-8065; RPMI 1640).
For transient expression, cells grown to 80% confluency in a 6-well plate (10 cm2/well) were incubated for 4 h with the transfection mix consisting of 2.0 µg plasmid, 10 µl FUGENE HD (Roche, Mannheim, Germany), and 100 µl Opti-MEM (Invitrogen) in 2 ml of standard medium. After 4 h, cells were washed and incubated for a further 20 h in 2 ml of standard medium without antibiotics before analysis. Control cells for biochemical experiments were transfected with pcDNA3.
Formation of lipid droplets was induced by incubation with 180 µM oleate bound to fatty acid free BSA (Sigma A-6003) with a molar ratio of 4:1 (Figs. 2B, 3, 4Figs. 2B, 3, 4) or with 600 µM oleate (6:1; Figs. 1, 5B, 6A, BFigs. 1, 5B, 6A, B). Protein synthesis was inhibited by 200 µM cycloheximide, which was added 30 min before the oleate treatment and kept during the oleate incubation.
Fig. 2.
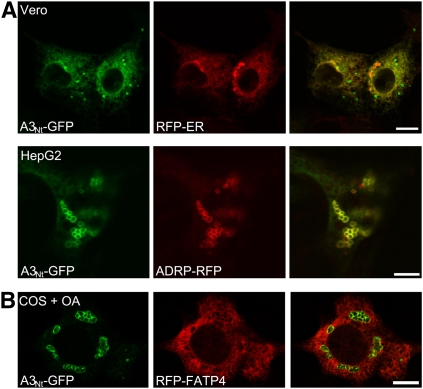
Localization of ACSL3 in different cell types. A: The distribution between ER and LDs is cell type specific and dependent on fatty acid supply. Transient expression of the N-terminus of ACSL3 fused to GFP (A3Nt-GFP) in Vero cells showed predominantly an ER distribution (compare with RFP-ER) with only some scattered LDs (top row; bar, 10 µm). In human hepatoma cells (HepG2), A3Nt-GFP was localized prominently on clusters of LDs, which costained with ADRP/perilipin 2 (ADRP-RFP; bottom row; bar, 5 µm). B: Compartmentalization of acyl-CoA synthetases. Comparison between A3Nt-GFP localized on lipid droplets and RFP-FATP4 present in the ER. COS-7 cells were treated overnight with oleic acid (+ OA). Confocal section; bar, 10 µm.
Fig. 3.
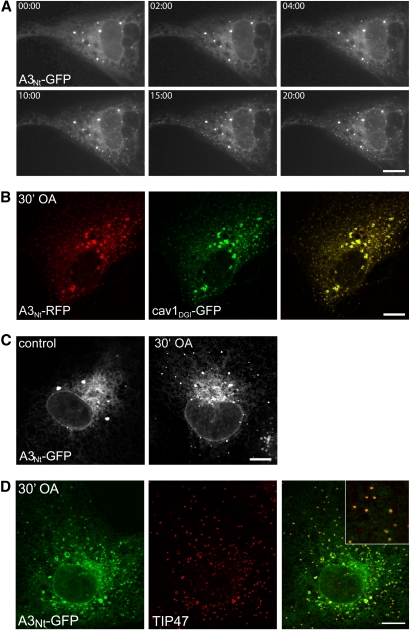
Localization of A3Nt-GFP to emerging lipid droplets. A: Time series of the localization of A3Nt-GFP after the addition of oleic acid. A3Nt-GFP emerges in tiny puncta, which are discernible 2 min after the addition of 180 µM oleic acid. The same Ptk2 cell was imaged over 20 min. Bar, 10 µm. B: Colocalization of A3Nt-RFP with caveolin-1DGI-GFP. Transiently expressing Ptk2 cells were treated for 30 min with 180 µM oleic acid. Caveolin and ACSL3 overlap strikingly in both the tiny new and the mature lipid droplets. Confocal sections; bar, 10 µm. C: Induction of new lipid droplets by the addition of fatty acid in COS-7 cells. Localization of A3Nt-GFP under standard culture conditions (control; left) and after the addition of 180 µM oleic acid (OA) for 30 min. Bar, 10 µm. D: A3Nt-GFP and TIP47/perilipin-3 costain on emerging lipid droplets. COS-7 cells expressing A3Nt-GFP were treated for 30 min with 180 µM OA, fixed, and treated with antibodies against endogenous TIP47 (red). Confocal sections; bar, 10 µm.
Fig. 4.
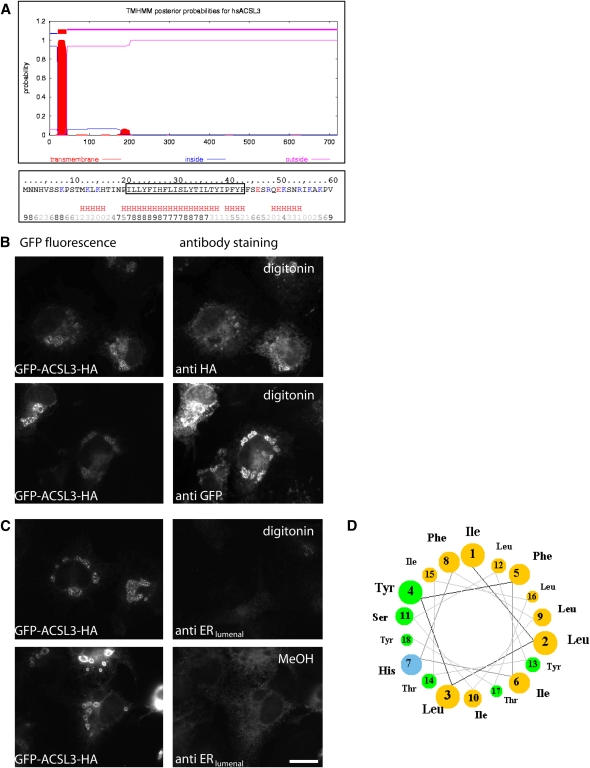
Membrane topology of ACSL3. A: In silico analysis of the N-terminal region of ACSL3. Software prediction suggested that the hydrophobic domain at the N-terminus of ACSL3 comprises 24 amino acids (I21 to F43) and may form an integral transmembrane domain (http://www.cbs.dtu.dk/services/TMHMM/). The hydrophobic region (boxed) has a high likelihood of being α-helical (red H). The bottom row numbers indicate the reliability of the prediction (0 low, 9 high; http://www.predictprotein.org/submit.php). B, C: Selective permeabilization. COS cells were transiently cotransfected with plasmids encoding GFP-ACSL3HA and an ER marker protein located inside the ER (anti ERlumenal corresponding to rat protein disulfide isomerase A6/CaBP1). After overnight incubation with oleic acid, cells were fixed with PFA and permeabilized with 10 µM digitonin for 10 min at RT (B and C, upper panel) or with methanol (MeOH) (C, lower panel) at −20°C for 2 min. Antibodies against the N-terminally located GFP (anti-GFP) and the C-terminal HA epitope (anti-HA) stained cells permeabilized with digitonin (B). Digitonin did not allow access of the antibodies against the lumenal ER protein (C, upper panel), but methanol treatment confirmed the presence of CaBP1 (C, lower panel). Bar, 20 µm. D: Helical wheel projection of the N-terminus of ACSL3. Amino acids I21 (Ile 1) to Y38 (Tyr 18) show an exceptional sidedness between nonpolar (light brown) and polar/basic (green/blue) amino acids. All six polar amino acids (Ser, Thr, Tyr) contain a hydroxyl group in their side chain.
Fig. 1.
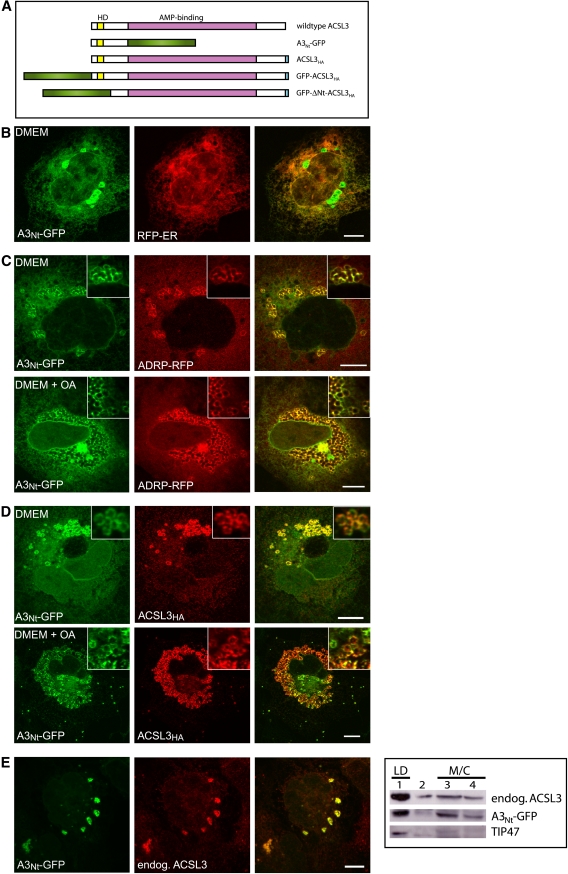
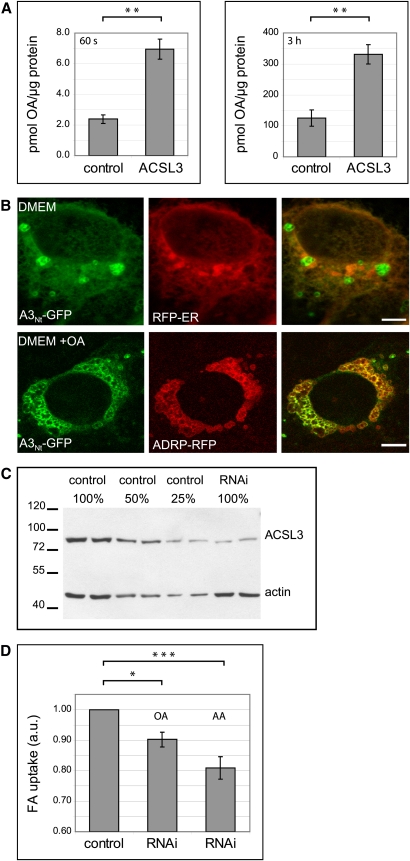
The N-terminus of ACSL3 confers localization to the ER and to lipid droplets. A: Domain organization of ACSL3 and overview of the constructs used. Wild-type human ACSL3 contains 720 amino acids, corresponding to 80.4 kDa (http://www.uniprot.org/uniprot/O95573). The AMP-binding domain spanning S136 to A612 (as defined by Pfam at http://smart.embl-heidelberg.de/) is colored in purple, and the hydrophobic domain (HD) is indicated in yellow. A3Nt-GFP contains the N-terminus of ACSL3 (M1-L135) fused to GFP (green). HA (light blue) is the hemagglutinin epitope tag. GFP-ΔNt-ACSL3HA lacks the N-terminus of ACSL3 (M1-Y68). B: Partial localization of A3Nt-GFP to the endoplasmic reticulum (ER). Transient coexpression of A3Nt-GFP and the red fluorescent membrane marker protein for the ER (RFP-ER) in COS-7 cells. Cells were fixed 24 h post transfection and analyzed by confocal laser scanning microscopy. A single representative section is shown. The colocalization is especially evident at the nuclear envelope, which is continuous with the ER. Bar, 10 µm. C: Partial localization of A3Nt-GFP to lipid droplets (LD). Transient expression of A3Nt-GFP and the LD marker protein (ADRP/perilipin 2) in COS-7 cells. The colocalization between A3Nt-GFP and ADRP-RFP is even more prominent if the culture medium (DMEM) is supplemented with oleic acid overnight (lower panel; DMEM + OA). Confocal sections; bar, 10 µm. D: Comparison of A3Nt-GFP to full length wild-type ACSL3. COS-7 cells were fixed and permeabilized 24 h after cotransfection with A3Nt-GFP and epitope-tagged human ACSL3. Indirect immunofluorescence staining of ACSL3 was achieved by sequential incubation with mouse anti-HA and donkey anti-mouse coupled to Cy3 (red). The overlap between both proteins is apparent under standard conditions (DMEM) as well as after oleic acid supplementation (DMEM + OA). Confocal sections; bar, 10 µm. E: Comparison of A3Nt-GFP to endogenous ACSL3. Left: Indirect immunofluorescence microscopy. The affinity purified antibody against ACSL3 (red) stains the same structures as A3Nt-GFP expressed transiently in COS-7 cells. Confocal sections; bar, 10 µm. Right: Subcellular fractionation. Postnuclear supernatants of homogenized COS-7 cells were applied to sucrose density gradients. Membrane (M) and cytosolic (C) proteins remain at the bottom of the gradient (fractions 3 and 4), but lipid droplet associated proteins float into the top fraction (fraction 1). Endogenous ACSL3 was detected by the antibody raised against the C-terminal half of recombinant human ACSL3. A3Nt-GFP was recognized by the GFP antiserum, and TIP47 served as a lipid droplet marker protein.
Fig. 5.
The expression level of ACSL3 influences fatty acid uptake. A: Expression of ACSL3 enhances fatty acid uptake. Transiently transfected COS cells were incubated for 60 s (left) or 3 h (right) with [3H]oleate bound to BSA. Results are from three independent experiments, with triplicate measurements each; ** P < 0.01 (Student's t-test). Error bars are SEM. B: Localization of A3Nt-GFP in A431 cells. Transiently expressing A431 cells were cultured under standard conditions (DMEM) or with oleic acid overnight (DMEM + OA). Confocal sections; bar, 10 µm. C: Stable knockdown of ACSL3 expression in A431 cells by RNAi. Total lysates from control cells were compared with RNAi cells. Actin served as a loading control. D: Depletion of ACSL3 by RNAi diminishes fatty acid uptake. Cells were incubated for 3 h with 200 µM [3H]labeled oleic (OA) or arachidonic (AA) acid bound to 100 µM BSA, and fatty acid uptake was quantified by scintillation counting. Results are from six independent experiments using cells with different passage numbers. * P < 0.05; *** P < 0.001. Error bars are SEM.
Fig. 6.
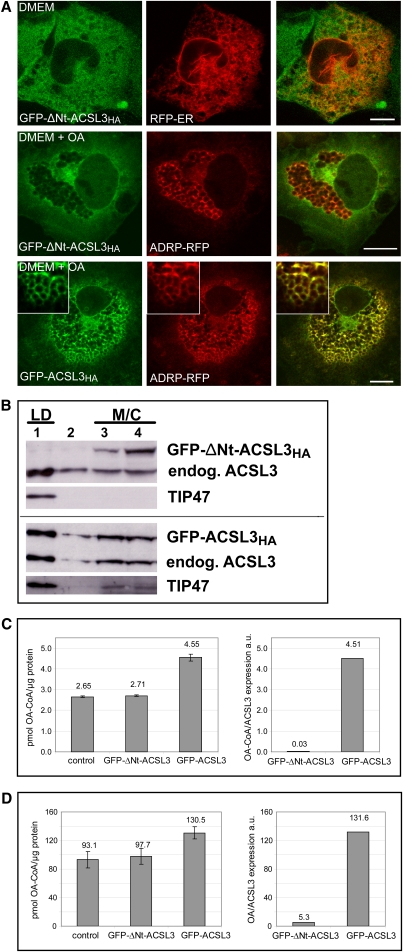
The N-terminus is required for the localization and function of ACSL3. A: Immunofluorescence localization of ACSL3 lacking the N-terminus. GFP-ΔNt-ACSL3HA localizes diffusely to the cytoplasm/cytosol (top and middle panel), reminiscent of but different from the endoplasmic reticulum (RFP-ER, top panel). It is excluded from lipid droplets marked by ADRP-RFP (middle panel). The corresponding control fusion protein GFP-ACSL3HA colocalizes with ADRP/perilipin-2 on lipid droplets like A3Nt-GFP or wild-type ACSL3. Confocal sections; bar, 10 µm. B: The N-terminus of ACSL3 is required for cofractionation with lipid droplets. Cell homogenates were fractionated on sucrose density gradients. GFP-ΔNt-ACSL3HA is present in the fractions 3 and 4, which contain membrane (M) and cytosolic (C) proteins. No significant amount is observed in the LD fraction (#1). GFP-ACSL3HA containing the hydrophobic N-terminus of ACSL3 cofractionates with lipid droplets marked by TIP47/ perilipin-3. C: Cells expressing GFP-ΔNt-ACSL3HA show no increase in enzyme activity. The oleoyl-CoA synthetase activities of cell lysates were determined in vitro. Left: Absolute values for the specific enzyme activity related to total protein amount. Error bars are SD of triplicates of a representative experiment. Right: Enzyme activities related to the amount of overexpressed GFP fusion protein (determined by Western blotting). D: GFP-ΔNt-ACSL3HA does not enhance fatty acid uptake. Left: Oleate uptake related to total protein. The amount of overexpressed ACSL3 is much lower than in Fig. 5A (data not shown; see Materials and Methods for details). Error bars are SD of triplicates of a representative experiment. Right: Oleate uptake calibrated to the amount of overexpressed ACSL3 determined by blotting.
Stable depletion of ACSL3 by RNAi was achieved by retroviral integration of the ACSL3RNAi plasmid into A431 cells. The generation of replication-deficient retrovirus pseudotyped with VSV-G from phoenix-gp cells, transduction, and antibiotic selection (2 days, 4.0 µg/ml puromycin for A431 cells) were as described earlier (28). Control cells were transduced with the plasmid pRVH1-puro containing no shRNA sequence.
GFP-ACSL3HA was expressed more efficiently than GFP-ΔNt-ACSL3HA. To enable a better comparison, we used 0.4 µg GFP-ACSL3HA plasmid together with 1.6 µg pcDNA3 and 2.0 µg of GFP-ΔNt-ACSL3HA (Fig. 6).
Fluorescence microscopy
Cells expressing fluorescent proteins were fixed for 20 min with 4% PFA, washed, and embedded directly in mowiol (4-88; Calbiochem, San Diego, CA) containing 1 µg/ml Hoechst 33342 for staining of nuclei.
For staining of endogenous ACSL3, fixed cells were permeabilized with 0.1% saponin and blocked with 0.5% gelatin and 0.5% BSA. Indirect immunofluorescence was performed as described (29).
Selective permeabilization of transiently transfected COS cells was achieved by using 10 µM digitonin for 10 min at room temperature. To stain the lumenal ER antigen CaBP1/PDI A6, fixed cells were treated for 2 min with methanol prechilled at −20°C.
For following the same cell over time, 12 mm cover slips were fitted into a metal slide holder. After addition of control or oleic acid containing medium, cells were imaged using a 60× oil immersion objective mounted on an Olympus BX41 equipped with a F-view II CCD camera controlled by the cell^D software (Olympus, Hamburg, Germany). This microscope was also used for Figs. 3A, 4B, and 4CFigs. 3A, 4B, and 4C. Figs. 1, 2B–E, 4B–D, 5B, and 6A were obtained with the confocal microscope Leica TCS SP2 as described (32).
Floatation of lipid droplets
Transiently transfected COS cells (A3Nt-GFP, GFP-ACSL3HA, and GFP-ΔNt-ACSL3HA) from one 55 cm2 dish and nontransfected COS cells from two 152 cm2 dishes were incubated for 39 h with 600 µM oleic acid bound to fatty acid free BSA in a molar ratio of 6:1. Cells were then collected with PBS, mixed, and centrifuged for 5 min at 500 g. Separation of lipid droplets from other cellular components was essentially done as described (23). Briefly, the cell pellet was resuspended in sucrose buffer A (50 mM Tris-HCl [pH 7.4], 20 mM sucrose) and homogenized by passing 15 times through a syringe fitted to a 22 gauge needle followed by 20 strokes in a glass Dounce homogenizer. Homogenization was confirmed by inspection under a microscope. The homogenate was centrifuged at 1,000 g for 10 min at 4°C, yielding a postnuclear supernatant, which was mixed with an equal amount of high-sucrose buffer (50 mM Tris-HCl [pH 7.4], 2 M sucrose) and overlaid with buffer A. The gradients were centrifuged in a SW40 rotor at 100,000 g for 3 h at 4°C. Four fractions with equal volume were collected: a top fraction containing lipid droplets (#1), an intermediate fraction (#2), and two bottom fractions (#3/4) containing cytosol and membranes. Proteins were concentrated by methanol-chloroform precipitation, separated by 8% SDS-PAGE, transferred by Western blotting, and detected by ECL.
Fatty acid uptake and TLC
COS-7 or A431 cells were preincubated for 1 h in DMEM supplemented with glutamine but without FCS. For short-term uptake, cells from one 10 cm2 well were collected with PBS, centrifuged at 200 g, and resuspended into 400 µl PBS. To three 100 µl aliquots, 100 µl of labeling mix (340 µM [3H]oleate 8 Ci/ mol, 340 µM BSA in PBS) were added for 60 s. Uptake was terminated by adding an excess of ice cold 0.5% w/v BSA in PBS. Cells were washed four times and solubilized in 1 M NaOH. Aliquots were measured in a β-counter (LS 6500; Beckman Coulter, Brea, CA) for oleate content and with Bradford solution (Biorad, Hercules, CA) for protein concentration. Long-term uptake was analyzed with adherent cells grown in 12-well plates. Eight hundred microliters of labeling mix (200 µM [3H]oleate/[3H]arachidonate 0.5 Ci/ mol, 100 µM BSA in DMEM) replaced the standard medium for 3 h. Preincubation, washing, and analysis was as for short-term uptake. Three aliquots of each cell population were analyzed for every experiment. Statistical analysis by Student's t-test was based on at least three independent experiments (SPSS 16.0; SPSS Inc., Chicago, IL). Errors bars correspond to SEM in Fig. 5 and to SD in Fig. 6. Radioactive fatty acids were from PerkinElmer (Waltham, MA).
Lipid extraction and thin layer chromatography (TLC) were performed as described (29), except that the running solvent was changed to chloroform:methanol:acetic acid:water 75:43:3:1 (33). The TLC was exposed to phosphorimaging plates, which were scanned by the BAS-1500 system (Fuji, Tokyo, Japan). Quantification was done with ImageJ.
Acyl-CoA synthetase activity
Oleoyl-CoA synthetase activity was determined from cell lysates as described (29). Briefly, cells were lysed for 30 min on ice with 1% Triton X-100, 130 mM KCl, and 25 mM Tris-HCl (pH 7.4). Lysates containing 5–10 µg of total protein were incubated for 10 min at 30°C in 100 mM Tris (pH 7.4), 5 mM MgCl2, 200 µM dithiothreitol, 10 mM ATP, 0.2% Triton X-100, 20 µM [14C]oleate (specific activity 10 Ci/mol), and 200 µM CoA. The incubation was terminated by the addition of Dole's solution (isopropanol-n-heptane-H2SO4, 40:10:1), and unreacted oleate was extracted four times with n-heptane. The remaining oleoyl-CoA in the aqueous phase was quantified by scintillation counting.
Quantitative real-time PCR
Measurement of mRNA expression was by calibrator-normalized efficiency corrected relative quantification using the LightCycler system (Roche, Mannheim, Germany) as described (25). The reference gene was human β-actin (primer, 5′–3′ orientation: AGGATGCAGAAGGAGATCACT; GGGTGTAACGCAACTAAGTCATAG). Calibration plasmids FATP1, FATP2, and FATP4 were provided by Paul A. Watkins, Kennedy Krieger Institute, Baltimore, MD and FATP3 by Johannes Berger, Medical University Vienna, Austria. For human ACSL1 the PCR product obtained with the RT-PCR primers was cloned into the pGEM-T vector (Promega).
Primers
The following primers were used for qPCR (in 5′–3′ orientation): ACSL1: CTTCTGGTACGCCACGAGAC; GTCGCTGTCAAGTAGTGCG; ACSL3: GATGTTGGGTCAGAAACCAAAGA; ATGGCTGGACCTCCTAGAGTG; ACSL4: CTGGCCGACCTAAGGGAGT; ACATGAGCCAAAGGCAAGTAG; FATP1: GGACCCCAACGCGATATAC; GCCTCGTCTTCTGGATCTTG; FATP2: TGGTGTCGCCAGAACTACAAG; GAAAGAGTCAATCCCATCTGTGT; FATP3: ACGCTTTGGACCTTATGCAG; CTCATTTGCCATCCGAACTT; and FATP4: CGGTTCTGGGACGATTGTAT; AACCTGGTGCTGGTTTTCTG.
Software
Helical wheel projection was at http://cti.itc.virginia.edu/~cmg/Demo/scriptFrame.html. For the prediction of transmembrane helices, the TMHMM Server was used (http://www.cbs.dtu.dk/services/TMHMM/). The secondary structure of the N-terminus was analyzed by the PROF software (http://www.predictprotein.org/submit.php) (34).
Images were arranged with Adobe Photoshop and labeled with Adobe Illustrator (Adobe Systems Inc., Mountain View, CA).
Densitometric analysis of TLC and Western blots was done with ImageJ 1.37v software (http://rbs.info.nih.gov/iJ).
RESULTS
The N-Terminus of ACSL3 contains sorting information for lipid droplets and the endoplasmic reticulum
We observed before that the N-terminal domains of the mammalian acyl-CoA synthetases ACSL1 and FATP4/ACSVL4 were sufficient to target reporter proteins to specific subcellular compartments and that their localization was identical to the full-length wild-type proteins (29). Analysis of human ACSL3 by the Pfam protein family database (35) showed a large AMP-binding domain flanked by shorter N-terminal and C-terminal regions (Fig. 1A). We designed a cDNA coding for a fusion protein containing the N-terminal amino acids of ACSL3 followed by GFP and expressed this plasmid in mammalian COS-7 cells (A3Nt-GFP) (Fig. 1BndashE). Immunofluorescence microscopy showed a fine reticular staining in the cytoplasm, which colocalized with a red fluorescent marker protein for the endoplasmic reticulum (ER) (Fig. 1B). Several circular objects (typically 2 µm in diameter) did not correspond to the ER but overlapped with the lipid droplet (LD) marker adipose differentiation-related protein (ADRP/perilipin 2) (Fig. 1C), which is consistent with the localization of endogenous ACSL3 to LDs in human hepatoma cells (19, 20). The localization of A3Nt-GFP to LDs was especially striking after overnight treatment of cells with oleic acid, which stimulated the formation of large LD clusters in the cytoplasm (Fig. 1C, bottom). A3Nt-GFP showed the same staining pattern as epitope–tagged, full-length ACSL3 (Fig. 1D) and endogenous ACSL3 (Fig. 1E). Floatation of LDs on sucrose density gradients confirmed the presence of A3Nt-GFP on LDs and demonstrated the cofractionation with endogenous ACSL3 (Fig. 1D). LD fractions contained TIP47/perilipin-3, a long established marker protein (36). This suggests that the essential targeting information for localization to LDs and the ER is contained within the N-terminus of ACSL3.
The dual localization of ACSL3 was further investigated in two different cell lines. Kidney-derived epithelial Vero cells showed predominantly a cytoplasmic reticular ER staining of A3Nt-GFP, with only a few scattered LDs (Fig. 2A, top panel). In HepG2 hepatoma cells, A3Nt-GFP marked predominantly clusters of LDs colocalizing with ADRP/perilipin 2 (Fig. 2A, bottom panel). This indicates that the distribution of ACSL3 is cell type specific, in addition to the dependence on fatty acid supply.
The localization of A3Nt-GFP to LDs was unique among mammalian acyl-CoA synthetases. We also analyzed nine other GFP fusion proteins containing the N-termini of the remaining ACSL and ACSVL/FATP family proteins, but none of these was found on LDs (37). A striking example of the differential localization of acyl-CoA synthetases is given by RFP-FATP4 located at the ER and A3Nt-GFP on LDs after fatty acid supplementation (Fig. 2B). This hints at compartmentalized metabolism of acyl-CoA synthetases even though it is commonly assumed that they are all oriented toward the cytosolic side of cellular membranes (38).
Emerging lipid droplets
Overnight incubation with fatty acids leads to the appearance of numerous clustered A3Nt-GFP labeled LDs with an apparent size range from 1.5 to 2.0 µm (Figs. 1C, D and ). To determine how the formation of these lipid droplets was initiated, we used Ptk2 cells, which feature a large and flat cell body, allowing the easy discrimination of subcellular structures by light microscopy. Already 2 min after the addition of oleic acid, A3Nt-GFP appeared in numerous tiny punctate entities that increased in fluorescence intensity over time (Fig. 3A). A dominant-negative caveolin mutant had been suggested as an expressable marker to follow LD biogenesis (4, 39). We compared cav1DGI-GFP with a red fluorescent A3Nt variant and found an exceptionally high degree of colocalization after the addition of fatty acid to the medium (Fig. 3B). To further verify that the tiny puncta are indeed emerging LDs, we demonstrated that A3Nt-GFP showed the same behavior in COS-7 cells (Fig. 3C). These cells also allowed the labeling of endogenous TIP47/perilipin-3 by indirect immunofluorescence, confirming that A3Nt-GFP is present on early lipid droplets (Fig. 3D). The localization shift of A3Nt-GFP from the ER to newly emerging LDs was due to the translocation of already existing molecules because inhibition of protein synthesis with cycloheximide did not change the immunofluorescence pattern (data not shown).
Membrane topology of ACSL3
The endoplasmic reticulum has an aqueous lumen segregated from the cytosol by a classical membrane bilayer, but lipid droplets feature a hydrophobic core surrounded by a phospholipid monolayer. These fundamental differences led us to the question of which topological organization would allow ACSL3 to be tightly associated with both membrane systems.
As assumed in general for acyl-CoA synthetases, the enzyme domain of ACSL3 is expected to face the cytosol. In silico predictions suggested that amino acids Ile21-Phe43 could form an integral transmembrane domain (Fig. 4A), which would put the first 20 amino acids into the lumen of the ER. We tested this by selective permeabilization using an ACSL3 variant containing two antibody epitopes, one at each end of the protein (GFP-ACSL3HA). The detergent digitonin permeabilizes the plasma membrane efficiently, allowing access of antibodies to cytoplasmically oriented epitopes. The ER membrane, however, is not sufficiently permeabilized, which we verified by antibodies against a luminal ER protein (Fig. 4C). HA antibodies stained GFP-ACSL3HA as expected. GFP antibodies also gave a distinct pattern with digitonin, regardless whether GFP-ACSL3HA was localized to the ER or to LDs (Fig. 4B). This suggests that ACSL3 has both the N- and the C-terminus oriented toward the cytosol. The hydrophobic domain at the N-terminus likely forms an α-helical secondary structure (Fig. 4A), and the corresponding projection showed a striking segregation between hydrophobic and polar side chains (Fig. 4D). This amphipathic helix would allow equal anchoring in the cytoplasmic leaflet of the ER and the phospholipid monolayer of lipid droplets.
Overexpression and knockdown of ACSL3 correlate with fatty acid uptake
Overexpression of ACSL3 increases the cellular acyl-CoA synthetase activity (18, 40) (see Fig. 6C). Building on the general concept that metabolism contributes to fatty acid uptake (41) and our previous observations regarding ACSVL4/FATP4 and ACSL1 (29), we analyzed the connection between ACSL3 and the cellular incorporation of fatty acids. Overexpression of ACSL3 in COS-7 cells resulted in an increased uptake of oleic acid when measured after 60 s or 3 h (Fig. 5A). Because ACSL3 is localized intracellularly to the ER and LDs, this effect is indirect and is best explained by assuming that fatty acids are metabolically trapped as CoA derivatives.
COS-7 cells allow high transfection rates but are not of human origin, which complicates DNA sequence-specific experimental approaches like RNAi and qRT-PCR. ACSL3 is the second most abundant protein on LDs prepared from human A431 cells (42). Therefore, we chose this cell model to analyze the functional consequences of a reduced amount of ACSL3. We confirmed that A3Nt-GFP localized to LDs and the ER in A431 cells (Fig. 5B), similar to what we observed for COS-7 cells. Stable depletion of ACSL3 by RNA interference was achieved by retroviral integration of an shRNA plasmid. Quantification of transcription by qRT-PCR gave a knockdown efficiency of 81%, and the expression of the ACSL3 protein was diminished correspondingly (Fig. 5C). The uptake of arachidonic acid was reduced by 19%; the reduction for oleate was 10% (Fig. 5D). The stronger reduction for arachidonic acid coincides with the substrate preferences reported for ACSL3 (16, 18). The expression of ACS enzymes is redundant: The A431RNAi cells do not only contain the remaining ACSL3 (relative expression level by qRT-PCR: 1.90) but also ACSL1 (14.19), ACSL4 (6.58), and several FATP acyl-CoA synthetases (ACSVL4/FATP4 0.88, FATP3 0.61, and FATP1 0.26). Therefore, even the efficient depletion of ACSL3 leaves a considerable amount of enzyme activity with the A431RNAi cells.
Many observations suggest that mammalian acyl-CoA synthetases influence lipid metabolism beyond the esterification of fatty acids with CoA (13). We therefore analyzed lipid extracts of A431RNAi cells by TLC after labeling with radioactive fatty acids. We observed a relative decrease of phosphatidylethanolamine (−11%**) and a relative increase of phosphatidylcholine (PC) (+9.3%*) when oleic acid was used for labeling. With arachidonic acid, these changes were less pronounced and did not reach statistical significance. However, phosphatidylserine showed a relative decrease (−14%*).
Functional requirement for the N-terminus of ACSL3
The N-terminus of ACSL3 is sufficient for the targeting of GFP to lipid droplets and the ER (Figs. 1, 2), but it was not clear if ACSL3 would contain additional sorting information. Therefore, we constructed GFP-ΔNt-ACSL3HA lacking the first 68 amino acids, including the hydrophobic domain. The corresponding protein localized to the cytoplasm but was excluded from LDs (Fig. 6A). Subcellular fractionation confirmed that GFP-ΔNt-ACSL3HA was not associated with LDs, in contrast to GFP-ACSL3HA containing the full length ACSL3 (Fig. 6B). Cells expressing GFP-ΔNt-ACSL3HA did not show increased acyl-CoA synthetase activity (Fig. 6C), suggesting that the proper membrane anchoring by the N-terminus is functionally required. In line with this, there was no effect on the cellular uptake of fatty acids when GFP-ΔNt-ACSL3HA was overexpressed in COS-7 cells (Fig. 6D).
DISCUSSION
Lipid droplets have become an intensely studied intracellular organelle in recent years because of their high relevance for lipid homeostasis and associated metabolic diseases. Here, we have characterized ACSL3, an enzyme of the acyl-CoA synthetase family, which is localized to lipid droplets.
Targeting of ACSL3 to LDs
We show that the N-terminal region of ACSL3 is sufficient to confer localization of fluorescent reporter proteins to emerging and mature lipid droplets. In addition, ACSL3 is localized to ER membranes, which is especially apparent when cells contain only few LDs. Some LD-associated proteins feature more than one targeting region (e.g., perilipin A [43]). We therefore analyzed the localization of a mutant ACSL3 lacking the N-terminus, demonstrating that this region is essential for sorting to LDs. We found that the enzyme activity of ACSL3 is also dependent on the N-terminal region. This suggests that the membrane association and proper orientation of this enzyme at LD and ER membranes is functionally required.
Proteomics of purified lipid droplets has greatly contributed to our understanding of this organelle (7), but some reservations remain (9, 44). In addition to ACSL3, ACSL1 and ACSL4 have been identified repeatedly in these studies. However, we found ACSL1 on mitochondria by immunofluorescence microscopy (29), and this localization did not change after induction of LDs (B. Rudolph, R. Großmann, and J. Füllekrug, unpublished observations). ACSL4 cloned from HepG2 cells also did not colocalize with LDs (R. Großmann and J. Füllekrug, unpublished observations). We are therefore confident that ACSL3 is the only long-chain fatty acyl-CoA synthetase significantly associated with lipid droplets.
Lipid droplet biogenesis
A3Nt-GFP rapidly shifts from the ER to emerging lipid droplets when triglyceride synthesis is boosted by the external addition of fatty acids. This movement from one membrane system to another is different from the behavior of the constitutive LD proteins of the perilipin family (5, 6). For instance, ADRP/perilipin 2 and TIP47/perilipin 3 rapidly exchange between the cytosol and the LD surface (45, 46). How membrane proteins like ACSL3 translocate from the ER to LDs is not obvious because these organelles are organized in a strikingly dissimilar fashion: Whereas the ER features an aqueous lumen segregated from the cytoplasm by a phospholipid bilayer, cytosolic LDs have a hydrophobic core that is surrounded by a lipid monolayer only.
ACSL3 displays a hairpin topology with both the N- and the C-terminus facing the cytosol, and this orientation is shared by DGAT2 (47) and caveolins (48), which are also localized partially to LDs. However, for these proteins, the stretch of hydrophobic amino acids is much longer than for ACSL3 and may give rise to two α-helical transmembrane domains (8, 49). The hydrophobic region of ACSL3 could in principle allow the formation of an integral transmembrane domain, but our data are more consistent with an amphipathic helix embedded only in the cytosolic leaflet of the ER double membrane. Similar to ACSL3, CTP:phosphocholine cytidylyltransferase, viperin, and two viral proteins (hepatitis C core protein and NS5A) were recently suggested to be bound to LDs by an amphipathic helix (50–53). Assuming that the outer leaflet of the ER is at least initially continuous with the lipid monolayer of the emerging lipid droplet (10, 54), the proposed topology would allow the trafficking of ACSL3 molecules from the ER to nascent LDs by simple diffusion.
The association of proteins with early lipid droplets has not been widely investigated. Among the PAT/perilipin family proteins, TIP47/perilipin 3 and S3-12/perilipin 4 appear within the first 10 min after induction (45, 55, 56). ALDI/methyltransferase-like protein 7B was found after 5 min on tiny LDs, significantly earlier than ADRP/perilipin 2 (57). Although dominant-negative caveolin (cav3DGV) was found on early LDs already after 2 min (39), a similar localization has not been reported for wild-type caveolins.
Regarding how ACSL3 is translocated so efficiently to emerging lipid droplets, we think it unlikely that there is a proteinaceous ACSL3 receptor because this putative protein would need to be directed to nascent LDs even earlier than ACSL3. Because the surface lipid composition of LDs differs substantially from the ER membrane (58), it is conceivable that ACSL3 simply has a higher affinity for the lipid monolayer of LDs. Lipid-mediated sorting has been primarily associated with the plasma membrane and the trans-Golgi network (59), but it seems certain that other organelles would also take advantage of this organizing principle.
Role of ACSL3 in fatty acid uptake and metabolism
Depletion of ACSL3 reduced fatty acid uptake, whereas overexpression strongly increased incorporation of fatty acids. Although this experimental range of expression levels is unlikely to be realized under physiological circumstances, the results are nevertheless suggesting that the acyl-CoA synthetase activity of ACSL3 is a critical factor for the cellular uptake of fatty acids. In general, the extent of uptake is determined by extracellular supply as well as intracellular metabolic demand, with plasma membrane proteins facilitating and regulating the transport process (41, 60, 61). The role of ACSL3 and other acyl-CoA synthetases would not only be to provide activated fatty acids for lipid metabolism, but at the same time could prevent the loss of fatty acids to the outside. The intracellular metabolic trapping (or vectorial acylation [14]) by esterification with CoA might be the essential driving force for uptake of free fatty acids from the extracellular space. In line with this, exogenous expression of mammalian ACSL1, -4, and -6 rescued the fatty acid transport activity of yeast cells (62). Overexpression or depletion of acyl-CoA synthetases from the ACSVL (FATP) family also leads to corresponding changes in fatty acid uptake, but the interpretation of these findings is complicated by the assertion from some laboratories that these proteins are fatty acid transporters localized to the plasma membrane (15, 61). However, FATP4/ACSVL4 was shown by us to be resident in the endoplasmic reticulum, and this localization was sufficient to drive the uptake of fatty acids in an enzyme-dependent manner (29). The intracellular localization of ACSL3 is not contentious, and the effect on fatty acid transport therefore necessarily indirect.
Key regulators of lipid metabolism modify the expression of ACSL3: The LXR nuclear receptors increased ACSL3 levels in placental trophoblasts (which corresponded to fatty acid uptake [63]), and PPARδ regulated the expression of ACSL3 in hepatoma HepG2 cells (64). RNAi of ACSL3 in primary hepatocytes decreased the activity of PPARγ and other lipogenic transcription factors (65). Although the incorporation of external fatty acids was not investigated, this offers an alternative explanation for the reduced uptake observed by us: Less active PPARγ could decrease the overall capacity for cellular lipid metabolism and, consequently, fatty acid uptake. However, in the same study, other acyl-CoA synthetases (ACSL1, -4, -5 and FATP2, -4, -5) did not change the activity of PPARγ, suggesting that the impact of these enzymes on fatty acid uptake (summarized in References 13 and 66) is likely independent of transcriptional regulation.
Functional relevance of ACSL3 on lipid droplets
Activated fatty acids are rapidly metabolized, and excessive acyl-CoAs are used for the synthesis of triglycerides, which are stored in lipid droplets. It is tempting to speculate that fatty acids taken up with the help of ACSL3 are used preferentially for this storage pathway as opposed to membrane synthesis or β-oxidation. The double immunofluorescence localization of cytoplasmic ER-bound ACSVL4/FATP4 and LD-localized ACSL3 is highly reminiscent of compartmentalized metabolism (Fig. 2B). Although differing in kinetics and substrate affinities, both enzymes catalyze the same reaction, the esterification of long chain fatty acids (e.g., oleate) with CoA. The enzyme part of these membrane-bound proteins, however, is not lumenal but faces the cytosolic side of their respective subcellular organelles. It is not easy to see how the different localizations would have functional relevance because it is in common tacitly assumed that fatty acids and acyl-CoAs are equilibrated throughout the cytoplasm. The differences in subcellular localization combined with the apparently redundant expression of ACSL isoforms within the same cell has led to the hypothesis that channeling by acyl-Co synthetases may determine the metabolic fate of fatty acids (38, 67). Over the years, considerable evidence has accumulated (reviewed in References 13, 68, and 69), but the molecular mechanism remains unclear. We observed a phospholipid class switch from phosphatidylethanolamine to PC in A431RNAi cells stably depleted for ACSL3. A different study reported that transient knockdown of ACSL3 in Huh7 hepatoma cells led to a specific decrease of fatty acid incorporation into PC (70), suggesting that, even when the same acyl-CoA synthetase is depleted, there are still cell type-specific effects.
Local synthesis of triglycerides at the LD surface has been demonstrated (20, 23), although it is not clear if this process would be efficient enough to account for a sustained growth of lipid droplets independent from the endoplasmic reticulum. The acyl-CoA needed for the TG synthesis is very likely provided mostly by ACSL3 (20). Even more intriguing is the idea that the local synthesis of phosphatidylcholine could enable the rapid growth of the phospholipid monolayer to keep up with an expanding hydrophobic core when TG synthesis rate is high. The role of ACSL3 would be to provide activated fatty acids for the synthesis of PC by lyso-PC acyltransferases, which have recently been shown to be partially localized to LDs (42). When cellular ACSL activity is inhibited pharmacologically, LDs decrease in size and number (20). This suggests another possible function of LD-localized ACSL3 in counteracting basal lipolysis by promoting reesterification of fatty acids into triacylglycerol, preventing the efflux of fatty acids, which are constantly liberated even under unstimulated conditions.
In conclusion, the N-terminal region of ACSL3 displays a high affinity for LDs and enables the smooth and efficient movement from the ER to just-emerging lipid droplets. At the same time, ACSL3 activates fatty acids for lipid metabolism and contributes indirectly to cellular fatty acid uptake.
The machinery for the formation of LDs presumably has a long evolutionary history. It remains for future studies to determine if the biogenesis of lipid droplets is in essence a simple self-organizing process, relying on the segregation of triglyceride-enriched lipid membranes from the parental ER organelle. This primordial LD membrane would accumulate diffusing molecules with an inherent affinity (like ACSL3), and these will in turn sustain and regulate the maturation to lipid droplets. There is little doubt that the relevance of LDs for lipid homeostasis and overall metabolic regulation is still underestimated, with many surprises ahead.
Acknowledgments
We thank Simone Staffer for help with fatty acid uptake assays, Sabine Tuma for assistance with tissue culture, Julia Wunsch for cloning RFP-FATP4, and Wolfgang Stremmel for continuous support. Paul A. Watkins (Kennedy Krieger Institute, Baltimore, MD) and Johannes Berger (Medical University Vienna, Austria) kindly provided calibration plasmids for qRT-PCR.
Footnotes
Abbreviations:
- ADRP
- adipose differentiation-related protein
- ER
- endoplasmic reticulum
- FATP
- fatty acid transport protein
- HA
- hemagglutinin epitope
- LD
- lipid droplet
- PC
- phosphatidylcholine
- RFP
- red fluorescent protein
This work was supported by the German research foundation DFG grants FU 340/5-1 (J.F.) and EH 196/5-1 (R.E.).
REFERENCES
- 1.Murphy D. J. 2001. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40: 325–438 [DOI] [PubMed] [Google Scholar]
- 2.Walther T. C., Farese R. V., Jr 2009. The life of lipid droplets. Biochim Biophys Acta 1791: 459–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohsaki Y., Cheng J., Suzuki M., Shinohara Y., Fujita A., Fujimoto T. 2009. Biogenesis of cytoplasmic lipid droplets: from the lipid ester globule in the membrane to the visible structure. Biochim Biophys Acta Mol. Cell. Biol. 1791: 399–407 [DOI] [PubMed] [Google Scholar]
- 4.Martin S., Parton R. G. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7: 373–378 [DOI] [PubMed] [Google Scholar]
- 5.Brasaemle D. L. 2007. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 48: 2547–2559 [DOI] [PubMed] [Google Scholar]
- 6.Bickel P. E., Tansey J. T., Welte M. A. 2009. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta 1792: 419–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodges B. D. M., Wu C. C. 2010. Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J. Lipid Res. 51: 262–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiele C., Spandl J. 2008. Cell biology of lipid droplets. Curr. Opin. Cell Biol. 20: 378–385 [DOI] [PubMed] [Google Scholar]
- 9.Digel M., Ehehalt R., Füllekrug J. 2010. Lipid droplets lighting up: insights from live microscopy. FEBS Lett. 584: 2168–2175 [DOI] [PubMed] [Google Scholar]
- 10.Murphy D. J., Vance J. 1999. Mechanisms of lipid-body formation. Trends Biochem. Sci. 24: 109–115 [DOI] [PubMed] [Google Scholar]
- 11.Watkins P. A. 1997. Fatty acid activation. Prog. Lipid Res. 36: 55–83 [DOI] [PubMed] [Google Scholar]
- 12.Watkins P. A., Maiguel D., Jia Z., Pevsner J. 2007. Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. J. Lipid Res. 48: 2736–2750 [DOI] [PubMed] [Google Scholar]
- 13.Mashek D. G., Li L. O., Coleman R. A. 2007. Long-chain acyl-CoA synthetases and fatty acid channeling. Future Lipidol. 2: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Black P. N., DiRusso C. C. 2003. Transmembrane movement of exogenous long-chain fatty acids: proteins, enzymes, and vectorial esterification. Microbiol. Mol. Biol. Rev. 67: 454–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doege H., Stahl A. 2006. Protein-mediated fatty acid uptake: novel insights from in vivo models. Physiology (Bethesda). 21: 259–268 [DOI] [PubMed] [Google Scholar]
- 16.Fujino T., Kang M-J., Suzuki H., Iijima H., Yamamoto T. 1996. Molecular characterization and expression of rat Acyl-CoA synthetase 3. J. Biol. Chem. 271: 16748–16752 [DOI] [PubMed] [Google Scholar]
- 17.Mashek D. G., Li L. O., Coleman R. A. 2006. Rat long-chain acyl-CoA synthetase mRNA, protein, and activity vary in tissue distribution and in response to diet. J. Lipid Res. 47: 2004–2010 [DOI] [PubMed] [Google Scholar]
- 18.Van Horn C. G., Caviglia J. M., Li L. O., Wang S., Granger D. A., Coleman R. A. 2005. Characterization of recombinant long-chain rat acyl-CoA synthetase isoforms 3 and 6: identification of a novel variant of isoform 6. Biochemistry. 44: 1635–1642 [DOI] [PubMed] [Google Scholar]
- 19.Fujimoto Y., Itabe H., Sakai J., Makita M., Noda J., Mori M., Higashi Y., Kojima S., Takano T. 2004. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim. Biophys. Acta. 1644: 47–59 [DOI] [PubMed] [Google Scholar]
- 20.Fujimoto Y., Itabe H., Kinoshita T., Homma K. J., Onoduka J., Mori M., Yamaguchi S., Makita M., Higashi Y., Yamashita A., et al. 2007. Involvement of long chain acyl-CoA synthetase in local synthesis of neutral lipids in cytoplasmic lipid droplets in human hepatocyte HuH7. J. Lipid Res. 48: 1280–1292 [DOI] [PubMed] [Google Scholar]
- 21.Ehehalt R., Keller P., Haass C., Thiele C., Simons K. 2003. Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts. J. Cell Biol. 160: 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fullekrug J., Sonnichsen B., Wunsch U., Arseven K., Nguyen Van P., Soling H., Mieskes G. 1994. CaBP1, a calcium binding protein of the thioredoxin family, is a resident KDEL protein of the ER and not of the intermediate compartment. J. Cell Sci. 107: 2719–2727 [DOI] [PubMed] [Google Scholar]
- 23.Kuerschner L., Moessinger C., Thiele C. 2008. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic. 9: 338–352 [DOI] [PubMed] [Google Scholar]
- 24.Kuerschner L., Ejsing C. S., Ekroos K., Shevchenko A., Anderson K. I., Thiele C. 2005. Polyene-lipids: a new tool to image lipids. Nat. Methods. 2: 39–45 [DOI] [PubMed] [Google Scholar]
- 25.Weiss K. H., Lozoya J. C., Tuma S., Gotthardt D., Reichert J., Ehehalt R., Stremmel W., Fullekrug J. 2008. Copper-induced translocation of the Wilson disease protein ATP7B independent of Murr1/ COMMD1 and Rab7. Am. J. Pathol. 173: 1783–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 99: 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brummelkamp T. R., Bernards R., Agami R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science. 296: 550–553 [DOI] [PubMed] [Google Scholar]
- 28.Schuck S., Manninen A., Honsho M., Fullekrug J., Simons K. 2004. Generation of single and double knockdowns in polarized epithelial cells by retrovirus-mediated RNA interference. Proc. Natl. Acad. Sci. USA. 101: 4912–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milger K., Herrmann T., Becker C., Gotthardt D., Zickwolf J., Ehehalt R., Watkins P. A., Stremmel W., Fullekrug J. 2006. Cellular uptake of fatty acids driven by the ER-localized acyl-CoA synthetase FATP4. J. Cell Sci. 119: 4678–4688 [DOI] [PubMed] [Google Scholar]
- 30.Pol A., Luetterforst R., Lindsay M., Heino S., Ikonen E., Parton R. G. 2001. A caveolin dominant negative mutant associates with lipid bodies and induces intracellular cholesterol imbalance. J. Cell Biol. 152: 1057–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pelkmans L., Kartenbeck J., Helenius A. 2001. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat. Cell Biol. 3: 473–483 [DOI] [PubMed] [Google Scholar]
- 32.Fullekrug J., Suganuma T., Tang B. L., Hong W., Storrie B., Nilsson T. 1999. Localization and Recycling of gp27 (hp24gamma 3): complex formation with other p24 family members. Mol. Biol. Cell. 10: 1939–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allan D., Cockcroft S. 1982. A modified procedure for thin-layer chromatography of phospholipids. J. Lipid Res. 23: 1373–1374 [PubMed] [Google Scholar]
- 34.Rost B., Sander C. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232: 584–599 [DOI] [PubMed] [Google Scholar]
- 35.Finn R. D., Mistry J., Schuster-Bockler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., et al. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34: D247–D251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolins N. E., Rubin B., Brasaemle D. L. 2001. TIP47 associates with lipid droplets. J. Biol. Chem. 276: 5101–5108 [DOI] [PubMed] [Google Scholar]
- 37.Becker M., Wunsch J., Rudolph B., Stremmel W., Fuellekrug J. 2007. Subcellular targeting of acyl-CoA synthetases. FEBS J. 274: 111 [Google Scholar]
- 38.Coleman R. A., Lewin T. M., Van Horn C. G., Gonzalez-Baro M. R. 2002. Do long-chain acyl-CoA synthetases regulate fatty acid entry into synthetic versus degradative pathways? J. Nutr. 132: 2123–2126 [DOI] [PubMed] [Google Scholar]
- 39.Pol A., Martin S., Fernandez M. A., Ferguson C., Carozzi A., Luetterforst R., Enrich C., Parton R. G. 2004. Dynamic and regulated association of caveolin with lipid bodies: modulation of lipid body motility and function by a dominant negative mutant. Mol. Biol. Cell. 15: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujino T., Man-Jong K., Minekura H., Suzuki H., Yamamoto T. T. 1997. Alternative translation initiation generates acyl-CoA synthetase 3 isoforms with heterogeneous amino termini. J. Biochem. 122: 212–216 [DOI] [PubMed] [Google Scholar]
- 41.Mashek D. G., Coleman R. A. 2006. Cellular fatty acid uptake: the contribution of metabolism. Curr. Opin. Lipidol. 17: 274–278 [DOI] [PubMed] [Google Scholar]
- 42.Moessinger C., Kuerschner L., Spandl J., Shevchenko A., Thiele C. 2011. Human lysophosphatidylcholine acyltransferases 1 and 2 are located in lipid droplets where they catalyze the formation of phosphatidylcholine. J. Biol. Chem. 286: 21330–21339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian V., Garcia A., Sekowski A., Brasaemle D. L. 2004. Hydrophobic sequences target and anchor perilipin A to lipid droplets. J. Lipid Res. 45: 1983–1991 [DOI] [PubMed] [Google Scholar]
- 44.Welte M. A. 2007. Proteins under new management: lipid droplets deliver. Trends Cell Biol. 17: 363–369 [DOI] [PubMed] [Google Scholar]
- 45.Wolins N. E., Quaynor B. K., Skinner J. R., Schoenfish M. J., Tzekov A., Bickel P. E. 2005. S3–12, adipophilin, and TIP47 package lipid in adipocytes. J. Biol. Chem. 280: 19146–19155 [DOI] [PubMed] [Google Scholar]
- 46.Wang H., Hu L., Dalen K., Dorward H., Marcinkiewicz A., Russel D., Gong D., Londos C., Yamaguchi T., Holm C., et al. 2009. Activation of hormone-sensitive lipase requires two steps: protein phosphorylation and binding to the PAT-1 domain of lipid droplet coat proteins. J. Biol. Chem. 284: 32116–32125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stone S. J., Levin M. C., Farese R. V. 2006. Membrane topology and identification of key functional amino acid residues of murine Acyl-CoA:diacylglycerol acyltransferase-2. J. Biol. Chem. 281: 40273–40282 [DOI] [PubMed] [Google Scholar]
- 48.Dupree P., Parton R. G., Raposo G., Kurzchalia T. V., Simons K. 1993. Caveolae and sorting in the trans-Golgi network of epithelial cells. EMBO J. 12: 1597–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parton R. G., Hanzal-Bayer M., Hancock J. F. 2006. Biogenesis of caveolae: a structural model for caveolin-induced domain formation. J. Cell Sci. 119: 787–796 [DOI] [PubMed] [Google Scholar]
- 50.Hinson E. R., Cresswell P. 2012. The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic α-helix. Proc. Natl. Acad. Sci. USA (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shavinskaya A., Boulant S., Penin F., McLauchlan J., Bartenschlager R. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282: 37158–37169 [DOI] [PubMed] [Google Scholar]
- 52.Boulant S., Montserret R., Hope R. G., Ratinier M., Targett-Adams P., Lavergne J. P., Penin F., McLauchlan J. 2006. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J. Biol. Chem. 281: 22236–22247 [DOI] [PubMed] [Google Scholar]
- 53.Krahmer N., Guo Y., Wilfling F., Hilger M., Lingrell S., Heger K., Newman H. W., Schmidt-Supprian M., Vance D. E., Mann M., Farese R. V., Jr, Walther T. C. 2011. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 14: 504–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jacquier N., Choudhary V., Mari M., Toulmay A., Reggiori F., Schneiter R. 2011. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 124: 2424–2437 [DOI] [PubMed] [Google Scholar]
- 55.Bulankina A. V., Deggerich A., Wenzel D., Mutenda K., Wittmann J. G., Rudolph M. G., Burger K. N. J., Honing S. 2009. TIP47 functions in the biogenesis of lipid droplets. J. Cell Biol. 185: 641–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolins N. E., Skinner J. R., Schoenfish M. J., Tzekov A., Bensch K. G., Bickel P. E. 2003. Adipocyte protein S3–12 coats nascent lipid droplets. J. Biol. Chem. 278: 37713–37721 [DOI] [PubMed] [Google Scholar]
- 57.Turro S., Ingelmo-Torres M., Estanyol J. M., Tebar F., Fernandez M. A., Albor C. V., Gaus K., Grewal T., Enrich C., Pol A. 2006. Identification and characterization of associated with lipid droplet protein 1: A novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 7: 1254–1269 [DOI] [PubMed] [Google Scholar]
- 58.Bartz R., Li W. H., Venables B., Zehmer J. K., Roth M. R., Welti R., Anderson R. G., Liu P., Chapman K. D. 2007. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J. Lipid Res. 48: 837–847 [DOI] [PubMed] [Google Scholar]
- 59.Lingwood D., Simons K. 2010. Lipid rafts as a membrane-organizing principle. Science. 327: 46–50 [DOI] [PubMed] [Google Scholar]
- 60.Hamilton J. A. 2007. New insights into the roles of proteins and lipids in membrane transport of fatty acids. Prostaglandins Leukot. Essent. Fatty Acids. 77: 355–361 [DOI] [PubMed] [Google Scholar]
- 61.Glatz J. F. C., Luiken J. J. F. P., Bonen A. 2010. Membrane fatty acid transporters as regulators of lipid metabolism: implications for metabolic disease. Physiol. Rev. 90: 367–417 [DOI] [PubMed] [Google Scholar]
- 62.Tong F., Black P. N., Coleman R. A., Dirusso C. C. 2006. Fatty acid transport by vectorial acylation in mammals: roles played by different isoforms of rat long-chain acyl-CoA synthetases. Arch. Biochem. Biophys. 447: 46–52 [DOI] [PubMed] [Google Scholar]
- 63.Weedon-Fekjaer M. S., Dalen K. T., Solaas K., Staff A. C., Duttaroy A. K., Nebb H. I. 2010. Activation of LXR increases acyl-CoA synthetase activity through direct regulation of ACSL3 in human placental trophoblast cells. J. Lipid Res. 51: 1886–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao A., Li H., Zhou Y., Wu M., Liu J. 2010. Long chain Acyl-CoA synthetase-3 is a molecular target for peroxisome proliferator-activated receptor delta in HepG2 hepatoma cells. J. Biol. Chem. 285: 16664–16674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bu S. Y., Mashek M. T., Mashek D. G. 2009. Suppression of long chain acyl-CoA synthetase 3 (ACSL3) decreases hepatic de novo fatty acid synthesis through decreased transcriptional activity. J. Biol. Chem. 284: 30474–30483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ellis J. M., Frahm J. L., Li L. O., Coleman R. A. 2010. Acyl-coenzyme A synthetases in metabolic control. Curr. Opin. Lipidol. 21: 212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muoio D. M., Lewin T. M., Wiedmer P., Coleman R. A. 2000. Acyl-CoAs are functionally channeled in liver: potential role of acyl-CoA synthetase. Am. J. Physiol. Endocrinol. Metab. 279: E1366–E1373 [DOI] [PubMed] [Google Scholar]
- 68.Ellis J. M., Li L. O., Wu P-C., Koves T. R., Ilkayeva O., Stevens R. D., Watkins S. M., Muoio D. M., Coleman R. A. 2010. Adipose Acyl-CoA synthetase-1 directs fatty acids toward [beta]-oxidation and is required for cold thermogenesis. Cell Metab. 12: 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Digel M., Ehehalt R., Stremmel W., Fullekrug J. 2009. Acyl-CoA synthetases: fatty acid uptake and metabolic channeling. Mol. Cell. Biochem. 326: 23–28 [DOI] [PubMed] [Google Scholar]
- 70.Yao H., Ye J. 2008. Long chain Acyl-CoA synthetase 3-mediated phosphatidylcholine synthesis is required for assembly of very low density lipoproteins in human hepatoma Huh7 Cells. J. Biol. Chem. 283: 849–854 [DOI] [PubMed] [Google Scholar]