Abstract
The DAZ family of genes are important fertility factors in animals, including humans. The family consists of Y-linked DAZ, and autosomal homologs Boule and Dazl. All three genes encode RNA-binding proteins that are nearly exclusively expressed in germ cells. The DAZ family is highly conserved, with ancestral Boule present in sea anemones through humans, Dazl conserved among vertebrates, and DAZ present only in higher primates. Here we review studies on DAZ family genes from multiple organisms, and summarize the common features of each DAZ gene and their roles during spermatogenesis in animals. DAZ family proteins are thought to activate the translation of RNA targets, but recent work has uncovered additional functions. Boule, Dazl, and DAZ likely function through similar mechanisms, and we present known functions of the DAZ family in spermatogenesis, and discuss possible mechanisms in addition to translation activation.
Keywords: BOULE, DAZ, DAZL, evolution, RNA-binding proteins, translation regulation
Introduction
Infertility affects approximately 10% of couples, with half of these cases attributable to the male partner.1 Though many of these cases are idiopathic, a high proportion of men with non-obstructive azoospermia (no sperm produced) have a microdeletion on the Y chromosome.2 The discovery of such deletions led to the proposal of an “Azoospermia Factor” (AZF) as a genetic cause for some cases of infertility.2 The AZF region has been further mapped into the three candidate regions AZFa, AZFb and AZFc,3,4 each deleted in subsets of infertile men. Among the handful of genes in these regions, Deleted in Azoospermia (DAZ) was found to be deleted in 12–15% of a cohort of azoospermic men, making it a strong candidate for the AZFc gene.5 DAZ is part of a large gene family (DAZ family) with autosomal homologs Dazl (DAZ-Like) and Boule, all of which encode RNA-binding proteins.6-9 DAZ family genes are reproduction-specific and present in nearly all animals,10 making them an important gene family in reproduction. The identification of the DAZ family has led to research into genetic causes of infertility, and expansive work into understanding how the DAZ family regulates fertility.
RNA-binding proteins are abundant during spermatogenesis, largely involved in post-transcriptional regulation. During spermatogenesis, extensive translational regulation is used to control the proper timing of differentiation, particularly during spermiogenesis, the differentiation of round spermatids into mature sperm (reviewed in refs. 11, 12). Many genes are transcribed several days before translation occurs, necessitating a network of mRNA storage and translational control. In addition to mRNA regulation, multiple species of non-coding small RNAs have been identified in the testis. These include miRNAs, piRNAs and MSY-RNAs, though how they intersect with translation regulation and sperm differentiation is unclear.13-18 Some of this RNA storage and control has been proposed to occur at the chromatoid body, a perinuclear structure most prevalent in round spermatids that contains mRNA, miRNA and several RNA-binding proteins (reviewed in ref. 19). The presence of such a structure and the abundance of multiple classes of small RNAs highlight the importance of RNA binding proteins during spermatogenesis.
The DAZ family of proteins is thought to be involved in the translation activation of mRNA targets.20,21 In recent years, relevant candidate targets have been identified, and the mechanism underlying this regulation is becoming clearer. Additionally, novel roles for DAZ family genes in mRNA transport and stability have been discovered. Here we review the functions of the DAZ family of genes during spermatogenesis, and discuss the various models of their action.
Evolution of the DAZ Gene Family
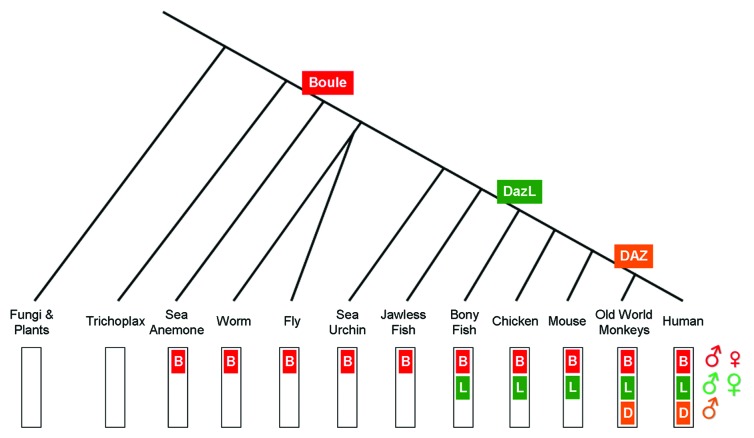
After the discovery of DAZ as a candidate gene for AZF,5 the identification of homologs in other species revealed a larger gene family. DAZ family genes have a common structure consisting of a RNA-Recognition Motif (RRM) and at least one copy of a motif rich in basic amino acids termed the DAZ repeat.5,6,8,22-24 Boule is the ancestral member of the family, and is widely conserved across Metazoan, from the sea anemone through humans (Fig. 1).10,23,25 Boule is autosomal and has a single RRM and one DAZ repeat.8,23,25 The RRM is highly conserved among all Boule homologs, with a distinct signature in the RNP1 and RNP2 motifs within the RRM. Boule is not found in fungi or plants, indicating that the DAZ family is an animal specific family of reproduction genes.10
Figure 1.DAZ family evolution. Boule is the ancestral member of the family, and is conserved from the sea anemone through humans, but is not present in Trichoplax, fungi or plants. A duplication of Boule during early vertebrate evolution led to Dazl. Dazl was then duplicated and transposed onto the Y chromosome in the evolution of old world monkeys. It further expanded into a cluster of multiple DAZ genes in the evolution of human lineage. Symbols at right indicate sex-specific roles. Boule has predominantly testis functions with occasional ovarian roles, Dazl functions in testes and ovaries, while DAZ is testis-specific.
Dazl arose from a duplication of Boule during vertebrate evolution (Fig. 1).8,10Dazl homologs are also autosomal with only one RRM and one DAZ repeat,6,9,22,24 and are distinguishable from Boule homologs by unique sequences in the RNP1 and RNP2 motifs.8,10Dazl arose around the time of vertebrate radiation, and homologs are conserved from bony fish through humans,6,9,10,22,24,26-28 but are not present in cartilaginous or jawless fish.10
During primate evolution, a duplication and transposition of Dazl onto the Y chromosome led to DAZ (Fig. 1).6,7,29 Subsequent duplication and gene pruning led to four DAZ genes in two clusters, each with multiple numbers of DAZ repeats and two with duplications of the RRM.30 The number of DAZ repeats among the DAZ genes is polymorphic both between and within individuals.30,31 DAZ homologs are only present in humans and catarrhine primates (old world monkeys).7,24,29,32
Surprisingly, sequence analysis has shown that the presence of DAZ has had little effect on either Dazl or Boule gene evolution in primates, indicating strong functional constraint on these two genes.33 DAZ itself has a higher rate of genetic changes,34 but neither nonsense nor frameshift mutations affecting the ORF have been detected, suggesting positive selection on DAZ.35 Dazl homologs have a higher rate of change than Boule,33 while Boule homologs have been shown to be under purifying selection.10 Indeed, no polymorphisms within the Boule coding region were detected among more than 200 fertile and infertile men examined in two different studies,8,36 further indicating a strong functional constraint. Such a high level of conservation is rare among reproductive genes, suggesting that Boule has an essential germ cell role in animals. Similarly, the continued maintenance of multiple gene duplications suggests that all DAZ family genes are critical regulators of fertility.
DAZ Family Gene Expression
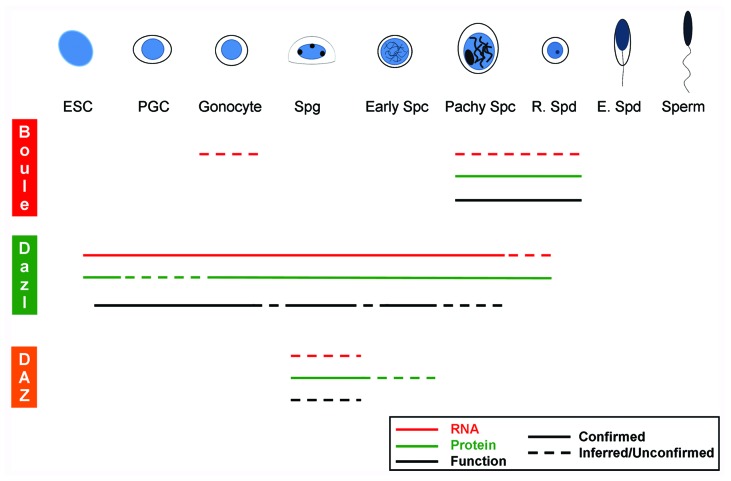
Though each DAZ family gene has a unique expression pattern, the whole family is restricted to germ cells in nearly all animals. Despite the presence of newer members Dazl and DAZ, reproduction-specific expression has been preserved for all three DAZ family genes. While there is some species specific expression for each DAZ family homolog, each gene has maintained the same general pattern across species. Gene families may often show similarities in expression among species, but such clear conservation of homolog-specific expression is unusual. This phenomenon allows a composite picture of common RNA and protein expression to be constructed for each DAZ family homolog, summarized in Figure 2 (red and green lines, respectively). This summary view does not represent the data from any single species, but rather the common expression patterns seen in multiple organisms. Data from specific species is discussed below, with a focus on each homolog’s common pattern of expression.
Figure 2. Common expression and functions of DAZ family genes. Data from multiple species is combined to present a picture of common expression patterns and functions for each DAZ gene during spermatogenesis. A schematic of different steps of germ cell specification and spermatogenesis is shown at top, and mRNA expression (red lines) and protein expression (green lines) relative to these steps is shown for each gene. Black lines represent steps in spermatogenesis where each gene is known to have a function. Solid lines represent data confirmed in at least two studies, while dashed lines are either unconfirmed, or inferred from known expression or function data. Boule protein is present in pachytene spermatocytes through round spermatids (solid red line), and functions in both meiosis and spermiogenesis (solid black line). Boule mRNA is presumed to be present (dashed red line) in cells with Boule protein. Dazl homologs are expressed continuously from embryonic stem cells through round spermatids (red and green lines), and are known to function in ESCs, PGCs, gonocytes, spermatogonia and early spermatocytes (solid black lines). Dazl likely functions in other cells where it is expressed (dashed black line), but this has not been shown. DAZ is known to be expressed in spermatogonia (solid green line), and presumably functions in those cells, though it has not been shown (dashed black line). For details, see text. ESC, embryonic stem cell; PGC, primordial germ cell; Spg, spermatogonia; Early Spc, leptotene/zygotene spermatocyte; Pachy Spc, pachytene spermatocyte; R. Spd, round spermatid; E. Spd, elongating spermatid.
At least one DAZ family homolog is expressed in nearly every stage of spermatogenesis. Though spermatogenesis begins when spermatogonia differentiate, it can be traced back to the differentiation of a subset of embryonic stem cells (ESCs) into primordial germ cells (PGCs). These migrate to the embryonic gonad and become gonocytes (also called prospermatogonia), which proliferate further and eventually become spermatogonia, containing the adult stem cell population. Spermatogonia proliferate further and give rise to primary spermatocytes, which undergo meiosis to produce haploid round spermatids. Through a process called spermiogenesis, round spermatids undergo dramatic morphological changes and first become elongating spermatids, and are finally released from the testis as mature spermatozoa.
Boule homologs are predominantly transcribed in the testes of fruit flies, sea urchins, chickens, mice and primates,10 though mRNA expression is reported in the ovaries of C. elegans, medaka fish and at low levels in mice.10,25,37,38 However, in these three species, testis transcription is also observed. Furthermore, since the only instance of Boule protein in ovaries is in C. elegans,37Boule transcriptions in the ovaries of fish and mice may not lead to protein. Similarly, we have seen low levels of Boule mRNA in embryonic gonocytes in mice,10 but do not detect protein (our unpublished observations). Additionally, boule mRNA is detected in the brains of flies,39,40 but this has not been reported in any other animals.
In the testis, Boule proteins are first present in mid-pachytene spermatocytes and remain through metaphase spermatocytes, with peak levels occurring just before the metaphase of cell division. Boule protein then persists into round spermatids, but is gone by the time elongation begins (Fig. 2, Boule green line).8,41,42 This is true in flies, mice and humans.8,41 Cell-type specific Boule mRNA expression has only been examined in medaka fish, where it is similarly present in both spermatocytes and spermatids.38 This mRNA pattern is likely the same in both flies and mammals, given the similar protein expression patterns, but it has not been confirmed.
Dazl homolog expression has diverged from Boule, and homologs are expressed in both males and females in all species so far examined.10Dazl homologs are initially expressed in ESCs through PGC specification in frogs, fish and mammals (Fig. 2, Dazl red and green lines). In Xenopus, Xdazl mRNA and protein are both present in the embryonic germ plasm,26,43 and zebrafish zDazl mRNA is detected in the vegetal pole of embryos,28 a region that later gives rise to germ cells.44 Similarly, mouse Dazl mRNA and human DAZL mRNA and protein are present in ESCs through PGCs.45-47 Together, these expression data indicate common expression of Dazl during PGC determination of early embryogenesis.
After germ cells are specified, Dazl homologs continue to be expressed in gonocytes, through spermatogonia and spermatocytes, and into early round spermatids (Fig. 2). Such protein expression is seen in humans, mice and frogs,8,48-50 while underlying mRNA expression has been confirmed in mammals only through pachytene spermatocytes.22,51-53 Though reports of post-meiotic Dazl expression in mammals have differed,8,33,48 we have detected Dazl protein in both mouse and human spermatids (our unpublished observations), and Dazl is present post-meiotically in Xenopus.50 Additionally, a transgenic reporter driven by the Dazl promoter in mice has shown a similar transcription pattern.54 Furthermore, one study found human DAZL protein in sperm tails,55 but this result has not been repeated. Such discrepancies in the reports of Dazl expression are likely due to the use of different antibodies recognizing varying antigens that are differentially accessible during spermatogenesis. Despite the absence of confirmed protein and mRNA data at each stage and slight variations in species-specific Dazl expression,32 a common pattern of continuous Dazl expression from ESCs through haploid spermatids is clear (Fig. 2).
While several papers have reported DAZ gene expression, a consensus pattern is not yet clear. In addition to differing antibodies, cross-reactivity with Dazl proteins was sometimes unavoidable. Habermann et al. found DAZ2 protein in mature sperm tails,56 but this was not repeated in two other studies.48,57 Using several antibodies to control for cross-reactivity with Dazl, Reijo et al. showed that DAZ is present only in spermatogonia and spermatocytes, with rare expression in spermatids.48 DAZ protein has been confirmed in human spermatogonia, though not in spermatocytes (Fig. 2, DAZ green line).57 All four DAZ genes are transcribed in humans,57 further complicating expression studies of DAZ.
Interestingly, though DAZ family proteins are predominantly present in the cytoplasm, occasional nuclear localization is also observed. In flies, Boule protein is initially in the nucleus of early spermatocytes, and then transits to the cytoplasm just before metaphase,41 though this is not seen in mammals.8 Similarly, human and mouse Dazl is nuclear in gonocytes,48,58 and may translocate from the nuclei of spermatogonia into the cytoplasm of spermatocytes.48 This translocation of Dazl has not been confirmed, but nonetheless raises an intriguing question of why DAZ family proteins localize to the nuclei of certain cell types. DAZ family proteins may sequester certain transcripts in the nucleus, or could be involved in mRNA processing. However, nuclear localization of Drosophila Boule is dispensable for its function, raising the possibility that Boule is simply stored in the nucleus prior to its function in the cytoplasm.41 However, the common presence of Dazl in the nuclei of spermatogonia in multiple species suggests important functionality. What this role is remains to be seen, but will likely be important in fertility.
Functions of DAZ Genes During Spermatogenesis
Because of the broad evolutionary conservation of the DAZ family, the functions of DAZ genes have been examined in a number of species, revealing requirements for DAZ family genes at multiple points during spermatogenesis (summarized in Figure 2, black lines). DAZ is known to be important in human spermatogenesis since its deletion is associated with azoospermia.5 However, causative point mutations in infertile men have yet to be identified, and some DAZ deleted men still produce low levels of sperm.59 Indeed, men with DAZ deletions have fathered children both through the use of reproductive technologies60,61 and, though rare, naturally,3,62 indicating that DAZ is not absolutely required for spermatogenesis. However, the presence of DAZ within the AZFc region as well as studies detailed below about spermatogenesis requirements of other DAZ family members strongly suggest that DAZ plays a critical role in normal spermatogenesis.
In accordance with its broad expression pattern, Dazl has been shown to have multiple roles throughout spermatogenesis. In frogs and mice, Dazl is initially important for PGC proliferation and development. Knockdown of Xenopus Xdazl leads to few surviving PGCs, and those that do survive fail to migrate.43 Similarly, Dazl knockout mice have few germ cells that survive into the adult, in both males and females.49 This defect is first evident at the gonocyte stage in embryonic testes. In a mixed genetic background, Dazl null testes are sparsely populated with germ cells by embryonic day 19 (E19),49 while increased germ cell apoptosis is seen by E14.5 in a pure C57/Bl6 background.63 Additionally, Dazl null ESCs fail to differentiate to PGCs in vitro, while PGCs in vivo fail to properly erase genomic methylation marks.45 These defects together indicate a problem in PGC development and differentiation, similar to those seen in Xenopus. Though few PGCs are present in Dazl null mice and frogs, the presence of germ cells indicates that germ cell specification is occurring in the absence of Dazl. However, the in vitro ESC differentiation defect may hint at a role in germ cell specification.
Further studies on Dazl null mice on a mixed genetic background have shown additional roles for Dazl in both spermatogonia and early spermatocytes. Though most germ cells are absent at birth, some As (A-single) and Apr (A-paired) spermatogonia survive, but most do not progress beyond the Aal (A-aligned) stages, revealing a function for Dazl in spermatogonia differentiation.64 The few cells that do pass this block are able to enter meiosis, but synaptonemal complexes necessary for homologous recombination fail to form in postnatal day 19 (p19) knockout mice, and spermatocytes cannot progress beyond leptonema.65 Null germ cells in the pure C57/Bl6 background fail to induce meiosis genes in response to the meiotic signal retinoic acid, showing a further requirement for Dazl at the onset of meiosis.66 This range of defects is specific to germ cells, as wild type spermatogonia can colonize and repopulate a Dazl null testis.67 Taken together, Dazl has functions during PGC development and migration, spermatogonia differentiation and the onset and progression of meiosis (Fig. 2). Dazl presumably also functions between these known steps, corresponding to known expression, but explicit demonstrations of such roles have not yet been shown.
Boule functions complement those of Dazl, and homologs are important for meiotic division and spermatid differentiation. In Drosophila, boule mutant flies have a male-specific arrest at pachynema, prior to metaphase.23 However, meiosis completes normally in Boule knockout mice, and haploid round spermatids are abundant.42 Instead, there is a global arrest at step 6 of spermiogenesis, with varying defects in acrosome biogenesis and a complete lack of elongating spermatids.42 Despite the lack of a meiosis phenotype in Boule null mice, Boule regulation of meiosis is likely conserved among animals. A pachytene arrest similar to that seen in flies occurs in C. elegans with a mutation in the Boule homolog daz-1, but only in females.25 In addition, a human BOULE transgene can restore meiosis in boule mutant flies,68 and a lack of BOULE protein has been associated with meiotic arrest in men.36 We therefore proposed that Dazl and Boule redundantly regulate the progression to meiotic metaphase in mice, and that Dazl can compensate for the loss of Boule in spermatocytes.42 While this model has not yet been tested, the accumulating evidence suggests that Boule regulation of meiosis is conserved in mammals.
Additionally, though knockout mice revealed a novel role for Boule in spermatid differentiation, this function may also be present in flies. In boule mutant flies, it was noted that the pachytene-arrested spermatocytes did not differentiate,23 a phenotype not common to other meiosis-arrest mutants. For example, flies with a mutation in the putative Boule target, twine, have a similar meiotic arrest, but many meiosis-arrested spermatocytes in those flies begin to elongate.69,70 Since differentiation was also disrupted in Drosophila boule mutants, this suggests that the spermiogenesis function of Boule is also conserved.
Regardless of which specific spermatogenesis roles are conserved, the male-fertility requirement of Boule is the same between flies and mice. Despite more than 500 million years of evolution separating mice and flies, Boule mutations in both species lead to a complete lack of sperm due to a global arrest in spermatogenesis, and the presence of similar-looking multinucleate cysts in the testis (Fig. 3). Furthermore, these testes defects are the only phenotype reported in Boule null animals of either species,10,23,39,40,42 highlighting the conservation of a male fertility requirement of Boule.
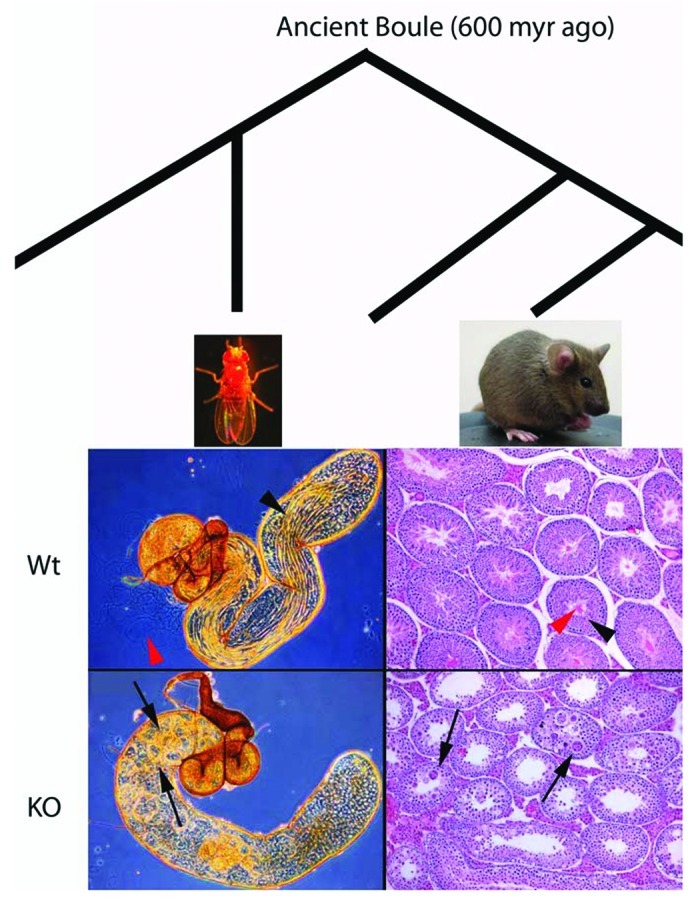
Figure 3.Boule testis function is conserved in flies and mice. Despite the wide divergence of flies and mice, Boule mutations in each species leads to male-only sterility due to a global arrest of spermatogenesis. Mature sperm (red arrowhead) and elongating spermatids (black arrowhead) are abundant in the testis tube of wild type flies, the same is true for the wild type mouse testis section. Mature sperm (red arrowhead) are seen in the lumen of seminiferous tubules and elongating spermatids are often located next to the lumen (black arrowhead). However both mature sperm and elongating spermatids are completely absent in Boule knockout testes of both species. Multinucleate cysts are prevalent throughout null testes of both animals (arrows), further highlighting the conserved spermatogenesis function of Boule. Fly testes are from 1-d old males, and images taken at 10x. Mouse testes are from 3 mo old mice, and images taken at 10x. Myr-million years.
Many experiments have shown a remarkable ability of the DAZ family genes to functionally replace each other. Both human DAZ and DAZL can partially restore germ cell numbers in Dazl null mice, though the rescue was moderate and variable among animals.71,72 These experiments showed that human DAZ can function during mammalian spermatogenesis, despite the lack of direct evidence that DAZ is necessary for human spermatogenesis. Interestingly, Xenopus Xdazl can restore meiosis in boule mutant flies,26 similar to the human BOULE rescue discussed above,68 further supporting the model of Boule and Dazl redundancy during mammalian meiosis.42
Finally, all three DAZ family genes have been shown to enhance human ESC differentiation into germ cells in vitro, with overexpression of each DAZ family gene alone or in combination leading to varying degrees of enhancement.46 When all three were expressed together, ESCs were able to differentiate into germ cells with molecular features of spermatids, highlighting the wide range of functions DAZ family genes play in spermatogenesis. A similar transient overexpression of Dazl in mouse ESCs was also able to promote germ cell differentiation.73 These ectopic expression studies may not reflect in vivo functions, however. For example, Boule overexpression enhanced PGC differentiation in XX (female) ESCs, but not XY (male) ESCs.46 However, Boule expression has not been reported in ESCs of either sex, and Boule null female mice have no germ cell defects.10,42 While these experiments suggest in vivo roles for the ESC-expressed Dazl, the results for Boule and DAZ underscore the ability for compensation among the DAZ family when expressed in the right time and place. Together with the data that different homologs can functionally replace each other, the similar in vitro results for each DAZ family gene suggest that all DAZ family genes can function through similar mechanisms.
Candidate RNA Targets for DAZ Family Proteins
Though many functions of DAZ family genes are known (discussed below), relevant and validated RNA targets are less clear. Drosophila boule enhances translation of a Lac-Z reporter carrying the 3′ UTR of the Cdc25 homolog twine in vivo, suggesting that the fly germ cell-specific Cdc25 homolog is the downstream target of Boule.74 In addition, a twine transgene with a tubulin 3′ UTR was able to rescue meiosis in boule mutant flies, indicating that twine translation is absent in mutants. Though this model nicely explains the observed meiosis arrests in both Drosophila and C. elegans,8,25,74 no direct interaction between any Boule and Cdc25 homologs has been shown.
Subsequent research into targets has focused on Dazl homologs. In homopolymeric binding studies, frog, mouse and human Dazl preferentially bind polyU RNA.26,75 A SELEX approach identified a (G/CUn)n motif which is found in the 5′UTR of mouse Cdc25c,76 and zebrafish zDazl binds to GUUC, a site found in the 3′UTR of Drosophila twine RNA.77 Using GST-Dazl bound to a column, a 26bp motif was identified that is present in the Cdc25a 3′ UTR.78 Another study identified targets bound by both Dazl and Pumilio2 (Pum2),79 an RNA-binding protein shown to interact with Dazl.80 Follow-up studies confirmed Dazl binding to a U-rich motif in Sdad1 mRNA, another cell-cycle regulator first identified in yeast.79
However, despite multiple reports of candidate targets, there is no overlap between lists, and a discrepancy in the Dazl binding site, perhaps due to the in vitro approaches used in these experiments. Binding conditions are unlikely to match those found in vivo, mRNAs normally in separate cells from Dazl are inappropriately brought together, or legitimate targets may be tightly bound by endogenous protein, therefore preventing their binding to Dazl in vitro. To determine in vivo targets, Reynolds et al. used endogenous immunoprecipitation from whole mouse testes followed by a microarray on co-precipitating RNA, and identified 15 targets with high confidence.81 Targets were further validated by IP followed by RT-PCR on UV-crosslinked testes to reduce non-specific interactions. Dazl binding to Mvh (Mouse vasa homolog) and Sycp3 (Synaptonemal complex protein 3) has been confirmed, and translation defects for both of these targets occurs in Dazl null animals.81,82
Additionally, the presence of the proposed binding sites in the 15 target genes was analyzed.81 The initial 26bp motif78 was only found in six targets, and was not present significantly more than predicted by chance.81 The SELEX defined (G/CUn)n motif, however, was statistically over-represented in the 3′UTRs of targets, and was found to be in evolutionarily conserved regions of the transcripts. This motif was also found in the eight targets previously reported by Jiao et al.,81 providing strong support for a common U2–10(G/C)U2–10 binding motif among Dazl targets. It is not known how prevalent this motif is among testis transcripts, but the flexible nature of this motif suggests that Dazl may bind a wide range of mRNAs.
Notably, using in vivo UV-crosslinking followed by IP and RT-PCR, Reynolds et al. failed to detect Dazl binding to Prm2,81 a target identified by an in vitro screening method.79 This experiment showed that while in vitro binding studies can correctly uncover a particular binding motif, the specific targets identified may not be relevant in vivo. Therefore, while multiple studies have examined targets of Dazl, only a few candidates have in vivo significance.
The most promising in vivo targets are Mvh, Sycp3, and Cdc25 homologs (Table 1). Three reports showing Dazl binding to Cdc25 homologs76-78 together with meiosis rescue studies in flies26,68,74 is strong evidence for in vivo Cdc25 binding. Mice have three Cdc25 genes,83,84 and Venables et al. only detected binding to Cdc25c,76 while Jiao et al. could only detect binding to Cdc25a.78 Both of these Cdc25 genes are abundantly expressed in the testis,83,84 and whether the differential binding is due to the different techniques used, or represents artificial binding due to the in vitro systems remains to be seen.
Table 1. Candidate in vivo mRNA targets.
| Gene | Candidate Target | Motif Bound | References |
|---|---|---|---|
| Fly boule, Zebrafish zDazl |
twine |
GUUC |
74, 77 |
| Mouse Dazl |
Cdc25 |
|
|
| |
Cdc25c 5′ UTR |
GU7GU10GU10GU7 |
76 |
| |
Cdc25a 3′UTR |
U/AA/GUUC/UAGUAU/AAANAACUUUG/UGAAU/AUG/A |
78 |
| |
Mvh |
UUCUUCUGUUCUU |
81 |
| Sycp3 | U6GU3GU3GU4 | 81, 82 |
Though many mRNA targets have been reported, the in vivo candidates shown fulfill three criteria: (1) demonstrated in vivo binding, (2) reported translational defect in Boule or Dazl null animal model and (3) functional relevance to Boule or Dazl null phenotype.
Mvh and Sycp3 were both identified by the in vivo approach,81 and are related to known Dazl functions. Mvh is highly expressed in all germ cells, similar to Dazl, and Vasa homologs have conserved roles in PGC differentiation.85 In addition, the male sterile phenotype in Mvh knockout mice is due to a final arrest at zygonema,85 close to the reported leptotene arrest seen in mixed background Dazl null mice.65 Similarly, Sycp3 is an essential part of the synaptonemal complex that forms during the early stages of meiosis, a time when Dazl has been shown to function.45,65,66 A demonstrated in vivo interaction, translation defects in knockouts and relevance to the observed phenotype together make these genes the best candidate targets so far reported, though none of these targets are a “magic bullet” that explains the primary Dazl null phenotype. Similar physiologically relevant data are needed for other reported targets in order to confirm their in vivo regulation by Dazl.
Using a similar in vivo immunoprecipitation approach, we have identified the first candidate targets for Boule in mice (VanGompel and Xu, in preparation). We were able to detect interactions between Boule and Prm1 and Prm2 mRNAs, genes important for round spermatid differentiation. While not a complete list, these targets are directly relevant to the major phenotype of spermiogenesis arrest in Boule null mice.
The DAZ Family as Translational Activators
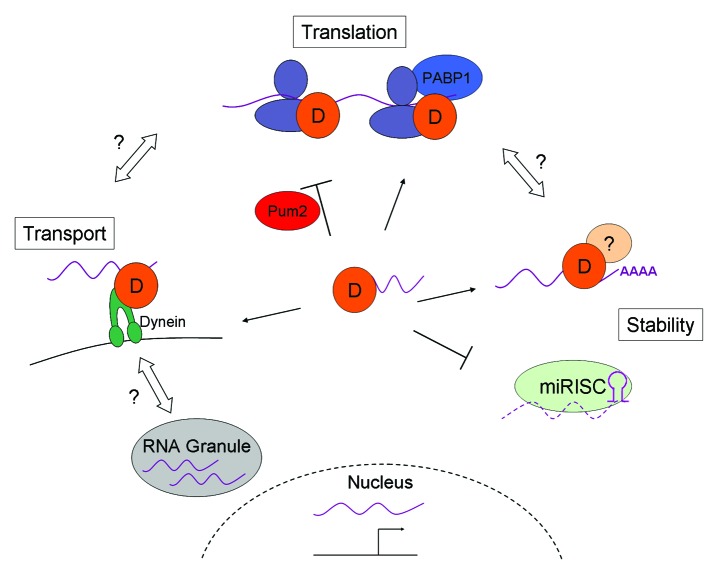
Most studies have focused on the DAZ family as translational activators. This model was first established through the studies in Drosophila discussed above, implicating boule in the translational regulation of twine.74 Similarly, zebrafish zDazl can stimulate translation of a luciferase reporter fused to the twine 3′ UTR in cell culture.77 Using a tethered assay in Xenopus oocytes in which proteins were forced into proximity of reporter mRNAs, Dazl homologs stimulated translation through enhanced recruitment of 80S ribosomes.86 This translation activation was dependent on an interaction with Poly(A) Binding Protein 1 (PABP1), but still occurred in the absence of poly(A) tails on reporter constructs. This led to a model in which Dazl recruits PABP1 to mRNAs in the absence of an adequate poly(A) tail, and thus promotes translation.86 This model is particularly intriguing in the context of mammalian spermatogenesis because several transcripts are known to be deadenylated prior to translation.87,88 Xdazl was similarly shown to activate translation of RINGO/Spy mRNA, but in a Pum2 dependent manner.89 Dazl bound target mRNA, but could not activate translation until the translational repressor Pum2 dissociated from the transcript. This shows that the function of the DAZ family is context dependent, and may vary depending on the stage of spermatogenesis and what other proteins are present at any given time (Fig. 4). Additionally, while the general mechanisms are likely to be broadly conserved, the specific contexts and players involved may differ among species. Further studies into how other interacting proteins regulate DAZ family function will prove fruitful.
Figure 4. Model for DAZ family gene functions. DAZ family proteins likely function through similar mechanisms and a composite model representing known cytoplasmic functions of DAZ family proteins is presented. A generic DAZ family protein (orange circle labeled “D”) bound to RNA is represented in the middle of the figure. DAZ family proteins have multiple functions, likely dependent on which protein partner they are bound to. Binding to PABP1 promotes association with ribosomes and translation, while binding to the repressor Pum2 inhibits translation. Interactions with Dynein may mediate transport of mRNA targets. Where mRNA is transported is unknown, but DAZ family proteins may transport targets to and from RNA granules, such as the chromatoid body, or to polysomes for translation activation. DAZ family proteins may promote mRNA stability either through the inhibition of miRNA-mediated degradation, or the promotion of polyadenylation through binding to an unknown factor (beige circle labeled “?”). Increased stability may enhance translation, or vice versa. Solid lines represent known mechanisms, while open double-sided arrows represent speculated links between known roles.
While these studies examined the mechanism of DAZ family function in vitro, others have shown a similar translational role in vivo. Mammalian Dazl is present in active polysomes in mouse testes,75 and Drosophila boule enhances the translation of a transgenic reporter through the twine 3′ UTR.74 As discussed previously, translation of the two Dazl candidate targets, Mvh and Sycp3, was reduced in Dazl null testes, further suggesting that Dazl is a translational activator in vivo.81,82 Protein of these targets was still detectable in Dazl knockouts, however, indicating that Dazl is acting as an enhancer of translation, and not an essential activator as proposed in the Boule-Cdc25 model. While the combined evidence for the translational regulation of mRNA targets is strong, it is important to note that these data cannot yet account for the dramatic loss of germ cells in Dazl null mice. Key targets that regulate germ cell numbers may not yet be identified, or alternate functions that have broader effects may cause the observed phenotype.
Non-Translational Roles for the DAZ Family
In addition to translation activation, DAZ family genes have other roles. Several binding partners have been identified (Table 2, Figure 4), and interactions with other RNA-binding proteins is a common theme. The mammalian proteins can form homo- and heterodimers,8,58,80,90 further indicating similar functions, and suggesting a possible mechanism for the flexibility in functionality observed in ectopic rescue studies. The translational repressor Pum2 can bind all three DAZ members in humans.80,90 PABP1 can interact with Boule, Dazl and DAZ homologs from frog, mouse and humans,86 while Xdazl also interacts with ePABP (embryonic PABP),86,89 and C. elegans DAZ-1 interacts with the CPEB (Cytoplasmic Polyadenylation Element Binding protein) homolog Cpb-3.91 Both human DAZ and DAZL have also been shown to interact with RNA-binding proteins hQK3 and DAZAP1.80,92 In addition, novel, non-RNA-binding proteins DZIP1, and DAZAP2 have been identified as binding partners through interaction screens with DAZ and DAZL.80,92,93 Such a variety of interactions further supports a model of context-dependent DAZ family function, where specific protein partners mediate a range of roles (Fig. 4). Indeed, which RNA targets DAZ family proteins are bound to may also depend on the context of other protein partners. The Pumilio family of RNA-binding proteins has been shown to differentially bind RNA targets based on what protein partners they are bound to,90 and DAZ family proteins may utilize a similar mechanism for binding different targets.
Table 2. DAZ family interacting proteins.
| Gene | Category | Partners | References |
|---|---|---|---|
| Boule |
DAZ Family |
Boule, Dazl, DAZ |
8, 80, 90 |
| |
RNA-Binding Proteins |
PABP1, Pum2, Cpb-3 |
86, 90, 91 |
| Dazl |
DAZ Family |
Boule, Dazl, DAZ |
8, 55, 80, 90 |
| |
RNA-Binding Proteins |
PABP1, ePABP, Pum2, hQK3,
DAZAP1 |
80, 86, 90, 92 |
| |
Other |
Dynein, Dzip1, DAZAP2 |
80, 92, 95 |
| DAZ |
DAZ Family |
Boule, Dazl |
55, 80 |
| |
RNA-Binding Proteins |
PABP1, Pum2, hQK3, DAZAP1 |
80, 86, 92 |
| Other | Dzip1, DAZAP2 | 80, 92, 93 |
Mouse Dazl was also found to interact with dynein light chain in mouse testes, and can move on the microtubule network in cell culture (Fig. 4).94 In a dynein-dependent manner in vitro, Dazl can transport mRNA carrying putative binding sites, including those found in candidate targets Tpx-1, Cdc25c and Mvh, on microtubules. These mRNAs formed perinuclear aggregates, at structures presumed to be stress granules, where ectopic Dazl also accumulated.94 Active mRNA transport in male germ cells is not well-studied, but has been reported. The testis specific kinesin KIF17b can shuttle protein-RNA complexes in and out of the nucleus,95 and also associates with Miwi and the chromatoid body (CB),96 suggesting transport of mRNA to and from the CB. In other cell types, mRNA is stored in stress granules to protect transcripts from degradation,97,98 a parallel the authors propose occurs with Dazl-bound targets.94 Further studies are needed to determine if Dazl transports targets to the CB or other RNA granules in germ cells, and whether other DAZ family proteins are similarly involved in transport.
Could the DAZ family transport RNA for safe storage? If protecting mRNA from degradation is important in spermatogenesis, what is the targeting mechanism? A likely candidate is through specific miRNAs (Fig. 4). miRNAs are known to inhibit translation of targets, and this inhibition is often due to miRNA-mediated mRNA degradation.99 Zebrafish zDazl was recently shown to prevent miRNA mediated decay of nanos1 and tdrd7 transcripts,100though direct binding to these mRNAs was not shown. Using injections into zebrafish embryos, the authors showed that zDazl prevents miRNA mediated inhibition of reporters, dependent on the presence of the GUUC binding motif. Furthermore, this motif does not overlap with the miRNA binding site, but was necessary for zDazl to stabilize the mRNA.100 Since miRNA is present in the chromatoid body, it is an intriguing possibility that DAZ family proteins are either protecting targets from miRNA within the CB, or are involved in transporting them away from miRISC (microRNA Induced Silencing Complex) in germ cells.
While surprising, reduction of mRNA levels of Dazl targets has also been reported. Though reduced translation was noted for Mvh and Sycp3 in Dazl null mice, transcripts were reduced in postnatal day 5 (P5) null testes, a result that contributed to their identification as targets.81 This instability was presumed to be a consequence of reduced translation, but a direct stability effect could not be ruled out.81 Furthermore, quantitative RT-PCR using Dazl null embryonic testes has also shown a reduction in mRNA of both of these targets.45,66 Those experiments focused on the ability of Dazl null germ cells to respond to meiosis signals, so the reduction was noted only as a failure to initiate meiosis. Additionally, a microarray study on P7 wild type and Dazl null testes found a large number of transcripts that were reduced in knockouts.101 A similar result was obtained in a human microarray study on men with DAZ deletions.102 These studies hint at a potential role for the DAZ family in maintaining RNA levels, though a direct role for this in vivo has not yet been shown. Determining if reductions in transcript levels are due to a direct loss of DAZ family genes will clarify these new data.
Finally, in the zebrafish miRNA study described above, zDazl induced polyadenylation of transcripts, a novel function for the DAZ family. This polyadenylation was independent of translation, indicating that mRNA stability is independent from translation activation. Furthermore, polyadenylation may be an alternate method of PABP recruitment and subsequent translation activation. How zDazl mediates polyadenylation is not known, but it is likely through an as yet unidentified binding partner (Fig. 4). Cytoplasmic polyadenylation is well described during oogenesis (reviewed in ref. 103), and is beginning to be appreciated in spermatogenesis.104 In one well-studied mechanism in females, CPEB binds to cytoplasmic polyadenylation elements and recruits the polyadenylation apparatus. As mentioned, C. elegans DAZ-1 interacts with a CPEB homolog,91 suggesting a role for DAZ family genes in polyadenylation in worms. In mice, knockout of the testis-specific cytoplasmic poly(A) polymerase Tpap leads to a spermiogenesis arrest similar to that seen in Boule knockouts.42,105 Whether Boule and Tpap interact is not known, but the similar knockout phenotypes suggest that they may function in the same pathway, perhaps through regulation of mRNA stability. It is also possible that translation activation is a consequence of increased mRNA stability through polyadenylation, and not a direct function of the DAZ family. Determining how Boule regulates targets will help determine if such mechanisms are broadly used, and what roles they play in spermatogenesis.
Conclusion
Recent findings are painting a new picture for DAZ family-mediated regulation of targets, beyond the simple model of translation activation. Their roles in translation activation have been well-established using many systems, but likely represent only one of many functions. Specific mechanisms may differ in the broad range of cell types in which this family functions, and DAZ family genes may play multiple roles within the same cells. This range of functions may be determined in part by which proteins the DAZ family is bound to at any given time. Yet despite the variety of functions and mechanisms, the DAZ proteins have been highly conserved, and can still functionally replace each other in limited contexts. Such strong selective pressure on reproductive genes is rare, and suggests an essential role for these genes in the germ cells of animals. While possible mechanisms are emerging, why these functions are required in germ cells of all animals, and why humans require more DAZ family genes than other species, are puzzles that remain. Much work is needed to address these interesting questions.
Acknowledgments
We would like to thank Dr. Jane Wu for comments on the manuscript. This work was funded by NIH U01 HD045871 and Northwestern Memorial Hospital EAM grants (E.Y.X.); M.J.W.V. was supported by the Cell and Molecular Basis of Disease training grant, funded by NIH T32 GM08061.
Footnotes
Previously published online: www.landesbioscience.com/journals/spermatogenesis/article/14659
References
- 1.de Kretser DM. Male infertility. Lancet. 1997;349:787–90. doi: 10.1016/S0140-6736(96)08341-9. [DOI] [PubMed] [Google Scholar]
- 2.Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum Genet. 1976;34:119–24. doi: 10.1007/BF00278879. [DOI] [PubMed] [Google Scholar]
- 3.Vogt PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, et al. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet. 1996;5:933–43. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- 4.Vogt PH. Human chromosome deletions in Yq11, AZF candidate genes and male infertility: history and update. Mol Hum Reprod. 1998;4:739–44. doi: 10.1093/molehr/4.8.739. [DOI] [PubMed] [Google Scholar]
- 5.Reijo R, Lee TY, Salo P, Alagappan R, Brown LG, Rosenberg M, et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet. 1995;10:383–93. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- 6.Saxena R, Brown LG, Hawkins T, Alagappan RK, Skaletsky H, Reeve MP, et al. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat Genet. 1996;14:292–9. doi: 10.1038/ng1196-292. [DOI] [PubMed] [Google Scholar]
- 7.Shan Z, Hirschmann P, Seebacher T, Edelmann A, Jauch A, Morell J, et al. A SPGY copy homologous to the mouse gene Dazla and the Drosophila gene boule is autosomal and expressed only in the human male gonad. Hum Mol Genet. 1996;5:2005–11. doi: 10.1093/hmg/5.12.2005. [DOI] [PubMed] [Google Scholar]
- 8.Xu EY, Moore FL, Pera RA. A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. Proc Natl Acad Sci U S A. 2001;98:7414–9. doi: 10.1073/pnas.131090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yen PH, Chai NN, Salido EC. The human autosomal gene DAZLA: testis specificity and a candidate for male infertility. Hum Mol Genet. 1996;5:2013–7. doi: 10.1093/hmg/5.12.2013. [DOI] [PubMed] [Google Scholar]
- 10.Shah C, Vangompel MJ, Naeem V, Chen Y, Lee T, Angeloni N, et al. Widespread presence of human BOULE homologs among animals and conservation of their ancient reproductive function. PLoS Genet. 2010;6:e1001022. doi: 10.1371/journal.pgen.1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun RE. Post-transcriptional control of gene expression during spermatogenesis. Semin Cell Dev Biol. 1998;9:483–9. doi: 10.1006/scdb.1998.0226. [DOI] [PubMed] [Google Scholar]
- 12.Kleene KC. Patterns, mechanisms, and functions of translation regulation in mammalian spermatogenic cells. Cytogenet Genome Res. 2003;103:217–24. doi: 10.1159/000076807. [DOI] [PubMed] [Google Scholar]
- 13.Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, et al. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc Natl Acad Sci U S A. 2006;103:2647–52. doi: 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–14. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–7. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 16.Maatouk DM, Loveland KL, McManus MT, Moore K, Harfe BD. Dicer1 is required for differentiation of the mouse male germline. Biol Reprod. 2008;79:696–703. doi: 10.1095/biolreprod.108.067827. [DOI] [PubMed] [Google Scholar]
- 17.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–4. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 18.Xu M, Medvedev S, Yang J, Hecht NB. MIWI-independent small RNAs (MSY-RNAs) bind to the RNA-binding protein, MSY2, in male germ cells. Proc Natl Acad Sci U S A. 2009;106:12371–6. doi: 10.1073/pnas.0903944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotaja N, Sassone-Corsi P. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat Rev Mol Cell Biol. 2007;8:85–90. doi: 10.1038/nrm2081. [DOI] [PubMed] [Google Scholar]
- 20.Reynolds N, Cooke HJ. Role of the DAZ genes in male fertility. Reprod Biomed Online. 2005;10:72–80. doi: 10.1016/S1472-6483(10)60806-1. [DOI] [PubMed] [Google Scholar]
- 21.Yen PH. Putative biological functions of the DAZ family. Int J Androl. 2004;27:125–9. doi: 10.1111/j.1365-2605.2004.00469.x. [DOI] [PubMed] [Google Scholar]
- 22.Cooke HJ, Lee M, Kerr S, Ruggiu M. A murine homologue of the human DAZ gene is autosomal and expressed only in male and female gonads. Hum Mol Genet. 1996;5:513–6. doi: 10.1093/hmg/5.4.513. [DOI] [PubMed] [Google Scholar]
- 23.Eberhart CG, Maines JZ, Wasserman SA. Meiotic cell cycle requirement for a fly homologue of human Deleted in Azoospermia. Nature. 1996;381:783–5. doi: 10.1038/381783a0. [DOI] [PubMed] [Google Scholar]
- 24.Reijo R, Seligman J, Dinulos MB, Jaffe T, Brown LG, Disteche CM, et al. Mouse autosomal homolog of DAZ, a candidate male sterility gene in humans, is expressed in male germ cells before and after puberty. Genomics. 1996;35:346–52. doi: 10.1006/geno.1996.0366. [DOI] [PubMed] [Google Scholar]
- 25.Karashima T, Sugimoto A, Yamamoto M. Caenorhabditis elegans homologue of the human azoospermia factor DAZ is required for oogenesis but not for spermatogenesis. Development. 2000;127:1069–79. doi: 10.1242/dev.127.5.1069. [DOI] [PubMed] [Google Scholar]
- 26.Houston DW, Zhang J, Maines JZ, Wasserman SA, King ML. A Xenopus DAZ-like gene encodes an RNA component of germ plasm and is a functional homologue of Drosophila boule. Development. 1998;125:171–80. doi: 10.1242/dev.125.2.171. [DOI] [PubMed] [Google Scholar]
- 27.Johnson AD, Bachvarova RF, Drum M, Masi T. Expression of axolotl DAZL RNA, a marker of germ plasm: widespread maternal RNA and onset of expression in germ cells approaching the gonad. Dev Biol. 2001;234:402–15. doi: 10.1006/dbio.2001.0264. [DOI] [PubMed] [Google Scholar]
- 28.Maegawa S, Yasuda K, Inoue K. Maternal mRNA localization of zebrafish DAZ-like gene. Mech Dev. 1999;81:223–6. doi: 10.1016/S0925-4773(98)00242-1. [DOI] [PubMed] [Google Scholar]
- 29.Seboun E, Barbaux S, Bourgeron T, Nishi S, Agulnik A, Egashira M, et al. Gene sequence, localization, and evolutionary conservation of DAZLA, a candidate male sterility gene. Genomics. 1997;41:227–35. doi: 10.1006/geno.1997.4635. [DOI] [PubMed] [Google Scholar]
- 30.Saxena R, de Vries JW, Repping S, Alagappan RK, Skaletsky H, Brown LG, et al. Four DAZ genes in two clusters found in the AZFc region of the human Y chromosome. Genomics. 2000;67:256–67. doi: 10.1006/geno.2000.6260. [DOI] [PubMed] [Google Scholar]
- 31.Yen PH, Chai NN, Salido EC. The human DAZ genes, a putative male infertility factor on the Y chromosome, are highly polymorphic in the DAZ repeat regions. Mamm Genome. 1997;8:756–9. doi: 10.1007/s003359900560. [DOI] [PubMed] [Google Scholar]
- 32.Gromoll J, Weinbauer GF, Skaletsky H, Schlatt S, Rocchietti-March M, Page DC, et al. The Old World monkey DAZ (Deleted in AZoospermia) gene yields insights into the evolution of the DAZ gene cluster on the human Y chromosome. Hum Mol Genet. 1999;8:2017–24. doi: 10.1093/hmg/8.11.2017. [DOI] [PubMed] [Google Scholar]
- 33.Tung JY, Luetjens CM, Wistuba J, Xu EY, Reijo Pera RA, Gromoll J. Evolutionary comparison of the reproductive genes, DAZL and BOULE, in primates with and without DAZ. Dev Genes Evol. 2006;216:158–68. doi: 10.1007/s00427-005-0039-2. [DOI] [PubMed] [Google Scholar]
- 34.Agulnik AI, Zharkikh A, Boettger-Tong H, Bourgeron T, McElreavey K, Bishop CE. Evolution of the DAZ gene family suggests that Y-linked DAZ plays little, or a limited, role in spermatogenesis but underlines a recent African origin for human populations. Hum Mol Genet. 1998;7:1371–7. doi: 10.1093/hmg/7.9.1371. [DOI] [PubMed] [Google Scholar]
- 35.Bielawski JP, Yang Z. Positive and negative selection in the DAZ gene family. Mol Biol Evol. 2001;18:523–9. doi: 10.1093/oxfordjournals.molbev.a003831. [DOI] [PubMed] [Google Scholar]
- 36.Luetjens CM, Xu EY, Rejo Pera RA, Kamischke A, Nieschlag E, Gromoll J. Association of meiotic arrest with lack of BOULE protein expression in infertile men. J Clin Endocrinol Metab. 2004;89:1926–33. doi: 10.1210/jc.2003-031178. [DOI] [PubMed] [Google Scholar]
- 37.Maruyama R, Endo S, Sugimoto A, Yamamoto M. Caenorhabditis elegans DAZ-1 is expressed in proliferating germ cells and directs proper nuclear organization and cytoplasmic core formation during oogenesis. Dev Biol. 2005;277:142–54. doi: 10.1016/j.ydbio.2004.08.053. [DOI] [PubMed] [Google Scholar]
- 38.Xu H, Li Z, Li M, Wang L, Hong Y. Boule is present in fish and bisexually expressed in adult and embryonic germ cells of medaka. PLoS One. 2009;4:e6097. doi: 10.1371/journal.pone.0006097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoopfer ED, Penton A, Watts RJ, Luo L. Genomic analysis of Drosophila neuronal remodeling: a role for the RNA-binding protein Boule as a negative regulator of axon pruning. J Neurosci. 2008;28:6092–103. doi: 10.1523/JNEUROSCI.0677-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joiner ML, Wu CF. Nervous system function for the testis RNA-binding protein boule in Drosophila. J Neurogenet. 2004;18:341–63. doi: 10.1080/01677060490477435. [DOI] [PubMed] [Google Scholar]
- 41.Cheng MH, Maines JZ, Wasserman SA. Biphasic subcellular localization of the DAZL-related protein boule in Drosophila spermatogenesis. Dev Biol. 1998;204:567–76. doi: 10.1006/dbio.1998.9098. [DOI] [PubMed] [Google Scholar]
- 42.VanGompel MJ, Xu EY. A novel requirement in mammalian spermatid differentiation for the DAZ-family protein Boule. Hum Mol Genet. 2010;19:2360–9. doi: 10.1093/hmg/ddq109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Houston DW, King ML. A critical role for Xdazl, a germ plasm-localized RNA, in the differentiation of primordial germ cells in Xenopus. Development. 2000;127:447–56. doi: 10.1242/dev.127.3.447. [DOI] [PubMed] [Google Scholar]
- 44.Hashimoto Y, Maegawa S, Nagai T, Yamaha E, Suzuki H, Yasuda K, et al. Localized maternal factors are required for zebrafish germ cell formation. Dev Biol. 2004;268:152–61. doi: 10.1016/j.ydbio.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 45.Haston KM, Tung JY, Reijo Pera RA. Dazl functions in maintenance of pluripotency and genetic and epigenetic programs of differentiation in mouse primordial germ cells in vivo and in vitro. PLoS One. 2009;4:e5654. doi: 10.1371/journal.pone.0005654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kee K, Angeles VT, Flores M, Nguyen HN, Reijo Pera RA. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature. 2009;462:222–5. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clark AT, Bodnar MS, Fox M, Rodriquez RT, Abeyta MJ, Firpo MT, et al. Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum Mol Genet. 2004;13:727–39. doi: 10.1093/hmg/ddh088. [DOI] [PubMed] [Google Scholar]
- 48.Reijo RA, Dorfman DM, Slee R, Renshaw AA, Loughlin KR, Cooke H, et al. DAZ family proteins exist throughout male germ cell development and transit from nucleus to cytoplasm at meiosis in humans and mice. Biol Reprod. 2000;63:1490–6. doi: 10.1095/biolreprod63.5.1490. [DOI] [PubMed] [Google Scholar]
- 49.Ruggiu M, Speed R, Taggart M, McKay SJ, Kilanowski F, Saunders P, et al. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389:73–7. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- 50.Mita K, Yamashita M. Expression of Xenopus Daz-like protein during gametogenesis and embryogenesis. Mech Dev. 2000;94:251–5. doi: 10.1016/S0925-4773(00)00295-1. [DOI] [PubMed] [Google Scholar]
- 51.Brekhman V, Itskovitz-Eldor J, Yodko E, Deutsch M, Seligman J. The DAZL1 gene is expressed in human male and female embryonic gonads before meiosis. Mol Hum Reprod. 2000;6:465–8. doi: 10.1093/molehr/6.5.465. [DOI] [PubMed] [Google Scholar]
- 52.Niederberger C, Agulnik AI, Cho Y, Lamb D, Bishop CE. In situ hybridization shows that Dazla expression in mouse testis is restricted to premeiotic stages IV-VI of spermatogenesis. Mamm Genome. 1997;8:277–8. doi: 10.1007/s003359900409. [DOI] [PubMed] [Google Scholar]
- 53.Seligman J, Page DC. The Dazh gene is expressed in male and female embryonic gonads before germ cell sex differentiation. Biochem Biophys Res Commun. 1998;245:878–82. doi: 10.1006/bbrc.1998.8530. [DOI] [PubMed] [Google Scholar]
- 54.Nicholas CR, Xu EY, Banani SF, Hammer RE, Hamra FK, Reijo Pera RA. Characterization of a Dazl-GFP germ cell-specific reporter. Genesis. 2009;47:74–84. doi: 10.1002/dvg.20460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin YM, Chen CW, Sun HS, Tsai SJ, Lin JS, Kuo PL. Presence of DAZL transcript and protein in mature human spermatozoa. Fertil Steril. 2002;77:626–9. doi: 10.1016/S0015-0282(01)03226-5. [DOI] [PubMed] [Google Scholar]
- 56.Habermann B, Mi HF, Edelmann A, Bohring C, Bäckert IT, Kiesewetter F, et al. DAZ (Deleted in AZoospermia) genes encode proteins located in human late spermatids and in sperm tails. Hum Reprod. 1998;13:363–9. doi: 10.1093/humrep/13.2.363. [DOI] [PubMed] [Google Scholar]
- 57.Huang WJ, Lin YW, Hsiao KN, Eilber KS, Salido EC, Yen PH. Restricted expression of the human DAZ protein in premeiotic germ cells. Hum Reprod. 2008;23:1280–9. doi: 10.1093/humrep/den099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruggiu M, Cooke HJ. In vivo and in vitro analysis of homodimerisation activity of the mouse Dazl1 protein. Gene. 2000;252:119–26. doi: 10.1016/S0378-1119(00)00219-5. [DOI] [PubMed] [Google Scholar]
- 59.Reijo R, Alagappan RK, Patrizio P, Page DC. Severe oligozoospermia resulting from deletions of azoospermia factor gene on Y chromosome. Lancet. 1996;347:1290–3. doi: 10.1016/S0140-6736(96)90938-1. [DOI] [PubMed] [Google Scholar]
- 60.Kamischke A, Gromoll J, Simoni M, Behre HM, Nieschlag E. Transmission of a Y chromosomal deletion involving the deleted in azoospermia (DAZ) and chromodomain (CDY1) genes from father to son through intracytoplasmic sperm injection: case report. Hum Reprod. 1999;14:2320–2. doi: 10.1093/humrep/14.9.2320. [DOI] [PubMed] [Google Scholar]
- 61.Mau Kai C, Juul A, McElreavey K, Ottesen AM, Garn ID, Main KM, et al. Sons conceived by assisted reproduction techniques inherit deletions in the azoospermia factor (AZF) region of the Y chromosome and the DAZ gene copy number. Hum Reprod. 2008;23:1669–78. doi: 10.1093/humrep/den124. [DOI] [PubMed] [Google Scholar]
- 62.Calogero AE, Garofalo MR, Barone N, Longo GA, De Palma A, Fichera M, et al. Spontaneous transmission from a father to his son of a Y chromosome microdeletion involving the deleted in azoospermia (DAZ) gene. J Endocrinol Invest. 2002;25:631–4. doi: 10.1007/BF03345088. [DOI] [PubMed] [Google Scholar]
- 63.Lin Y, Page DC. Dazl deficiency leads to embryonic arrest of germ cell development in XY C57BL/6 mice. Dev Biol. 2005;288:309–16. doi: 10.1016/j.ydbio.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 64.Schrans-Stassen BH, Saunders PT, Cooke HJ, de Rooij DG. Nature of the spermatogenic arrest in Dazl -/- mice. Biol Reprod. 2001;65:771–6. doi: 10.1095/biolreprod65.3.771. [DOI] [PubMed] [Google Scholar]
- 65.Saunders PT, Turner JM, Ruggiu M, Taggart M, Burgoyne PS, Elliott D, et al. Absence of mDazl produces a final block on germ cell development at meiosis. Reproduction. 2003;126:589–97. doi: 10.1530/rep.0.1260589. [DOI] [PubMed] [Google Scholar]
- 66.Lin Y, Gill ME, Koubova J, Page DC. Germ cell-intrinsic and -extrinsic factors govern meiotic initiation in mouse embryos. Science. 2008;322:1685–7. doi: 10.1126/science.1166340. [DOI] [PubMed] [Google Scholar]
- 67.Rilianawati SR, Speed R, Taggart M, Cooke HJ. Spermatogenesis in testes of Dazl null mice after transplantation of wild-type germ cells. Reproduction. 2003;126:599–604. doi: 10.1530/rep.0.1260599. [DOI] [PubMed] [Google Scholar]
- 68.Xu EY, Lee DF, Klebes A, Turek PJ, Kornberg TB, Reijo Pera RA. Human BOULE gene rescues meiotic defects in infertile flies. Hum Mol Genet. 2003;12:169–75. doi: 10.1093/hmg/ddg017. [DOI] [PubMed] [Google Scholar]
- 69.Courtot C, Fankhauser C, Simanis V, Lehner CF. The Drosophila cdc25 homolog twine is required for meiosis. Development. 1992;116:405–16. doi: 10.1242/dev.116.2.405. [DOI] [PubMed] [Google Scholar]
- 70.Alphey L, Jimenez J, White-Cooper H, Dawson I, Nurse P, Glover DM. twine, a cdc25 homolog that functions in the male and female germline of Drosophila. Cell. 1992;69:977–88. doi: 10.1016/0092-8674(92)90616-K. [DOI] [PubMed] [Google Scholar]
- 71.Slee R, Grimes B, Speed RM, Taggart M, Maguire SM, Ross A, et al. A human DAZ transgene confers partial rescue of the mouse Dazl null phenotype. Proc Natl Acad Sci U S A. 1999;96:8040–5. doi: 10.1073/pnas.96.14.8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vogel T, Speed RM, Ross A, Cooke HJ. Partial rescue of the Dazl knockout mouse by the human DAZL gene. Mol Hum Reprod. 2002;8:797–804. doi: 10.1093/molehr/8.9.797. [DOI] [PubMed] [Google Scholar]
- 73.Yu Z, Ji P, Cao J, Zhu S, Li Y, Zheng L, et al. Dazl promotes germ cell differentiation from embryonic stem cells. J Mol Cell Biol. 2009;1:93–103. doi: 10.1093/jmcb/mjp026. [DOI] [PubMed] [Google Scholar]
- 74.Maines JZ, Wasserman SA. Post-transcriptional regulation of the meiotic Cdc25 protein Twine by the Dazl orthologue Boule. Nat Cell Biol. 1999;1:171–4. doi: 10.1038/11091. [DOI] [PubMed] [Google Scholar]
- 75.Tsui S, Dai T, Warren ST, Salido EC, Yen PH. Association of the mouse infertility factor DAZL1 with actively translating polyribosomes. Biol Reprod. 2000;62:1655–60. doi: 10.1095/biolreprod62.6.1655. [DOI] [PubMed] [Google Scholar]
- 76.Venables JP, Ruggiu M, Cooke HJ. The RNA-binding specificity of the mouse Dazl protein. Nucleic Acids Res. 2001;29:2479–83. doi: 10.1093/nar/29.12.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maegawa S, Yamashita M, Yasuda K, Inoue K. Zebrafish DAZ-like protein controls translation via the sequence ‘GUUC’. Genes Cells. 2002;7:971–84. doi: 10.1046/j.1365-2443.2002.00576.x. [DOI] [PubMed] [Google Scholar]
- 78.Jiao X, Trifillis P, Kiledjian M. Identification of target messenger RNA substrates for the murine deleted in azoospermia-like RNA-binding protein. Biol Reprod. 2002;66:475–85. doi: 10.1095/biolreprod66.2.475. [DOI] [PubMed] [Google Scholar]
- 79.Fox M, Urano J, Reijo Pera RA. Identification and characterization of RNA sequences to which human PUMILIO-2 (PUM2) and deleted in Azoospermia-like (DAZL) bind. Genomics. 2005;85:92–105. doi: 10.1016/j.ygeno.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 80.Moore FL, Jaruzelska J, Fox MS, Urano J, Firpo MT, Turek PJ, et al. Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc Natl Acad Sci U S A. 2003;100:538–43. doi: 10.1073/pnas.0234478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reynolds N, Collier B, Maratou K, Bingham V, Speed RM, Taggart M, et al. Dazl binds in vivo to specific transcripts and can regulate the pre-meiotic translation of Mvh in germ cells. Hum Mol Genet. 2005;14:3899–909. doi: 10.1093/hmg/ddi414. [DOI] [PubMed] [Google Scholar]
- 82.Reynolds N, Collier B, Bingham V, Gray NK, Cooke HJ. Translation of the synaptonemal complex component Sycp3 is enhanced in vivo by the germ cell specific regulator Dazl. RNA. 2007;13:974–81. doi: 10.1261/rna.465507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wickramasinghe D, Becker S, Ernst MK, Resnick JL, Centanni JM, Tessarollo L, et al. Two CDC25 homologues are differentially expressed during mouse development. Development. 1995;121:2047–56. doi: 10.1242/dev.121.7.2047. [DOI] [PubMed] [Google Scholar]
- 84.Wu S, Wolgemuth DJ. The distinct and developmentally regulated patterns of expression of members of the mouse Cdc25 gene family suggest differential functions during gametogenesis. Dev Biol. 1995;170:195–206. doi: 10.1006/dbio.1995.1207. [DOI] [PubMed] [Google Scholar]
- 85.Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, et al. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14:841–53. [PMC free article] [PubMed] [Google Scholar]
- 86.Collier B, Gorgoni B, Loveridge C, Cooke HJ, Gray NK. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J. 2005;24:2656–66. doi: 10.1038/sj.emboj.7600738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kleene KC. Poly(A) shortening accompanies the activation of translation of five mRNAs during spermiogenesis in the mouse. Development. 1989;106:367–73. doi: 10.1242/dev.106.2.367. [DOI] [PubMed] [Google Scholar]
- 88.Kleene KC, Distel RJ, Hecht NB. Translational regulation and deadenylation of a protamine mRNA during spermiogenesis in the mouse. Dev Biol. 1984;105:71–9. doi: 10.1016/0012-1606(84)90262-8. [DOI] [PubMed] [Google Scholar]
- 89.Padmanabhan K, Richter JD. Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes Dev. 2006;20:199–209. doi: 10.1101/gad.1383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Urano J, Fox MS, Reijo Pera RA. Interaction of the conserved meiotic regulators, BOULE (BOL) and PUMILIO-2 (PUM2) Mol Reprod Dev. 2005;71:290–8. doi: 10.1002/mrd.20270. [DOI] [PubMed] [Google Scholar]
- 91.Hasegawa E, Karashima T, Sumiyoshi E, Yamamoto MC. C. elegans CPB-3 interacts with DAZ-1 and functions in multiple steps of germline development. Dev Biol. 2006;295:689–99. doi: 10.1016/j.ydbio.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 92.Tsui S, Dai T, Roettger S, Schempp W, Salido EC, Yen PH. Identification of two novel proteins that interact with germ-cell-specific RNA-binding proteins DAZ and DAZL1. Genomics. 2000;65:266–73. doi: 10.1006/geno.2000.6169. [DOI] [PubMed] [Google Scholar]
- 93.Moore FL, Jaruzelska J, Dorfman DM, Reijo-Pera RA. Identification of a novel gene, DZIP (DAZ-interacting protein), that encodes a protein that interacts with DAZ (deleted in azoospermia) and is expressed in embryonic stem cells and germ cells. Genomics. 2004;83:834–43. doi: 10.1016/j.ygeno.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 94.Lee KH, Lee S, Kim B, Chang S, Kim SW, Paick JS, et al. Dazl can bind to dynein motor complex and may play a role in transport of specific mRNAs. EMBO J. 2006;25:4263–70. doi: 10.1038/sj.emboj.7601304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chennathukuzhi V, Morales CR, El-Alfy M, Hecht NB. The kinesin KIF17b and RNA-binding protein TB-RBP transport specific cAMP-responsive element modulator-regulated mRNAs in male germ cells. Proc Natl Acad Sci U S A. 2003;100:15566–71. doi: 10.1073/pnas.2536695100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kotaja N, Lin H, Parvinen M, Sassone-Corsi P. Interplay of PIWI/Argonaute protein MIWI and kinesin KIF17b in chromatoid bodies of male germ cells. J Cell Sci. 2006;119:2819–25. doi: 10.1242/jcs.03022. [DOI] [PubMed] [Google Scholar]
- 97.Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30:963–9. doi: 10.1042/BST0300963. [DOI] [PubMed] [Google Scholar]
- 98.Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am J Physiol Cell Physiol. 2003;284:C273–84. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- 99.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takeda Y, Mishima Y, Fujiwara T, Sakamoto H, Inoue K. DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS One. 2009;4:e7513. doi: 10.1371/journal.pone.0007513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Maratou K, Forster T, Costa Y, Taggart M, Speed RM, Ireland J, et al. Expression profiling of the developing testis in wild-type and Dazl knockout mice. Mol Reprod Dev. 2004;67:26–54. doi: 10.1002/mrd.20010. [DOI] [PubMed] [Google Scholar]
- 102.Gatta V, Raicu F, Ferlin A, Antonucci I, Scioletti AP, Garolla A, et al. Testis transcriptome analysis in male infertility: new insight on the pathogenesis of oligo-azoospermia in cases with and without AZFc microdeletion. BMC Genomics. 2010;11:401. doi: 10.1186/1471-2164-11-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Radford HE, Meijer HA, de Moor CH. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta. 2008;1779:217–29. doi: 10.1016/j.bbagrm.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kashiwabara S, Nakanishi T, Kimura M, Baba T. Non-canonical poly(A) polymerase in mammalian gametogenesis. Biochim Biophys Acta. 2008;1779:230–8. doi: 10.1016/j.bbagrm.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 105.Kashiwabara S, Noguchi J, Zhuang T, Ohmura K, Honda A, Sugiura S, et al. Regulation of spermatogenesis by testis-specific, cytoplasmic poly(A) polymerase TPAP. Science. 2002;298:1999–2002. doi: 10.1126/science.1074632. [DOI] [PubMed] [Google Scholar]