Abstract
Dissecting the genetics of complex traits such as obesity allows the identification of causal genes for disease. Here, we show that the BALB/c mouse strain carries genetic variants that confer resistance to obesity induced by leptin-deficiency or a high-fat diet (HFD). We set out to identify the physiological and genetic bases underlying this phenotype. When compared with C57BL6/J ob/ob mice (B6), BALB/c ob/ob mice exhibited decreased food intake, increased thermogenic capacity, and improved fat catabolism, each of which can potentially modify obesity. Interestingly, analysis of F1 ob/ob (progeny of B6 ob/+ × BALB/c ob+) mice revealed that obesity resistance in BALB/c ob/ob mice principally relied upon improved fat mobilization. This was mechanistically explained by increased adipose triglyceride lipase (ATGL) content in adipocytes, along with increased lipolysis and fatty acid oxidation. We conducted a genome-wide scan and defined a quantitative trait locus (QTL) on chromosome 2. BALB/c alleles on chromosome 2 not only associated with the obesity resistance phenotype but also supported increased ATGL content in adipose tissue. In summary, our study provides evidence that leptin-independent control of adipocyte lipolysis rates directly modifies the balance of macronutrient handling and is sufficient to regulate fat mass in the absence of alterations in food intake and energy expenditure.—Marcelin, G., S-M. Liu, X. Li, G. J. Schwartz, and S. Chua.
Keywords: adipocytes, adipose triglyceride lipase, obesity, quantitative trait locus
Obesity, defined as the accumulation of triglycerides within adipocytes, leads to increased body fat content and exacerbates susceptibility to type 2 diabetes, cardiovascular dysfunction, and cancer (1–3). Individual susceptibility to obesity is variable and integrates both environmental and genetic factors (4). Because of the increasing prevalence of obesity worldwide, identification of tissues and molecules to target should be a high priority to expand the list of potential therapeutic modalities (5).
The adipose tissue-derived hormone leptin and its downstream signaling pathways play a central role in obesity, as leptin or leptin receptor deficiency provokes morbid obesity (6–9). In addition, leptin resistance has been implicated in obesity ontogenesis (10). Most leptin receptor-mediated pathways are active mainly within the central nervous system (11), principally localized within the hypothalamus, to decrease food intake, increase energy expenditure, inhibit lipogenesis, and maintain normal glucose homeostasis (12). Consistent with these pleiotropic functions of leptin, studies on the regulation of body composition has centered upon leptin and the mechanisms mediated by its receptor. Alternatively, identifying pathways that regulate fat mass independently of leptin would be of interest in the design of therapies aimed to combat obesity.
We and others have reported that mouse inbred strains carry specific sets of genetic variants promoting significant variation in the expression of the leptin-deficient phenotype (13, 14). Thus, regulatory pathways independent of leptin can control body composition and metabolism, and mapping of quantitative trait loci (QTL) using leptin-deficient inbred mice is an ideal method of identifying such pathways. In this setting, leptin-deficient mice on a BALB/c genetic background have reduced adiposity when compared with ob/ob mice of the prototypic C57BL/6J strain (15). However, the functional and genetic bases for these variant phenotypes were not thoroughly characterized. Here, we performed a comprehensive analysis of the physiological mechanism(s) that limit body fat content in BALB/c ob/ob mice and found that obesity resistance in these mice principally relied upon improved fat oxidation. This operated through enhanced adipocyte lipolysis due to increased intracellular content and activity of adipose triglyceride lipase (ATGL/PNPLA2/desnutrin). Thus, higher lipolysis rate in adipocytes sustains increased fatty acid oxidation in BALB/c ob/ob mice. In addition, a genome-wide QTL analysis uncovered a significant QTL Lipq1 (for lipolytic line QTL 1) on chromosome 2 associated with body fat fraction and ATGL adipocyte content. Interestingly, Lipq1 does not alter food intake. In conclusion, our study supports that leptin-independent control of adipocyte lipolysis rates through regulating ATGL directly modifies the balance of macronutrient handling and is sufficient to regulate fat mass in the absence of detectable changes in food intake and energy expenditure.
MATERIALS AND METHODS
Animals
We generated C57BL6/J ob/ob Agrp−/−, BALB/c ob/ob Agrp−/− (at generation N5), and F1 ob/ob Agrp−/− from a cross between C57BL6/J ob/+ Agrp−/− and BALB/c ob/+ Agrp−/−. For genome-wide scanning, we used backcross to the BALB/c strain (F1 ob/+ Agrp−/− × BALB/c ob/+ Agrp−/−) to produce N2 obese mice. For brevity, we have skipped the Agrp−/− from the designation of leptin-deficient mice. Mice were fed with chow diet (LabDiet 5001; kcal%: protein 28.5%, fat 13.5%, carbohydrates 58%; PMI Nutrition International). Lean C57BL6/J and BALB/c mice were obtained from the Jackson Laboratory and fed a chow diet or a high-fat diet (HFD) (D12451, kcal%: protein 20%, fat 45%, carbohydrates 35%; Research Diets). All procedures were reviewed and approved by the institution's Animal Care Committee, conforming to accepted standards of humane animal care.
Body composition and indirect calorimetry
Body composition was measured by magnetic resonance spectroscopy (MRS) using an EchoMRI (Echo Medical Systems). Fat-mass percentage was calculated as the ratio between fat mass and body weight. For indirect calorimetry, mice were individually housed in metabolic chambers maintained at 20–22°C on a 12 h light, 12 h dark cycle with lights on at 0800 h. Metabolic measurements [oxygen consumption, respiratory exchange ratio (RER), locomotor activity] were obtained continuously using a CLAMS (Columbus Instruments) open-circuit indirect calorimetry system. Calorimetry data were normalized to individual lean body mass. Cumulative food intake of pelleted chow diet (#F0163, precision dustless food pellet; Bio-Serv) was measured daily and collected over five days for each individual animal.
Histological analysis of adipose tissue and estimation of adipocyte number per fat pad
Fat samples were fixed in 10% buffered formalin, embedded in paraffin, cut into 6 μm sections, and stained with hematoxylin and eosin. Adipocyte sizes were determined with ImageJ software. To assess adipocyte number, total DNA content from adipose tissue was determined. Briefly, fat pads were resected, weighed, and immediately frozen in liquid nitrogen. About 50 mg of tissue was homogenized, and genomic DNA was extracted following the manufacturer's protocol (DNeasy Blood and Tissue Kit, 69504; Qiagen). DNA was measured by spectrophotometric method, and total DNA content from fat pads was determined by multiplying DNA concentration in each sample by the fat pad weight for each mouse (16, 17).
Glucose tolerance test
Mice were fasted overnight and glucose was given intraperitoneally (1 mg/g). Glycemia was measured at the different indicated time for 2 h from tail blood.
Cold tolerance assay
BALB/c, B6, and F1 ob/ob mice were placed at 4°C, and body temperature drop was measured with a rectal probe after 75 min. Water and food were removed during the experiment.
Ex-vivo lipolysis
Fat explants freshly isolated from fed mice were cut into small pieces and incubated at 37°C under agitation in Krebs-Ringer medium containing 2% of free fatty acid BSA with or without isoproterenol (Sigma). Glycerol was measured from supernatants after 2 h of incubation. ATGL-mediated lipolysis (i.e., non-HSL lipolysis) was determined in presence of the inhibitor CAY10499 (100 μM). Fat explants were preincubated for 1 h with or without CAY10499. Then the medium was changed with medium containing (or not) CAY10499, and FFA was measured from supernatants after 2 h of incubation. Six mice in each group were studied, and analysis was performed in triplicate.
Metabolites and hormone concentration determination
Blood samples for plasma or serum were always collected at the same time in the morning. Mice were either fed ad libitum (without previous fasting) or fasted overnight (15 h period of fast). Glycemia was determined using a glucometer (Abbot), serum insulin levels using ELISA (Linco Mouse Insulin kit), plasma NEFA (Wako), and serum glycerol (Cayman) using a colorimetric assay. The HOMA index uses the following formula: insulin (μU/ml) × [glucose (mmol/l) / 22.5].
Treadmill
Wild-type BALB/c and C57BL6/J males were familiarized with the calorimetric treadmill for 2–3 days preceding the actual treadmill test. The adaptation period consisted of sessions lasting 5 min at belt speed 9 m/min. The number of aversive stimulations received was recorded, and the adaptation session was repeated once after 20 min of recovery if mice received more than five stimulations. For the treadmill test, the ramping protocol of the belt speed was as follows: 0 m/min for 20min, 5 m/min for 5 min (warm-up phase), 9 m/min for 5 min, 12 m/min for 5 min, 15 m/min for 5 min, 17 m/min for 2 min, 19 m/min for 2 min, and 20 m/min until the end of the test. RER was measured every 4 min during treadmill running by CLAMS calorimetry.
Immunoblot analysis
White adipose tissue was homogenized in lysis buffer containing protease and phosphatase inhibitor. Protein extracts were run on Criterion gels (Bio-Rad) and blotted onto nitrocellulose membranes. After blocking, immunoblots were incubated with primary antibodies against hormone-sensitive lipase (HSL), phospho-HSL (Ser563 and Ser660), ATGL, β-actin (Cell Signaling Technology), perilipin A (Sigma) and CGI-58 (gift from Dawn Brasaemle, Rutgers University). Blots were then incubated with fluorescent secondary antibodies, and proteins were detected using the fluorescence-based Odyssey Infrared Imaging System (LI-COR Biosciences). Quantification was performed using the Odyssey Infrared Imaging Software.
Genotyping
DNA was isolated from ear clips. Allelic constitution at various microsatellite markers was determined by PCR and analysis of amplicon size. The markers (from the Mouse Phenome Database and the NCBI database) used for the chromosome 2 scanning were D2Mit37, D2Mit42, D2Mit30, and D2Mit456. SNPs were also examined as described in supplementary Table I. Briefly, we amplified by PCR the region containing the single nucleotide polymorphism (SNP) and digested the amplicon with the appropriate restriction enzyme to discriminate between the BALB and B6 alleles as predicted by the database (http://insilico.ehu.es/restriction/two_seq/).
Statistical analysis
Results are shown as average ± SEM. Comparisons between groups for indirect calorimetry were performed by two-way repeated measures ANOVA, with genotype and time as factors. One-way ANOVA and unpaired Student t-tests were performed as indicated in the figure legends. Significance was accepted at P < 0.05. For the whole-genome scanning, Fisher's exact test was performed, and statistical significance was accepted at P < 0.00125 after Bonferroni correction. Statistical analysis was performed with GraphPad Prism 5.
RESULTS
Protection against obesity and diabetes in BALB/c mouse strain
We studied the development of obesity in HFD-fed mice and in genetic obesity due to leptin deficiency in C57BL6/J (B6) and BALB/c strains. Of note, the leptin-deficient mice (ob/ob) were also Agrp−/−. Because insulin signaling could be a confounding factor in obesity study, we used Agrp deletion as a tool to lower glucose and insulin level (supplementary Fig. I) (18). Thus, all of our data were based on BALB/c ob/ob Agrp−/− mice compared with C57BL/6J ob/ob Agrp−/− mice (B6). For brevity, we skipped the Agrp−/− designation below.
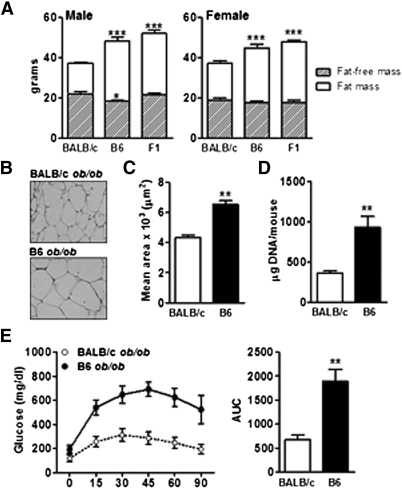
We observed that both B6 males and females fed a HFD for three months exhibited significantly increased body fat fraction compared with BALB/c animals (supplementary Fig. II-A). We next examined the impact of leptin deficiency in three different genetic backgrounds: BALB/c, B6, and F1 (BALB/c × B6). BALB/c ob/ob mice developed obesity but to a much lesser extent than the B6 ob/ob mice. Body composition analysis showed that three-month-old B6 ob/ob males and females had much greater fat mass (+40%) compared with BALB/c ob/ob mice (Fig. 1A). The F1 ob/ob mice developed obesity similar to the parental B6 strain with increased body weight and fat mass compared with BALB/c strain (Fig. 1A). Consequently, the BALB/c genome carries alleles recessive to B6 alleles that limit fat accumulation. Inguinal fat pad wet weight was reduced in BALB/c ob/ob (1.9 ± 0.1 g vs. 6.3 ± 0.6 g, P < 0.0001, in BALB/c and B6 ob/ob, respectively), and histological analysis of adipose tissue revealed a greater frequency of small adipocytes in subcutaneous fat depots from BALB/c ob/ob compared with B6 ob/ob (Fig. 1B), resulting in a significant decrease in mean adipocyte cross-sectional area (Fig. 1C). Moreover, the increased fat mass for a given fat pad was accompanied by a significant increase in total DNA content, which is consistent with an increased number of adipocytes in B6 ob/ob compared with BALB ob/ob (Fig. 1D). Similar characteristics were observed for the gonadal fat pad (data not shown). Thus, decreased adipose tissue mass in BALB/c ob/ob mice was the result of both reduced adipocyte triglyceride storage and decreased adipogenesis.
Fig. 1.
Leptin-deficient BALB/c mice are protected against obesity and diabetes. (A) Body composition (fat mass and fat-free mass) were analyzed in three-month-old leptin-deficient BALB/c, C57BL6/J (B6), and F1 males and females (n = 5–8). (B) Histology (hematoxylin and eosion staining) of adipose tissue of three-month-old BALB/c and B6 ob/ob mice. (C) The mean area of adipocytes was determined (>100 cells per animal, n = 6). (D) Cell number per fat pad and per mouse was expressed in DNA content (n = 6). (E) GTT was performed by measuring blood glucose concentration after an overnight fast at the indicated times after intraperitoneal injection of glucose, and area under the curve (AUC) was calculated in BALB/c and B6 ob/ob females (n = 4). Data are expressed as average ± SEM. One-way ANOVA (A) and unpaired t-test (C–E) were performed. *P < 0.05, **P < 0.005, ***P < 0.0005.
Additionally, decreased adiposity in the BALB/c strain was associated with changes in glucose homeostasis. Fasting glycemia of BALB/c ob/ob mice was lower with unchanged insulinemia compared with B6 ob/ob mice (Table 1). F1 ob/ob mice displayed hyperglycemia compared with BALB/c ob/ob, and F1 females exhibited lower insulinemia compared with BALB/c ob/ob (Table 1). On a HFD, fasting glucose was lowered in wild-type BALB/c relative to wild-type B6, and insulin level was decreased in BALB/c males (supplementary Fig. II-B). Glucose tolerance tests (GTT) performed on both ob/ob (Fig. 1E) and HFD mice (supplementary Fig. II-C) showed that glucose clearance was markedly improved in BALB/c versus B6 mice.
TABLE 1.
Metabolic and behavioral characteristics of leptin-deficient BALB/c, C57BL6/J, and F1 mice
| BALB/c ob/ob | B6 ob/ob | F1 ob/ob | ||
| Glucose (mg/ml) | MaleFemale | 151.2 ± 22.6a91.7 ± 7.6a | 226.2 ± 28.4b135.1 ± 12.8b | 305.8 ± 42.9b147.3 ± 21.06b |
| Insulin (ng/ml) | MaleFemale | 16.81 ± 2.917.44 ± 2.2a | 14.2 ± 5.1413.36 ± 3.3a | 8.18 ± 2.66.14 ± 0.75b |
| HOMA index | MaleFemale | 140.8 ± 28.290.3 ± 15.5 | 119.5 ± 40.476.2 ± 10.1 | 138.8 ± 33.056.3 ± 9.2 |
| Food intake (per mouse) | Daily food intake (g) | 6.78 ± 0.62a | 10.9 ± 0.21b | 8.3 ± 0.66a |
| Meal size (g) | 0.26 ± 0.03 | 0.33 ± 0.02 | 0.20 ± 0.01 | |
| Meal number | 23.5 ± 1.7a | 30.0 ± 2.0b | 23.7 ± 1.1c | |
| Cold resistance assay (Δ °C)b | −3.15 ± 0.48a | −6.66 ± 1.35b | −0.43 ± 0.71a | |
| Ambulatory activity | Day | 3397 ± 290a | 847 ± 192b | 2194 ± 391c |
| Night | 5731 ± 1148a | 1220 ± 271b | 3624 ± 928c | |
| Total | 9128 ± 1363a | 2068 ± 438b | 5818 ± 1267c | |
| VO2 (ml/kg/hr) | Day | 4538 ± 157 | 4520 ± 696 | 4842 ± 467 |
| Night | 4629 ± 145 | 4639 ± 998 | 4582 ± 436 | |
| Total | 4590 ± 148 | 4585 ± 855 | 4692 ± 442 |
Data are expressed as average ± SEM and statistical significance is determined for P < 0.05.
Determined after an over-night fast.
Δ°C is the difference between the initial temperature and the temperature measured after 75 min at 4°C.
Individual fat-free body mass measured by magnetic resonance spectroscopy was used for normalization.
Increased fatty acid oxidation is associated with reduced adiposity in BALB/c mice
Given our findings that F1 and B6 ob/ob mice developed similar degrees of obesity, we next compared BALB/c ob/ob mice with both B6 and F1 ob/ob mice in search of key phenotypic traits associated with obesity resistance in BALB/c ob/ob mice.
We monitored daily caloric intake and found that BALB/c and F1 ob/ob mice manifested reduced food intake compared with the B6 ob/ob mice. The BALB/c and F1 hypophagia was solely due to decreased meal frequency, as meal size was similar in both strains (Table 1). As F1 and B6 ob/ob mice exhibited the same level of adiposity, we concluded that caloric intake is unlikely to be the major contributor to fat-mass deposition in our models.
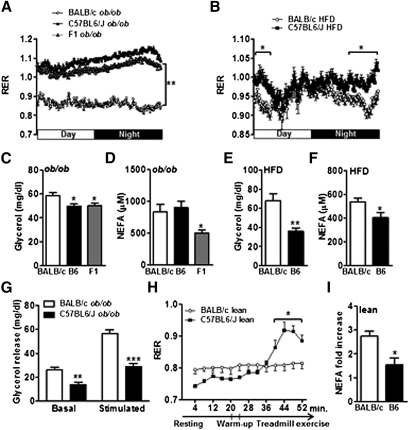
Assessment of cold tolerance to probe brown adipose tissue function, a critical modulator of metabolic imbalance (19), revealed that cold-induced thermogenesis was impaired in B6 ob/ob mice compared with BALB/c and F1 ob/ob mice (Table 1). As adiposity and cold tolerance were discordant in B6 ob/ob mice and F1 ob/ob mice, brown adipose tissue thermogenesis differences could not explain resistance to obesity in BALB/c ob/ob mice. Whole body energy expenditure evaluation using indirect calorimetry did not reveal any difference in oxygen consumption between strains despite decreased spontaneous activity in B6 and F1 compared with BALB/c ob/ob mice (Table 1). Similarly, no difference in oxygen consumption was found between HFD BALB/c and B6 mice (supplementary Fig. IV-A). Consequently, obesity resistance of BALB/c mice could not be ascribed to alterations in whole body energy expenditure. Interestingly, the revealing difference observed between BALB/c and both B6 and F1 ob/ob mice relates to the RER. B6 and F1 ob/ob mice exhibited higher RER values during both the light and the dark periods compared with BALB/c ob/ob mice (Fig. 2A). In B6 and F1 ob/ob mice, RER values were almost all greater than 1. This indicated that lipid metabolism is shifted toward net lipid synthesis with minor lipid oxidation in B6 and F1 ob/ob compared with BALB ob/ob (20). Moreover, lower RER values were also observed in HFD-fed BALB/c mice compared with B6 mice (Fig. 2B). Thus, our data reveal that the BALB/c strain is characterized by an elevation in fatty acid oxidation compared with the B6 strain, implying that increased fat catabolism confers protection against obesity.
Fig. 2.
Increased fatty acid oxidation and adipose lipolysis in BALB/c ob/ob mice. Respiratory exchange ratios (RER) were determined during light and dark cycles in (A) BALB/c, C57BL6/J (B6), and F1 ob/ob mice with free access to chow diet (n = 4) and in (B) wild-type BALB/c and B6 mice fed a HFD. (C) Determination of glycerol and (D) NEFA concentrations in serum from fed BALB/c, B6, and F1 ob/ob females (n = 8). (E) Glycerol and (F) NEFA concentrations in fed HFD BALB/c and B6 females (n = 6). (G) Glycerol release from adipose tissue explants of BALB/c and B6 ob/ob mice under basal condition and stimulated with isoproterenol (n = 6). (H) RER was assessed during resting and treadmill exercise of wild-type BALB/c and B6 males (n = 4–5). (I) Fold change of circulating NEFA between preexercise and postexercise. Data are expressed as average ± SEM. Two-way ANOVA (A, B, H), one-way ANOVA (C, D) and unpaired t-tests (E, F, G, I) were performed. *P < 0.05, **P < 0.005, ***P < 0.0005.
Increased basal lipolysis in BALB/c ob/ob mice
Lower RER values (Fig. 2A) and reduction in adipocyte lipid droplet size in BALB/c ob/ob mice (Fig. 1C) suggest that increased fatty acid oxidation may be a consequence of an increase in lipolysis. Consistent with this hypothesis, higher circulating glycerol concentrations were observed in BALB/c mice compared with B6 and F1 ob/ob mice (Fig. 2C). Nonesterified fatty acid (NEFA) concentrations were increased in BALB/c versus F1 ob/ob mice (Fig. 2D) while no difference was detected between BALB/c and B6 ob/ob mice (Fig. 2D). In addition, glycerol (Fig. 2E) and NEFA (Fig. 2F) were increased in BALB/c versus B6 HF fed mice.
Beta-3 adrenergic receptor (β3-AR), predominantly expressed in white and brown adipose tissues, stimulates sympathetic activation of adipose tissue to promote lipolysis. By qRT-PCR, we found that β3-AR expression was significantly increased in BALB/c versus B6 ob/ob white adipose tissue (supplementary Fig. IV-A). However, we observed that acute CL316,243 (a specific β3-AR agonist) administration raised serum glycerol concentration of BALB/c and B6 ob/ob mice to the same concentration (supplementary Fig. IV-B), suggesting similar degrees of β3-adrenergic signaling between the two strains (21). To ascertain whether the lipolysis rates were a cell-autonomous function rather than strictly regulated by the autonomic nervous system, we next quantified adipose tissue lipolysis by measuring glycerol release from adipose tissue explants. In both basal and stimulated conditions, we observed that glycerol release was significantly higher in explants derived from BALB/c ob/ob compared with B6 ob/ob (Fig. 2G). These data indicate that the increased rate of lipolysis of BALB/c adipocytes is intrinsic to adipocytes.
As fatty acid demand increases during exercise (22), we assessed the lipolytic capacity of wild-type B6 and BALB/c animals (fed a normal chow diet) during exercise involving treadmill running. No difference in RER was observed at rest. However, during exercise, whereas RER values remained stable for BALB/c mice, RER significantly increased for B6 mice, indicating increasing dependence upon carbohydrates for fuel (Fig. 2H). We observed that exercise-induced increase in plasma NEFA was low in B6 mice, whereas BALB/c animals significantly increased NEFA production by almost 3-fold (Fig. 2I). Thus, these data reveal an increased ability of BALB/c mice to perform lipolysis and oxidize fatty acids.
Increased amounts of ATGL in BALB/c adipocytes
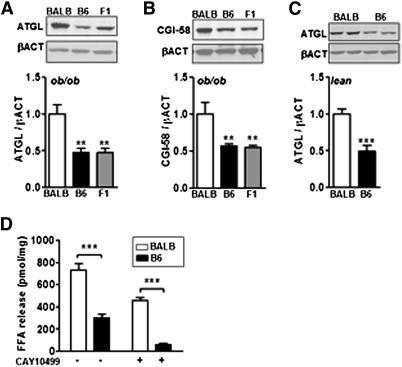
We further characterized BALB/c adipocytes by analyzing expression of key enzymes involved in triglyceride metabolism. Two main enzymes account for 95% of lipolytic activity in adipocytes, ATGL, and HSL (23). Interestingly, protein levels of ATGL and its coactivator CGI-58 (24) were increased in adipose tissue from BALB/c ob/ob mice compared with that from B6 and F1 ob/ob mice (Fig. 3A, B). Similarly, in HFD-fed mice, increased ATGL and CGI-58 protein levels were observed in BALB/c adipose tissue compared with that from B6 mice (supplementary Fig. III-B, C).
Fig. 3.
Increased adipose lipolysis is related to accumulation of ATGL in adipose tissue. Immunoblots and densitometry analysis of (A) ATGL and (B) CGI-58 in inguinal fat from BALB/c, C57BL6/J (B6), and F1 ob/ob mice (n = 6). Quantification of (C) ATGL in four-week-old BALB/c and B6 wild-type lean mice (n = 5). (D) FFA release from BALB and B6 ob/ob adipose tissue explants in presence or not of the HSL inhibitor CAY10499. Data are expressed as average ± SEM. One-way ANOVA (A, B) and unpaired t-tests (C, D) were performed. *P < 0.05, **P < 0.005, ***P < 0.0005.
We also checked HSL expression. We found that HSL protein level was increased in adipose tissue from BALB/c ob/ob mice compared with that from B6 and F1 ob/ob mice (supplementary Fig. V-A). However, there was no difference in the amount of the activated form of HSL (25, 26), phosphorylated HSL (supplementary Fig. V-B, C), or the lipolysis inhibitor perilipin A (27) (supplementary Fig. V-D). Hence, HSL may not participate in increased adipose lipolysis observed in BALB ob/ob. Accordingly, there was no change in total HSL protein expression or HSL activation in adipose tissue from HFD-fed mice (supplementary Fig. III-B, C). We further investigated adipose lipolysis in measuring ATGL-mediated lipolysis. To that aim, we measured free fatty acid (FFA) release from adipose tissue explants in presence or not of the HSL inhibitor CAY10499 (28, 29). We previously assessed the efficiency of CAY10499 in our experimental condition as we found that induced lipolysis with a selective β3-AR agonist (CL316,243) is blunted in presence of 100 μM CAY10499 (supplementary Fig. VI). Then we compared FFA release from freshly dissected adipose tissue explants from BALB/c and B6 ob/ob inguinal fat pad. We observed that FFA production was increased in explants from BALB/c mice relative to B6 ob/ob mice when HSL activity was inhibited by CAY10499 (Fig. 3D). Moreover, we calculated the same difference between BALB and B6 lipolysis (Δ−CAY = 431.3 ± 31.7 versus Δ+CAY = 397.1 ± 8.1, P = 0.31) without the HSL inhibitor (i.e., HSL + ATGL activity) or with CAY10499 (i.e., ATGL activity), respectively. Consequently, our data show that increased adipose lipolytic rate in BALB/c principally relied on increased ATGL activity. Of note, the increased ATGL activity was greater than the increase in ATGL protein content in adipose tissue. This may be due to the concomitant increase of CGI-58 expression that is known to enhance ATGL lipolytic activity.
Thus, we postulated that the increase in ATGL protein level is an intrinsic characteristic of the BALB/c strain. Indeed, ATGL protein levels were also differentially expressed in adipose tissue from four-week-old lean BALB/c and B6 mice with no difference in adiposity (Fig. 3C). Therefore, it is likely that the differences in ATGL protein between the BALB/c and B6 strains were not due to the development of obesity but may be the underlying cause of increased adipose lipolysis.
A genome-wide scan points to a chromosome 2 locus associated with fractional body fat
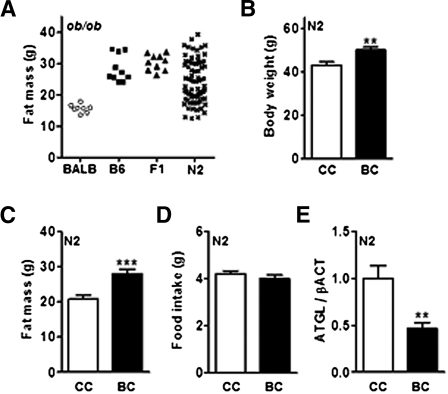
We generated obese N2 progeny (from F1 ob/+ Agrp−/− × BALB/c ob/+ Agrp−/−). Body composition studied for each mouse showed that N2 progeny displayed fat-mass values scattered over a broad range, from low adiposity similar to BALB ob/ob, to high adiposity comparable to B6 or F1 ob/ob mice (Fig. 4A). Previously, we did not report any gender difference in BALB or B6 ob/ob mice. Moreover, both N2 ob/ob males and females displayed the same distribution of fat-mass values (not shown). Consequently, N2 ob/ob males and females were combined for the genome-wide scan.
Fig. 4.
BALB/c alleles on chromosome 2 support lower fat accumulation and increased ATGL content in adipose tissue of N2 ob/ob mice. (A) Fat mass of BALB/c, C57BL6/J (B6), F1, and N2 ob/ob mice. (B) Body weight, (C) fat mass, (D) daily food intake (n = 16–27), and (E) ATGL protein content in inguinal adipose tissue (n = 10–14) were determined in N2 mice with homozygous BALB/c allele on chromosome 2 (CC) and heterozygous with BALB/c and B6 alleles on chromosome 2 (BC). Data are expressed as average ± SEM. Unpaired t-tests were performed. **P < 0.005, ***P < 0.0005.
To map the QTL associated with obesity resistance in BALB/c strain, we only genotyped N2 ob/ob mice at the two extremes of the distribution of body fat fraction: N2 mice with high fat-mass percentage (>60%), and N2 mice with low adiposity (<50%) (Table 2). On the basis of our findings on BALB/c, B6, and F1 ob/ob mice, we reasoned that mice with low adiposity should be enriched with homozygous BALB/c alleles (CC genotype) at loci that control fat-mass deposition, whereas mice with high adiposity should be enriched with heterozygous B6 and BALB/c alleles (BC genotype). A set of 40 microsatellite markers were used to perform the genome scan, and 49 N2 ob/ob progenies were analyzed. We found a significant association with the following markers on chromosome 2: rs4136610, D2Mit37, and rs3667007. No other marker exhibited a significant association. Consequently, we concluded that our genome-wide analysis revealed markers on chromosome 2 strongly associated with obesity resistance of BALB/c strain, and this QTL was named Lipq1 for lipolytic line QTL 1. We further analyzed the N2 ob/ob mice according to their chromosome 2 haplotype. As expected, BALB/c alleles on chromosome 2 confer lower body weight and fat mass of leptin-deficient animals (Fig. 4B, C). Moreover, Lipq1 does not alter caloric intake (Fig. 4D). Finally, we checked ATGL expression in adipose tissue and found that ATGL protein level is increased in N2 ob/ob mice with BALB/c alleles on chromosome 2 (Fig. 4E). Consequently, the genome-wide scan provides supportive evidence that a locus or a set of loci on chromosome 2 supports increased ATGL expression in adipose tissue and control of body fat fraction, independently of caloric intake.
TABLE 2.
Segregation of BALB/c and C57BL6/J alleles on mouse chromosome 2 with body fat fraction in leptin-deficient N2 progeny (F1xBALB/c)
| High Adiposity | Low Adiposity | P | ||
| Mean Fat Mass % | 61.3 ± 0.45 | 46.2 ± 0.79 | <0.0001a | |
| Markers | Position (Mb) | CC : BC Genotype Ratios | ||
| rs3678168 | 5.57 | 9 : 14 | 13 : 13 | 0.44 |
| rs4136610 | 70 | 3 : 22 | 18 : 9 | <0.0001b |
| D2Mit37 | 74.5 | 3 : 22 | 18 : 9 | <0.0001b |
| rs3667007 | 81 | 4 : 21 | 17 : 10 | 0.0007b |
| D2Mit42 | 104 | 6 : 19 | 16 : 11 | 0.010 |
| D2Mit30 | 124.7 | 10 : 15 | 14 : 13 | 0.39 |
| D2Mit456 | 168.7 | 11 : 11 | 11 : 13 | 0.77 |
Student t-test performed and statistical significance accepted at P < 0.05.
Fisher's exact tests performed and statistical significance determined for Bonferroni-corrected P < 0.00125.
SNP architecture of the chromosome 2 and candidate genes
C57BL6/J and BALB/c genome sequence strains are included in the Sanger sequencing database. Consequently, we studied the SNP architecture of the proximal chromosome 2 spanning from 5.5 to 68 Mb (i.e., ∼60 Mb) and found less than 20 SNPs for gene-associated location (supplementary File I). By contrast, the domain including the QTL (40 Mb spanning from 65 to 104 Mb) contains ∼500 SNPs for gene-associated location (supplementary File II). As a consequence, the low SNP occurrence in the proximal chromosome 2 indicated that BALB and B6 strain are not polymorphic at these loci. In addition to this in silico analysis, we examined SNPs defined as polymorphic between BALB and B6 (supplementary Table I), for which allele information is available online through the Mouse Genome Database. For the SNP genotyping, we used genomic DNA extracted from the BALB/c and B6 parental strains. Interestingly, the SNP analyzed from 24.7 Mb to 67.09 Mb were not polymorphic between BALB/c and B6 strain (supplementary Table I). Altogether, a nonpolymorphic region of 42 Mb extending from marker rs27233322 to marker rs27987286 is defined on chromosome 2. As QTLs are preferentially assigned to polymorphic loci between parental strains, we narrowed the critical interval to a polymorphic region of chromosome 2 spanning from 65 Mb to 104 Mb. Overall, our results support that genetic determinants on chromosome 2 are associated with expression level of ATGL and CGI-58. It is conceivable that a transcriptional or posttranslational mechanism, or both, through a direct or indirect mechanism could be responsible for increased protein expression. Thus, we established a list of the plausible candidates (Table 3) to these processes by studying SNPs in conjunction with the functional information available.
TABLE 3.
Putative candidate genes with SNPs in Lipq1
| Gene Symbol | Gene Name | Position (Mb) | SNP Conseq:uence |
| Transcription regulator | |||
| Csrnp3 | Cysteine-serine-rich nuclear protein 3 | 65.71 | ESS |
| Evx2 | Even skipped homeotic gene 2 homolog | 74.49 | SS |
| Signaling pathway | |||
| Fastkd1 | FAST kinase domains 1 | 69.54 | Cn |
| Rapgef4 | Rap guanine nucleotide exchange factor 4 | 71.99 | ESS, SS |
| Dusp19 | Dual specificity phosphatase 19 | 80.47 | Cn |
| Ptprj | Protein tyrosine phosphatase, receptor type, J | 90.39 | Cn, SS |
| Madd | MAP-kinase activating death domain | 91.00 | Cn, SS |
| Protein folding and degradation | |||
| Ppig | Peptidyl-prolyl isomerase G (cyclophilin G) | 69.58 | Cn, StopG |
| Ubr3 | Ubiquitin protein ligase E3 component n-recognin 3 | 69.73 | Cn, SS |
| Dnajc10 | DnaJ (Hsp40) homolog, subfamily C, member 10 | 80.18 | SS, Cn |
| Zswim2 | Zinc finger, SWIM domain containing 2 | 83.75 | Cn |
| Ube2l6 | Ubiquitin-conjugating enzyme E2L 6 | 84.64 | Cn |
| Psmc3 | Proteasome (prosome, macropain) 26S subunit, ATPase 3 | 90.89 | Cn, SS |
| Adipocyte biology/obesity | |||
| Frzb | Frizzled-related protein | 80.25 | Cn |
| Tnks1bp1 | Tankyrase 1 binding protein 1 | 84.89 | Cn |
| Nr1 h3 | Nuclear receptor subfamily 1, group H, member 3 | 91.03 | SS |
| Arhgap1 | Rho GTPase activating protein 1 | 91.50 | Cn |
Cn, coding nonsynonymous; ESS, essential splice site (in the first 2 or last 2 bp of intron); SS, splice site; StopG, stop gained.
DISCUSSION
Obesity is often associated with impaired response to the adipose-tissue derived hormone leptin, a central regulator of body fat content and metabolism. We proposed here to use genetics to unveil leptin-independent pathways that overcome obesity. Toward that aim, BALB/c mice, which have been shown previously to be more resistant to obesity induced by leptin deficiency, were compared with leptin-deficient C57BL6/J (B6) mice (15). To probe the primary function responsible for obesity resistance, we measured multiple metabolic and behavioral parameters. When compared with B6 ob/ob mice, BALB/c ob/ob mice exhibited decreased food intake, increased thermogenic capacity, and improved fat catabolism. However, analysis of F1 ob/ob (B6 ob/+ × BALB/c ob/+) mice revealed that obesity resistance in BALB/c ob/ob mice solely relied upon improved fat oxidation. We uncovered a leptin-independent increase in lipolysis intrinsic to adipocytes that contributed to the regulation of body fat content in BALB/c ob/ob mice. Interestingly, this process was also operative in lean mice fed a HFD, as we found similar increased fatty-acid oxidation; consequently, this mechanism supported by BALB/c modifier gene(s) was operative beyond leptin deficiency. Finally, we showed that increased lipolysis in these mice was associated with increased ATGL protein levels, a trait conferred by modifier genes present on the chromosome 2 of BALB/c mice.
Triglyceride (TG) hydrolysis, or lipolysis, is a sequential process that depends on ATGL (PNPLA2, desnutrin), which initiates TG breakdown and produces diacylglycerols (DG), whereas HSL preferentially generates monoglycerides from DGs (30). Thus, ATGL is rate-limiting for the first step of TG hydrolysis and may determine the maximal rate for fatty acid mobilization (31–33). In obese patients, defects in adipose tissue lipolytic activity have been reported, but it is not clear whether altered lipolysis is primary or secondary to obesity development (34–36). Here, we noticed that ATGL level was decreased in adipose tissue from B6 ob/ob mice compared with BALB/c ob/ob mice. In addition, comparison between BALB/c and B6 ob/ob mice revealed that protein levels of ATGL and its coactivator CGI-58 were elevated in the adipose tissue of BALB/c ob/ob mice, whereas active HSL levels were unchanged. Consequently, downregulation of ATGL level and activity occurred specifically in the obese-susceptible strain C57BL6/J, whereas the resistant strain BALB/c maintained higher levels of ATGL and its activators. In BALB/c ob/ob mice, the higher lipolysis rate was accompanied by increased whole body fatty acid oxidation. So, it is tempting to propose that even higher rates of adipocyte lipolysis could further reduce adipocyte lipid stores if fatty acid oxidation rates could accommodate the increased supply.
As lipolysis also supports fatty acid demand by exercising muscles, we further assessed the lipolytic capacity of lean BALB/c and B6 mouse using a mouse treadmill running model (22). In BALB/c mice, a flat RER during ramped exercise indicated a high fatty acid oxidation rate sufficient to efficiently provide ATP to supply exercising muscles. By contrast, C57BL/6J mice fail to increase fatty acid release during exercise and were apparently unable to increase fatty acid oxidation, leading to a rapid increase in RER during exercise ramp-up. Together, these findings imply that BALB/c alleles support a better lipolytic capacity in both obese and exercising mice, suggesting that increased TG hydrolysis is a principal mechanism protecting BALB/c mice from obesity.
We postulate that the increase in ATGL protein level is an intrinsic characteristic of the BALB/c strain. Indeed, ATGL protein levels were also differentially expressed in adipose tissue from young, lean BALB/c and B6 mice prior to the appearance of any differences in adiposity. Therefore, we argue that the differences in ATGL protein between the BALB/c and B6 strains are not secondary to obesity but, rather, are determined by BALB/c modifier genes. In humans, ATGL deficiency causes neutral lipid storage disease due to an inability to mobilize neutral lipids (37), and low body mass indexes (BMI) have been associated with higher levels of ATGL protein level in human adipose tissue (38). Consequently, identification of a regulatory mean of modulating ATGL might have important implications for obesity treatment. QTL analysis is commonly used to map chromosomal regions and genes that control obesity in mice. Having first identified lipolysis as the leading cellular mechanism that protects BALB/c from obesity, we wanted to find the genetic loci controlling body fat content and lipolysis in these mice. We identified a significant QTL named Lipq1 (for lipolytic line QTL 1) on chromosome 2 associated with body fat content. Furthermore, Lipq1 supports higher ATGL protein content in adipose tissue of N2 ob/ob mice. We narrowed the region to a critical interval that encompasses a chromosome 2 portion of ∼30 Mb. A previous study mapped an overlapping QTL Mob that confers increased adiposity in F2 (C57BL6/J × CAST/Ei) mice inheriting chromosomal regions from B6 and lower adiposity in mice inheriting the CAST chromosomal region (39, 40). Conversely, BALB/c alleles of Nidd5 on chromosome 2 have been shown to decrease obesity in Tsumura Suzuki obese diabetic (TSOD) mice (41). Underlying mechanisms were not described; however, analysis of lipolysis in those models would indicate whether a common locus was involved in obesity resistance. Interestingly, Lipq1 is syntenic to human chromosomal region 2q22-32 that contains loci related to Bardet-Biedl syndrome with manifestation of obesity (42), abdominal visceral fat (43), and higher BMI (44). Consequently, this observation supports that the mechanism we identified in the BALB/c strain could be relevant for a better understanding of human obesity.
In summary, our findings highlight a leptin-independent increase in adipose tissue lipolysis that restrains obesity and glucose intolerance. Recent advances in the understanding of the enzymology of lipolysis within adipocytes and other cell types have yielded insights into the process of triglyceride metabolism and the regulatory molecules involved (24, 45–47). Here, we identified a QTL on mouse chromosome 2 that confers resistance to obesity through modulation of lipolysis and the adipocyte content of ATGL. We propose that identification of Lipq1 will provide a novel target for the treatment of obesity and associated metabolic defects.
Supplementary Material
Footnotes
Abbreviations:
- ATGL
- adipose triglyceride lipase
- β3-AR
- beta-3 adrenergic receptor
- BMI
- body mass index
- CGI-58
- comparative gene identification-58
- GTT
- glucose tolerance test
- HFD
- high-fat diet
- HSL
- hormone-sensitive lipase
- QTL
- quantitative trait locus
- RER
- respiratory exchange ratio
- SNP
- single nucleotide polymorphism
- TG
- triglyceride
This work was supported in part by National Institutes of Health Grants RO1-DK-057621, PO1-DK-26687 (S.C.), HD-058155 (S.C.), DK-020541 (EINSTEIN DRTC), and DK-026687 (NYORC) (S.C. and G.J.S.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of six figures and one table.
REFERENCES
- 1.Behn A., Ur E. 2006. The obesity epidemic and its cardiovascular consequences. Curr. Opin. Cardiol. 21: 353–360 [DOI] [PubMed] [Google Scholar]
- 2.Renehan A. G., Tyson M., Egger M., Heller R. F., Zwahlen M. 2008. Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet. 371: 569–578 [DOI] [PubMed] [Google Scholar]
- 3.Olefsky J. M., Glass C. K. 2010. Macrophages, inflammation, and insulin resistance. Annu. Rev. Physiol. 72: 219–246 [DOI] [PubMed] [Google Scholar]
- 4.Chung W. K., Leibel R. L. 2008. Considerations regarding the genetics of obesity. Obesity (Silver Spring). 16(Suppl. 3): S33–S39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prentice A. M. 2006. The emerging epidemic of obesity in developing countries. Int. J. Epidemiol. 35: 93–99 [DOI] [PubMed] [Google Scholar]
- 6.Chua S. C., Jr, Chung W. K., Wu-Peng X. S., Zhang Y., Liu S. M., Tartaglia L., Leibel R. L. 1996. Phenotypes of mouse diabetes and rat fatty due to mutations in the OB (leptin) receptor. Science. 271: 994–996 [DOI] [PubMed] [Google Scholar]
- 7.Pelleymounter M. A., Cullen M. J., Baker M. B., Hecht R., Winters D., Boone T., Collins F. 1995. Effects of the obese gene product on body weight regulation in ob/ob mice. Science. 269: 540–543 [DOI] [PubMed] [Google Scholar]
- 8.Halaas J. L., Gajiwala K. S., Maffei M., Cohen S. L., Chait B. T., Rabinowitz D., Lallone R. L., Burley S. K., Friedman J. M. 1995. Weight-reducing effects of the plasma protein encoded by the obese gene. Science. 269: 543–546 [DOI] [PubMed] [Google Scholar]
- 9.Campfield L. A., Smith F. J., Guisez Y., Devos R., Burn P. 1995. Recombinant mouse OB protein: evidence for a peripheral signal linking adiposity and central neural networks. Science. 269: 546–549 [DOI] [PubMed] [Google Scholar]
- 10.Howard J. K., Cave B. J., Oksanen L. J., Tzameli I., Bjorbaek C., Flier J. S. 2004. Enhanced leptin sensitivity and attenuation of diet-induced obesity in mice with haploinsufficiency of Socs3. Nat. Med. 10: 734–738 [DOI] [PubMed] [Google Scholar]
- 11.de Luca C., Kowalski T. J., Zhang Y., Elmquist J. K., Lee C., Kilimann M. W., Ludwig T., Liu S. M., Chua S. C., Jr 2005. Complete rescue of obesity, diabetes, and infertility in db/db mice by neuron-specific LEPR-B transgenes. J. Clin. Invest. 115: 3484–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gautron L., Elmquist J. K. 2011. Sixteen years and counting: an update on leptin in energy balance. J. Clin. Invest. 121: 2087–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chua S., Jr, Liu S. M., Li Q., Yang L., Thassanapaff V. T., Fisher P. 2002. Differential beta cell responses to hyperglycaemia and insulin resistance in two novel congenic strains of diabetes (FVB- Lepr (db)) and obese (DBA- Lep (ob)) mice. Diabetologia. 45: 976–990 [DOI] [PubMed] [Google Scholar]
- 14.Stoehr J. P., Byers J. E., Clee S. M., Lan H., Boronenkov I. V., Schueler K. L., Yandell B. S., Attie A. D. 2004. Identification of major quantitative trait loci controlling body weight variation in ob/ob mice. Diabetes. 53: 245–249 [DOI] [PubMed] [Google Scholar]
- 15.Qiu J., Ogus S., Mounzih K., Ewart-Toland A., Chehab F. F. 2001. Leptin-deficient mice backcrossed to the BALB/cJ genetic background have reduced adiposity, enhanced fertility, normal body temperature, and severe diabetes. Endocrinology. 142: 3421–3425 [DOI] [PubMed] [Google Scholar]
- 16.Cariou B., Postic C., Boudou P., Burcelin R., Kahn C. R., Girard J., Burnol A. F., Mauvais-Jarvis F. 2004. Cellular and molecular mechanisms of adipose tissue plasticity in muscle insulin receptor knockout mice. Endocrinology. 145: 1926–1932 [DOI] [PubMed] [Google Scholar]
- 17.Satyanarayana A., Klarmann K. D., Gavrilova O., Keller J. R. 2012. Ablation of the transcriptional regulator Id1 enhances energy expenditure, increases insulin sensitivity, and protects against age and diet induced insulin resistance, and hepatosteatosis. FASEB J. 26: 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Israel D. D., Sheffer-Babila S., de Luca C., Jo Y. H., Liu S. M., Xia Q., Spergel D. J., Dun S. L., Dun N. H., Chua Jr S. C. Effects of leptin and melanocortin signaling interactions on pubertal development and reproduction. Endocrinology. epub ahead of print. Mar 9, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bachman E. S., Dhillon H., Zhang C. Y., Cinti S., Bianco A. C., Kobilka B. K., Lowell B. B. 2002. betaAR signaling required for diet-induced thermogenesis and obesity resistance. Science. 297: 843–845 [DOI] [PubMed] [Google Scholar]
- 20.Elia M., Livesey G. 1988. Theory and validity of indirect calorimetry during net lipid synthesis. Am. J. Clin. Nutr. 47: 591–607 [DOI] [PubMed] [Google Scholar]
- 21.Susulic V. S., Frederich R. C., Lawitts J., Tozzo E., Kahn B. B., Harper M. E., Himms-Hagen J., Flier J. S., Lowell B. B. 1995. Targeted disruption of the beta 3-adrenergic receptor gene. J. Biol. Chem. 270: 29483–29492 [DOI] [PubMed] [Google Scholar]
- 22.Marcaletti S., Thomas C., Feige J. N. 2011. Exercise performance tests in mice. Current Protocols in Mouse Biology. John Wiley & Sons, Inc; 141–154 [DOI] [PubMed] [Google Scholar]
- 23.Lass A., Zimmermann R., Oberer M., Zechner R. 2011. Lipolysis - a highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog. Lipid Res. 50: 14–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J. G., Gorkiewicz G., Zechner R. 2006. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 3: 309–319 [DOI] [PubMed] [Google Scholar]
- 25.Anthonsen M. W., Ronnstrand L., Wernstedt C., Degerman E., Holm C. 1998. Identification of novel phosphorylation sites in hormone-sensitive lipase that are phosphorylated in response to isoproterenol and govern activation properties in vitro. J. Biol. Chem. 273: 215–221 [DOI] [PubMed] [Google Scholar]
- 26.Garton A. J., Campbell D. G., Cohen P., Yeaman S. J. 1988. Primary structure of the site on bovine hormone-sensitive lipase phosphorylated by cyclic AMP-dependent protein kinase. FEBS Lett. 229: 68–72 [DOI] [PubMed] [Google Scholar]
- 27.Brasaemle D. L., Subramanian V., Garcia A., Marcinkiewicz A., Rothenberg A. 2009. Perilipin A and the control of triacylglycerol metabolism. Mol. Cell. Biochem. 326: 15–21 [DOI] [PubMed] [Google Scholar]
- 28.Grisouard J., Bouillet E., Timper K., Radimerski T., Dembinski K., Frey D. M., Peterli R., Zulewski H., Keller U., Muller B., et al. 2012. Both inflammatory and classical lipolytic pathways are involved in lipopolysaccharide-induced lipolysis in human adipocytes. Innate Immun. 18: 25–34 [DOI] [PubMed] [Google Scholar]
- 29.Reid B. N., Ables G. P., Otlivanchik O. A., Schoiswohl G., Zechner R., Blaner W. S., Goldberg I. J., Schwabe R. F., Chua S. C., Jr, Huang L. S. 2008. Hepatic overexpression of hormone-sensitive lipase and adipose triglyceride lipase promotes fatty acid oxidation, stimulates direct release of free fatty acids, and ameliorates steatosis. J. Biol. Chem. 283: 13087–13099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcelin G., Chua S., Jr 2010. Contributions of adipocyte lipid metabolism to body fat content and implications for the treatment of obesity. Curr. Opin. Pharmacol. 10: 588–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimmermann R., Strauss J. G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., et al. 2004. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 306: 1383–1386 [DOI] [PubMed] [Google Scholar]
- 32.Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., et al. 2006. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 312: 734–737 [DOI] [PubMed] [Google Scholar]
- 33.Ahmadian M., Duncan R. E., Varady K. A., Frasson D., Hellerstein M. K., Birkenfeld A. L., Samuel V. T., Shulman G. I., Wang Y., Kang C., et al. 2009. Adipose overexpression of desnutrin promotes fatty acid use and attenuates diet-induced obesity. Diabetes. 58: 855–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langin D., Dicker A., Tavernier G., Hoffstedt J., Mairal A., Ryden M., Arner E., Sicard A., Jenkins C. M., Viguerie N., et al. 2005. Adipocyte lipases and defect of lipolysis in human obesity. Diabetes. 54: 3190–3197 [DOI] [PubMed] [Google Scholar]
- 35.Large V., Reynisdottir S., Langin D., Fredby K., Klannemark M., Holm C., Arner P. 1999. Decreased expression and function of adipocyte hormone-sensitive lipase in subcutaneous fat cells of obese subjects. J. Lipid Res. 40: 2059–2066 [PubMed] [Google Scholar]
- 36.Hellström L., Langin D., Reynisdottir S., Dauzats M., Arner P. 1996. Adipocyte lipolysis in normal weight subjects with obesity among first-degree relatives. Diabetologia. 39: 921–928 [DOI] [PubMed] [Google Scholar]
- 37.Alsted T. J., Nybo L., Schweiger M., Fledelius C., Jacobsen P., Zimmermann R., Zechner R., Kiens B. 2009. Adipose triglyceride lipase in human skeletal muscle is upregulated by exercise training. Am. J. Physiol. Endocrinol. Metab. 296: E445–E453 [DOI] [PubMed] [Google Scholar]
- 38.Yao-Borengasser A., Varma V., Coker R. H., Ranganathan G., Phanavanh B., Rasouli N., Kern P. A. 2011. Adipose triglyceride lipase expression in human adipose tissue and muscle. Role in insulin resistance and response to training and pioglitazone. Metabolism. 60: 1012–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Estrada-Smith D., Castellani L. W., Wong H., Wen P. Z., Chui A., Lusis A. J., Davis R. C. 2004. Dissection of multigenic obesity traits in congenic mouse strains. Mamm. Genome. 15: 14–22 [DOI] [PubMed] [Google Scholar]
- 40.Mehrabian M., Wen P. Z., Fisler J., Davis R. C., Lusis A. J. 1998. Genetic loci controlling body fat, lipoprotein metabolism, and insulin levels in a multifactorial mouse model. J. Clin. Invest. 101: 2485–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizutani S., Gomi H., Hirayama I., Izumi T. 2006. Chromosome 2 locus Nidd5 has a potent effect on adiposity in the TSOD mouse. Mamm. Genome. 17: 375–384 [DOI] [PubMed] [Google Scholar]
- 42.Young T. L., Penney L., Woods M. O., Parfrey P. S., Green J. S., Hefferton D., Davidson W. S. 1999. A fifth locus for Bardet-Biedl syndrome maps to chromosome 2q31. Am. J. Hum. Genet. 64: 900–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rice T., Chagnon Y. C., Perusse L., Borecki I. B., Ukkola O., Rankinen T., Gagnon J., Leon A. S., Skinner J. S., Wilmore J. H., et al. 2002. A genomewide linkage scan for abdominal subcutaneous and visceral fat in black and white families: the HERITAGE Family Study. Diabetes. 51: 848–855 [DOI] [PubMed] [Google Scholar]
- 44.Rankinen T., Zuberi A., Chagnon Y. C., Weisnagel S. J., Argyropoulos G., Walts B., Perusse L., Bouchard C. 2006. The human obesity gene map: the 2005 update. Obesity (Silver Spring). 14: 529–644 [DOI] [PubMed] [Google Scholar]
- 45.Wang H., Bell M., Sreenevasan U., Hu H., Liu J., Dalen K., Londos C., Yamaguchi T., Rizzo M. A., Coleman R., et al. 2011. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J. Biol. Chem. 286: 15707–15715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmadian M., Abbott M. J., Tang T., Hudak C. S., Kim Y., Bruss M., Hellerstein M. K., Lee H. Y., Samuel V. T., Shulman G. I., et al. 2011. Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype. Cell Metab. 13: 739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang X., Lu X., Lombes M., Rha G. B., Chi Y. I., Guerin T. M., Smart E. J., Liu J. 2010. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab. 11: 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.