Abstract
The kakapo, a parrot endemic to New Zealand, is currently the focus of intense research and conservation efforts with the aim of boosting its population above the current ‘critically endangered’ status. While virtually nothing is known about the microbiology of the kakapo, given the acknowledged importance of gut-associated microbes in vertebrate nutrition and pathogen defense, it should be of great conservation value to analyze the microbes associated with kakapo. Here we describe the first study of the bacterial communities that reside within the gastrointestinal tract (GIT) of both juvenile and adult kakapo. Samples from along the GIT, taken from the choana (≈throat), crop and faeces, were subjected to 16 S rRNA gene library analysis. Phylogenetic analysis of >1000 16 S rRNA gene clones, derived from six birds, revealed low phylum-level diversity, consisting almost exclusively of Firmicutes (including lactic acid bacteria) and Gammaproteobacteria. The relative proportions of Firmicutes and Gammaproteobacteria were highly consistent among individual juveniles, irrespective of sampling location, but differed markedly among adult birds. Diversity at a finer phylogenetic resolution (i.e. operational taxonomic units (OTUs) of 99% sequence identity) was also low in all samples, with only one or two OTUs dominating each sample. These data represent the first analysis of the bacterial communities associated with the kakapo GIT, providing a baseline for further microbiological study, and facilitating conservation efforts for this unique bird.
Introduction
The kakapo (Strigops habroptilus) is one of the world's rarest bird species, with only 126 individuals remaining on two predator-free islands off New Zealand's south coast. Endemic to New Zealand, the kakapo possesses a range of behaviours and physiological characteristics that make it unique: it is the world's heaviest parrot, the only flightless parrot and the only parrot to carry out lek breeding [1]. Due to a combination of infrequent mating, low clutch numbers, and poor defense against mammalian predators the kakapo has been pushed to the verge of extinction [2], [3], though intensive conservation efforts by New Zealand's Department of Conservation have recently reversed the decline in numbers. Research programs into kakapo ecology, nutrition and genetics are well established and a management program has been enacted with the aim of restoring the kakapo population in New Zealand. Such practices as confining birds to predator-free islands, supplementary feeding, breeding programs and constant human supervision of both newborn chicks and adults have had a marked effect on the kakapo population – from just 62 remaining individuals in 1991 [4] to the current level. By contrast, the potentially important roles of symbiotic microorganisms in kakapo nutrition and pathogen defense remain unstudied, although positive bacterial influence on the gastrointestinal tract (GIT) was first observed in vertebrates almost 50 years ago [5].
The interactions between hosts and GIT-associated bacterial communities have been the subject of intense study in mammals, particularly humans [6], [7], with murine models often used to demonstrate causal links between microbes and aspects of host health [8]. Among avians, microbial research has mainly focused on either pathogen detection, or effects on weight gain in broiler chickens [9]. In the last decade, the study of microbial communities in the GIT of birds has become commonplace, with cultivation-dependent and -independent methods used to examine microbial presence and activity within avian gastrointestinal environments. The microbial communities associated with commercially farmed species such as turkey [10] and ostrich [11] have been investigated, as well as a range of wild birds, including parrots [12], [13] and the South American hoatzin [14], [15], [16], and their roles in bird fitness extend far beyond involvement in digestion and nutrient uptake. For example, studies on the effect of feather-degrading bacteria on mate selection and breeding fitness have revealed novel mechanisms through which bacteria can influence the lifecycle of their host [17], [18].
Links between microbial community structure and increased energy harvest from food have been demonstrated for a wide range of organisms by a variety of indirect techniques [19], [20], [21], [22]. In controlled murine models, these effects can be shown at a much more direct level, with gnotobiotic (germ-free) rodents used as controls in experiments that demonstrate the role of bacteria in regulating gene pathways in a range of organs [23], [24], [25], [26]. Microbes isolated from a particular host gut have been shown to be highly adapted to the host environment with the community being shaped by host-specific factors in a range of organisms [27], [28], [29]. Microbes transplanted into a new gnotobiotic host provide significantly reduced benefits to the new host [25]. Stimulation of the host immune system by GIT microbes has also been recognized in response to both viral and bacterial challenge [30], [31], [32], and development of gut-associated lymphoid tissues is increased in conventionally raised mice compared to their germ-free counterparts [33], [34].
With such varied and important roles being influenced by microbes, the lack of an accurate baseline description of kakapo-associated microbes represents a major gap in our knowledge of kakapo biology. Identification of the indigenous microbial community would be of great value to conservation efforts by enabling identification of allochthonous – potentially pathogenic – microbes. The existing literature surrounding kakapo-associated bacteria has so far focused on detecting and responding to pathogen outbreaks. Such an event occurred in 2004, when three kakapo died from erysipelas within 72 hours of translocation. The birds had been checked for known pathogens [35], and erysipelas had not previously been observed in kakapo [36]. While attacks from previously unidentified pathogens are unavoidable, this highlights an area in which molecular microbiology could play a key role in aiding kakapo recovery efforts, through the use of specific, high-sensitivity molecular probing techniques to detect pathogens before their numbers expand to levels that affect the bird.
Human interaction with wild birds can influence the composition of the GIT community [13], [37], and the potential for human impact on the kakapo GIT community is great (although unavoidable). In times of sickness, wild kakapo are taken into captivity and frequently treated with broad-spectrum antibiotics to combat pathogens. In captivity kakapo are fed a diet supplemented with fruit and pellets not available in the wild and hand-reared chicks are fed on bird formula exclusively until approximately 30 days of age [38]. A better understanding of kakapo microbiology carries clear potential for aiding conservation of this endangered bird, yet there are also sound academic reasons for researching this area. The kakapo diet consists mainly of shoots and leaves, and there has been speculation that kakapo may utilize microbes in the foregut to ferment ingested plant material [39]. While this process is common in ruminants (e.g. cattle and sheep) it is almost unknown among avians, with only the hoatzin known to use the foregut to facilitate fermentation [40]. The hoatzin, sole member of the family Opisthocomidae, exploits a diverse microbial community in its enlarged crop to aid in digestion, utilising up to 40 bacterial phyla as well as archaea to ferment plant material in the crop [15]. The kakapo has been suggested as a possible candidate for foregut fermentation due to its lack of a cecum, which is the primary site of hindgut fermentation [41], and its similar diet to the hoatzin.
The key aim of this study was to document the microbial community of the kakapo digestive tract in both newly hatched chicks and adults, using samples derived from both the fore- and hindgut to ensure maximum coverage of the GIT. 16 S rRNA gene analysis was used to identify bacteria at each sampling site, and the samples were compared to test for changes in community structure along the GIT. This study represents the first step in a wider investigation of the kakapo microbiome, with the ultimate goal of aiding conservation and management of this critically endangered bird.
Results
Bacterial community composition within the kakapo gastrointestinal tract
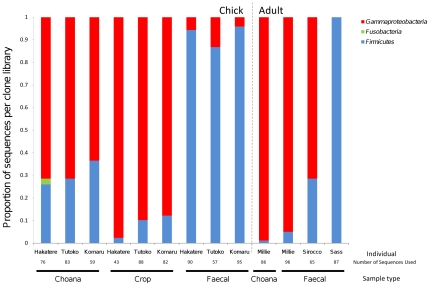
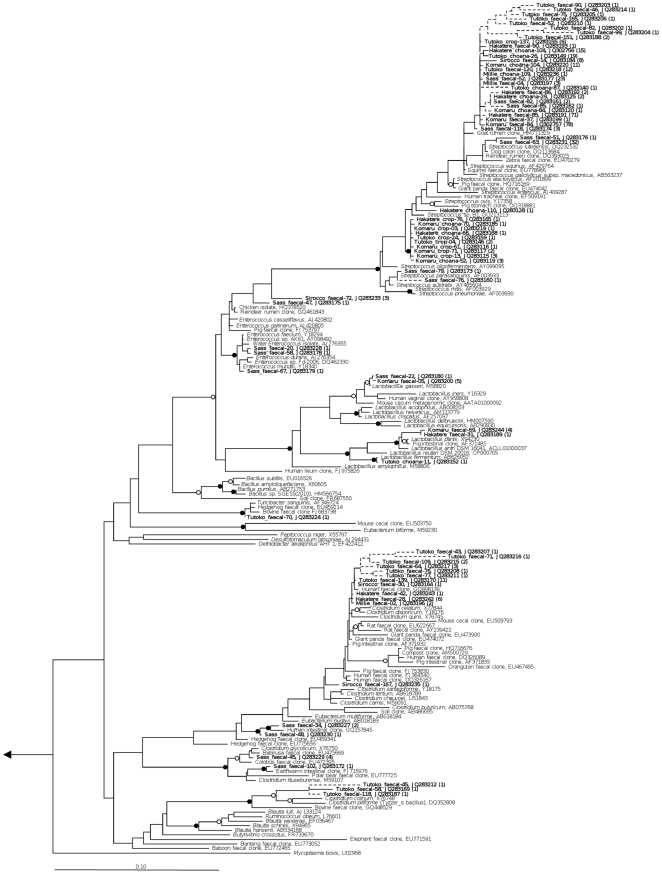
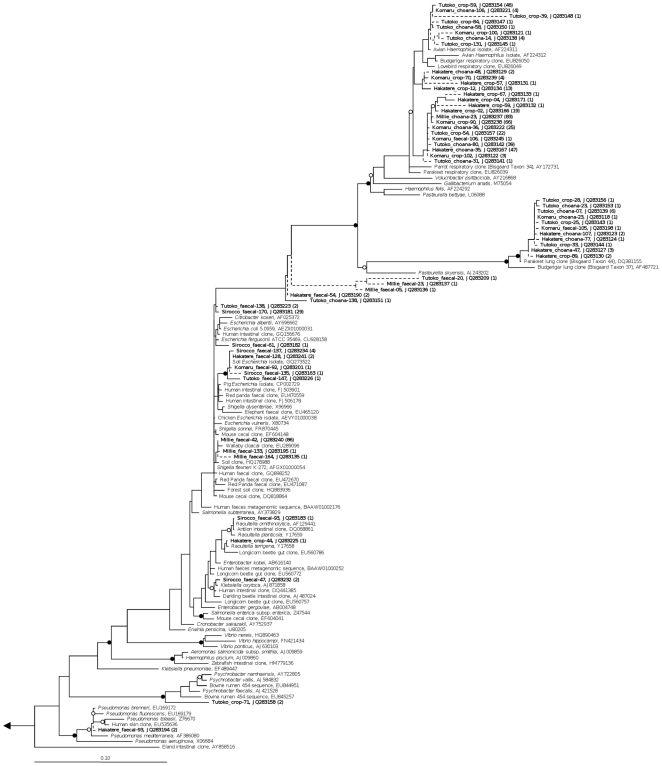
Bacterial 161S rRNA gene amplification was successful for all samples, whereas no archaea were amplified from any samples. A total of 1007 clones yielded high-quality sequence that passed chimera checking. The phylum Firmicutes was present in all libraries, and Gammaproteobacteria present in all except one (Sass). Slight representation from Fusobacteria was seen in a single chick choana sample (Fig. 1). When sequence data were dereplicated into 99% OTUs it was revealed that most of the sequences belonged to only a few key OTUs, such as Haemophilus felis and Streptococcus pasteurianus (Fig. 2). A Chao1 diversity estimator for each clone library was calculated at the 99% OTU level, and in almost all cases the expected number of OTUs per library was close to the observed number. The remainder of the diversity in each library was split among several low-abundance OTUs. Phylogenetic trees of kakapo-associated Firmicutes and Gammaproteobacteria are shown in Figs. 3 and 4, respectively.
Figure 1. Phylum-level distribution of kakapo-derived 16 S rRNA gene sequences.
Phylum-level affiliation of 16 S rRNA gene sequences obtained from the kakapo GIT. Samples to the left of the dotted line represent clone libraries derived from juveniles, and samples on the right represent adult-derived sequences.
Figure 2. OTU-level distribution of kakapo-derived 16 S rRNA gene sequences.
OTU-level affiliation of 16 S rRNA gene sequences obtained from the kakapo GIT. Values in this heatmap are scaled as a proportion of the total number of sequences per clone library. Observed numbers of OTUs at 99% sequence similarity are provided below the figure, as well as the estimated diversity for each library using the Chao1 estimator.
Figure 3. Phylogeny of Firmicutes-associated 16 S rRNA gene sequences derived from kakapo.
16 S rRNA gene-based phylogenetic analysis of Firmicutes recovered from kakapo samples. Solid junctions represent >90% bootstrap support, and hollow junctions >75%. Kakapo-derived sequences are in bold. Dashed lines indicate sequence length <1200 bp. Branch lengths were calculated using the maximum-likelihood method RAxML, using sequences >1200 bp in length, and short sequences were added subsequently using the Parsimony Interactive tool in ARB. Bootstrap values were calculated using maximum parsimony with 5000 resamplings. Scale bar, 10% sequence divergence.
Figure 4. Phylogeny of Gammaproteobacteria-associated 16 S rRNA gene sequences derived from kakapo.
16 S rRNA gene-based phylogenetic analysis of Gammaproteobacteria found within kakapo samples. Solid junctions represent >90% bootstrap support, and hollow junctions >75%. Kakapo-derived sequences are bolded. Dashed lines indicate sequence length <1200 bp. Branch lengths were calculated using the maximum-likelihood method RAxML, using sequences >1200 bp in length, and short sequences were added subsequently using the Parsimony Interactive tool in ARB. Bootstrap values were calculated using maximum parsimony with 5000 resamplings. Scale bar, 10% sequence divergence.
The extent of differences in bacterial community composition between samples was tested at the OTU level using an unweighted UniFrac analysis. Sequences obtained from kakapo chick, and adult faecal, samples were pooled according to sample type. Sequence data obtained from the single adult choana swab were not included in statistical comparison. Pairwise comparisons were made between sample types to test the null hypothesis that the bacterial community is homogeneous throughout the GIT. Significant differences in community structure were observed between the chick choana/crop (p<0.001) and crop/faeces (p = 0.002), but not between the choana/faeces. Between chick and adult faecal samples, no significant difference was seen. Given that Sass and Sirocco had been subject to considerable human intervention prior to sampling, their faecal samples were compared separately to the chick samples and wild adult sample (Millie). The faecal communities of Sass/Sirocco were significantly different from those of the wild adult (p = 0.002), but did not differ significantly from the chick samples. There was no significant difference between the wild adult and chick faecal samples.
Determination of Bacteroidetes and Archaea sensitivity
Bacteroidetes are common GIT-associated bacteria in many vertebrates, but were not detected in any of the kakapo samples. As certain DNA extraction techniques can lead to under-representation of Bacteroidetes in a sample [42], we tested whether our DNA extraction methods are able to detect the presence of Bacteroidetes within faecal and swab samples. Bacteroidetes DNA was successfully detected in all spiked faecal and swab samples (data not shown), down to approximately 0.15% of bacterial cell load, indicating that Bacteroidetes were not excluded by our DNA extraction protocols. Similarly, while Archaea were not detected in un-spiked kakapo faeces, the archaeal 16 S rRNA gene could be detected when swab and faecal samples were spiked with archaeal cells, down to approximately 0.4% of cell load.
Discussion
This paper describes the first molecular examination of the bacterial communities within the kakapo GIT, and provides evidence that qualitative differences exist between sites sampled throughout the GIT. The kakapo GIT appears to harbor a low-diversity community of microbes, with essentially only two phyla detected, Gammaproteobacteria and Firmicutes. Microbes in the kakapo GIT are abundant, with both cultivation-based measurements and DAPI cell counts indicating a microbial cell density in the order of 1010 cells per gram of faecal material (data not shown), yet each sample is dominated by only a few genera, typically Haemophilus, Streptococcus, and Clostridium. As the Fusobacteria discovered were only in a single sample, and found in low abundance, it is possible that their presence represents some form of contamination which occurred during sampling or DNA extraction. The Fusobacteria-associated sequences were similar to isolates and clones of the genus Leptotrichia, a bacterium commonly found in the human oral cavity [43].
At the phylum level, bacterial diversity is well conserved among all chicks sampled, but within the adults there is large variation in terms of relative abundance of each phylum. This may be explained by a range of factors regarding the adults, such as the frequent handling of Sirocco and, to a lesser extent, Sass, or the age difference between Sass and Millie/Sirocco. The bird Sass died several weeks after the collection of faecal samples, but not due to pathogen-related illness, and had not been treated with antibiotics prior to sampling (which can disrupt the GIT community [44], [45]). Subject age has been linked to a shift in the bacterial gut community in humans [46], [47] and mice [48], so it is conceivable that such a community change may be a natural phenomenon. While functional roles of the bacteria detected in this study can only be speculated upon, those bacteria encountered in the kakapo GIT correspond to genera commonly observed in other herbivores. In a study of the gut microbiota of deer it was recognized that Streptococcus played a role in degrading tannins ingested by the host animal, and many of the Streptococcus detected in the kakapo clone libraries were closely related to this species (Fig. 3, Streptococcus gallolyticus sub. macedonicus, AB563237) [49]. Most of the bacterial genera detected throughout the kakapo GIT are known anaerobic fermenters, capable of converting sugars such as glucose and cellulose into acids such as acetate, which are utilized by the host. Members of the genus Clostridium are frequently identified as cellulolytic [50], [51], [52], [53], and have been found to increase in proportion within the herbivore gut in the absence of starch [54]. The inability to detect Bacteroidetes in a parrot, using either 16 S rRNA-based techniques or cultivation, is not unique to our study [12], [13]. In addition to playing roles in butyrate production and bile acid metabolism [55], [56], Bacteroidetes are well-characterized degraders of starch and cellulose in the gut [57], [58], [59], [60]. Historically the kakapo have relied on a low-starch diet [61], which may have selected against Bacteroidetes colonization, as diet has been identified as one of the factors that shape gut microbiota [62], [63], [64], [65]. While the DNA extraction method utilized in this study is capable of extracting detectable levels of DNA from Bacteroidetes comprising less than 1% of the community, it is conceivable that the inability to detect Bacteroidetes stems from low sequence counts compared to those obtained using next-generation sequencing technologies.
Given the endangered status of the kakapo, destructive sampling (via dissection) is not possible. As such, our analyses were limited to swab and faecal samples rather than direct tissue and gut content samples. Although surface swabs may not give a perfect representation of the local bacterial community, they have been previously applied in a range of avian study systems [13], [17], [66], [67], [68] where dissection of the target organism was not feasible. There still exists the potential that mucosa-associated bacteria of the crop may not be recovered through the swabbing of live animals, indicating a potential blind spot in sampling. Nevertheless, it appears that swabbing of the kakapo crop is adequate for differentiating between microbial communities of the choana and crop, despite the fact that any probe into the crop risks potential contamination as the swab passes the choana. The use of faecal samples as a proxy for hindgut bacterial communities has been used extensively in a range of vertebrates, including humans and birds. While several studies have highlighted differences between bacterial recovery from mucosal biopsies and faecal samples [7], [69], this appears to be due to faecal samples containing not only mucosa-associated bacteria that have been shed into the faeces, but also bacteria that colonize the faecal substrate directly. Community data taken from faecal samples contains a reasonable representation of microbes within the hindgut, and differences in faecal microbiota (both at presence/absence and functional levels) have been shown to reflect differences in the intestinal tract of the host [48], [70], although it must be stressed that they do not provide an exact representation of microbial community structure and function within the intestine itself. Based on unweighted UniFrac analysis it appears that the faecal bacterial communities of adults and chick kakapo are not significantly different, which may indicate a vector for inoculation of kakapo chicks with their parents' microbiota. The lack of significant difference between choana and faecal communities in the chicks is not surprising considering the lifestyle of unfledged kakapo chicks. Essentially immobile, the chicks are unable to distance themselves from their own faeces. The chicks studied in this project have since been fledged and given the low population of kakapo and constant attention to the birds, these present an excellent opportunity for longitudinal studies throughout the lifespan of the birds.
Given the low-energy diet of the kakapo and its lack of cecum, it has been speculated that the kakapo may utilize microbially-mediated foregut fermentation to derive additional energy from its food. While this study was not targeted at confirming or rejecting the notion of kakapo foregut fermentation, the possibility that key microorganisms may be resident in the crop rather than hindgut was taken into consideration when planning this study. There are several frequently found bacterial phyla in the microbial community of foregut-fermenting mammals and the hoatzin, predominantly Firmicutes and Bacteroidetes, with representation from Verrucomicrobia, Actinobacteria and Spirochaetes commonly observed [15], [71]. Methanogenic archaea are also commonly found in the rumen or crop of foregut fermenters [14], [72], [73], [74], [75], [76]. With the exception of Firmicutes, none of the above-mentioned taxa were detected in the kakapo samples. In the hoatzin it has been shown that the microbial community of the foregut is similar to that of ruminants [16], but given the apparent absence of so many bacterial phyla in the kakapo crop it is unlikely that the kakapo shares this community structure and gut adaptation. Foregut fermentation is an adaptation to a diet rich in celluloses that the host cannot digest [77], but kakapo do not retain and digest cellulose in the manner seen in ruminants and the hoatzin, typically spitting away masticated plant material after extracting juices from the flesh [61], [78]. Although merely speculative at this stage, it thus seems unlikely that kakapo perform foregut fermentation in the traditional manner.
One observation from this study that may prove to be of future concern is the high number of Pasteurellaceae-like sequences within the choana and crop swabs. Many of the sequences were clustered with bacterial genera such as Haemophilus, or with several non-cultivated clades commonly detected in the avian respiratory tract, which are frequently found as respiratory pathogens in vertebrates [79]. It has been noted that certain Pasteurellaceae which were present in our libraries (Bisgaard Taxon 34, Bisgaard Taxon 44, Fig. 4) are frequently associated with respiratory disease in psittacine birds. Although not all bacterial species in these clades are causative agents of disease, their presence should be considered a warning, as they are often found in sick birds [80]. During the 2011 breeding season several chicks were removed from the nest due to respiratory problems, although this did not cause long-term health issues in the birds (D. Eason, personal communication). While there is no data to imply a causal link between the observed Pasteurellaceae and the illness, pathogens do appear to have been introduced to the kakapo population previously through avian vectors [36].
In summary, we performed the first 16 S rRNA-based microbial analysis of the bacteria that inhabit the kakapo GIT. We have shown that the GIT is inhabited by a few key organisms, and that the community composition changes throughout the GIT. Our results also provided preliminary evidence that the human influence on kakapo lifestyle appears to cause a shift in these bacterial communities, although whether this has a positive, negative, or neutral effect on the bird remains unknown.
Materials and Methods
Sampling
Samples were obtained from four kakapo on Codfish Island (46°47′S 167°38′E), off the coast of Stewart Island, New Zealand, during the nesting season, between 17th and 23rd April 2011. Two additional faecal samples had previously been collected from adult birds when they were brought to Auckland Zoo. A total of 13 samples were used in this analysis, collected from three unfledged chicks, and three adults. Samples of the upper GIT were taken by Department of Conservation staff using sterile rayon-tipped swabs (Copan, #170KS01), and samples were taken from the choana and crop of chicks, and choana of one adult. The choana is an opening in the roof of the mouth that joins to the nasal passage. Due to difficulties in restraining adults, crop samples could not be taken from adult birds. A faecal sample was collected from all six birds at the time swabbing was performed. As interference with nesting mothers can cause them to abandon the nest, samples from chick parents could not be obtained. Swabs were stored in sterile polypropylene tubes and kept on ice until they were frozen at the ranger hut on Codfish Island. Samples were shipped to The University of Auckland on dry ice, and stored at −20°C upon arrival.
DNA Extraction
Despite considerable efforts to standardize the DNA extraction procedure, it was necessary to use a different approach for extracting DNA from swab vs faecal samples due to unreliable DNA retrieval from hard-to-obtain swab samples and unwillingness to risk valuable samples. Genomic DNA was extracted from swabs using heat lysis. Swab tips were placed in a 1.5 mL cryotube containing 1 mL extraction buffer (20 mM EDTA, 0.1 M Tris (pH 8.0), 1% CTAB, 56 mM NaCl), briefly vortexed, then incubated at 94°C for 15 min in order to lyse both Gram-negative and Gram-positive cells [81], [82]. The solution was cooled briefly on ice, 300 µL of chloroform/isoamyl alcohol (24∶1 ratio) was added and the solution mixed by inversion, then centrifuged at 13,000 rpm for 5 min at room temperature. The supernatant was transferred to a 2 mL microcentrifuge tube, to which 0.6 volume (vol) isopropanol and 0.1 vol 3 M sodium acetate (pH 5.2) were added. Samples were incubated overnight at −20°C then centrifuged at 13,000 rpm at 4°C for 30 min. The supernatant was discarded and the pellet washed twice with ice-cold 70% ethanol followed by centrifugation at 13,000 rpm at 4°C for 10 min. Samples were dried and suspended in 50 µL UltraPure water (Invitrogen).
Genomic DNA was extracted from kakapo faecal samples using a variation on an extraction protocol previously described [83]. 100 mg of faeces were suspended in 1 mL of 70% ethanol with 200 mg of 0.1 mm zirconia/silica beads in a 1.5 mL cryotube. Samples were agitated using a FastPrep FP120 bead beater, at 5.5 ms−1 for 30 s, followed by centrifugation at 13,000 rpm for 5 min and removal of supernatant. 1 mL of extraction buffer was added to each tube in addition to 30 mg of polyvinylpolypyrrolidone (PVPP), before being agitated using the previous settings. Samples were then incubated at 65°C for 30 min, with gentle mixing every 10 min. Samples were centrifuged at 13,000 rpm for 5 min and the supernatant was transferred to a fresh 1.5 mL microcentrifuge tube containing 0.5 mL of chloroform/isoamyl alcohol solution (24∶1 ratio) and inverted to mix. Samples were centrifuged at 13,000 rpm for 5 min, before the supernatant (approx. 1 mL) was transferred to a 2 mL microcentrifuge tube containing 0.6 vol isopropanol and 0.1 vol 3 M sodium acetate (pH 5.2). Samples were mixed then incubated on ice for 1 h, then centrifuged at 13,000 rpm at 4°C for 1 min to remove any remaining sediment (presumed to be leftover SDS). The supernatant was transferred to a new microcentrifuge tube and centrifuged under the same conditions for 30 min. The supernatant was removed and the pellet washed twice using ice-cold 70% ethanol followed by 10 min centrifugation at 13,000 rpm, 4°C. The pellet was dried and resuspended in 50 µL UltraPure water (Invitrogen).
PCR and clone library construction
PCR was performed using the forward primer 616V and reverse primer 1492R, targeting Escherichia coli positions 8–27 and 1492–1513 respectively, to amplify bacterial 16 S rRNA genes, and 21F/958R for archaeal 16 S rRNA genes (Table 1). Reactions were conducted in 25 µL volumes, containing 20 mM Tris-HCl, 50 mM KCl (buffer), 1.5 mM MgCl2, 25 µM of each dNTP, 2.5 µM of each primer, 0.5 units Taq polymerase and 1.0 µL of extracted DNA template. Cycling conditions for the 616V/1492R primer pair were as follows: initial denaturing at 94°C for 5 min, 30 cycles of 94°C for 45 s, 57°C for 45 s and 72°C for 1.5 min, followed by a final elongation step at 72°C for 7 min. Cycling conditions for 21F/958R were described previously [84]. In order to successfully amplify from faecal samples, the addition of 2% bovine serum albumin per tube was required [85]. Cloning was performed using the P-GemT Easy Vector kit (Promega, Inc, Madison WI, USA) following the manufacturer's instructions. Approximately 96 clones from each of the 13 clone libraries were selected for sequencing (Macrogen Inc, Seoul, South Korea).
Table 1. Sequences for primers used in this study.
Phylogenetic Analysis
Sequences were analyzed using the Geneious software package [86] and low-quality data from the ends of each sequence removed. Chimeras were identified with the Pintail algorithm using the Mallard software package [87] and subsequently removed from the data set. Sequences were aligned via the SINA web aligner [88] and imported into ARB using the SILVA 108 database [89]. Sequence data were divided into operational taxonomic units (OTU) of 99% sequence identity using mothur [90] and one sequence to represent each OTU per sample was used in tree construction. Sequences representing each OTU were submitted to the DDBJ/EMBL/GenBank databases under accession numbers JQ283115–Q283245, JQ302756, and JQ302757. Phylogenetic trees were constructed in ARB using the maximum likelihood method RAxML. Bootstrap values were calculated using 5000 parsimony replications. Unweighted UniFrac analyses were performed in mothur to statistically compare bacterial community composition among different sample types.
Determination of Bacteroidetes and Archaea sensitivity
A pure culture of Chryseobacterium formosense (phylum Bacteroidetes), originally isolated from wastewater, was obtained from a colleague and cultivated at the original isolation conditions (R2A broth, 28°C for 48 h, C. Brown, personal communication). C. formosense cells were added to samples of kakapo chick faeces at proportions down to 0.15% total bacterial cell load, calculated through enumeration of C. formosense via plating counts, and DAPI staining of faecal samples, then subjected to both extraction methods detailed previously. A fragment of the 16 S rRNA gene was amplified using the 341-GC/518R primer pair. Cycling conditions consisted of an initial denaturing step at 94°C for 5 min, followed by 25 cycles at 94°C for 1 min, 60°C for 1 min, and 72°C for 30 s, then a final elongation step at 72°C for 5 min. The product was analyzed using denaturing gradient gel electrophoresis (DGGE) with a denaturing gradient of 40–70%. A positive control of pure C. formosense DNA was used as an indicator of a Bacteroidetes band in the gel pattern. A pure culture of Methanosarcina acetivorans (domain Archaea, strain DS2834) was obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ) and added to samples of kakapo chick faeces at proportions down to 0.4% total cell load. A fragment of the archaeal 16 S rRNA gene was amplified using the 21F/958R primer pair. Cycling conditions were described previously [84]. Samples were visualised on a 1% agarose gel and analysed using the BioRad Gel Doc imaging system.
Acknowledgments
We gratefully thank Daryl Eason and Jo Ledington (Department of Conservation), and John Potter (Auckland Zoo) for provision of all kakapo samples, and Caroline Brown for supplying the required Bacteroidetes strain. We would also like to thank Ron Moorhouse and Deidre Vercoe (Department of Conservation), and Richard Jakob-Hoff (Auckland Zoo) for their endorsement and support of this project, plus Jacqueline Beggs and Mick Clout (University of Auckland) for useful discussions on this topic.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by funding from the Department of Conservation as well as a University of Auckland Faculty Research Development Fund (grant 9841 3626187) to MWT, Feodor Lynen Research Fellowship from the Alexander von Humboldt Foundation to PD, and a University of Auckland Doctoral Scholarship to DW. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Merton DV, Morris RB, Atkinson IAE. Lek behaviour in a parrot: the Kakapo Strigops habroptilus of New Zealand. Ibis. 1984;126:277–283. [Google Scholar]
- 2.Lloyd BD, Powlesland RG. The decline of kakapo Strigops habroptilus and attempts at conservation by translocation. Biol Conserv. 1994;69:75–85. [Google Scholar]
- 3.Houston D, Mcinnes K, Elliott G, Eason D, Moorhouse R, et al. The use of a nutritional supplement to improve egg production in the endangered kakapo. Biol Conserv. 2007;138:248–255. [Google Scholar]
- 4.Elliott GP, Merton DV, Jansen PW. Intensive management of a critically endangered species: the kakapo. Biol Conserv. 2001;99:121–133. [Google Scholar]
- 5.Dubos R, Schaedler RW. The digestive tract as an ecosystem. Am J Med Sci. 1964;248:267–272. doi: 10.1097/00000441-196409000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev: Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–1615. doi: 10.1136/gut.2007.133603. [DOI] [PubMed] [Google Scholar]
- 8.Yi P, Li L. The germfree murine animal: An important animal model for research on the relationship between gut microbiota and the host. Vet Microbiol. 2011 doi: 10.1016/j.vetmic.2011.10.024. In press. [DOI] [PubMed] [Google Scholar]
- 9.Yegani M, Korver DR. Factors affecting intestinal health in poultry. Poultry Sci. 2008;87:2052–2063. doi: 10.3382/ps.2008-00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu J, Domingo JS. Turkey fecal microbial community structure and functional gene diversity revealed by 16 S rRNA gene and metagenomic sequences. J Microbiol. 2008;46:469–477. doi: 10.1007/s12275-008-0117-z. [DOI] [PubMed] [Google Scholar]
- 11.Matsui H, Kato Y, Chikaraishi T, Moritani M, Ban-Tokuda T, et al. Microbial diversity in ostrich ceca as revealed by 16 S ribosomal RNA gene clone library and detection of novel Fibrobacter species. Anaerobe. 2010;16:83–93. doi: 10.1016/j.anaerobe.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Pacheco MA, Garcia-Amado MA, Bosque C, Dominguez-Bello MG. Bacteria in the crop of the seed-eating Green-Rumped Parrotlet. Condor. 2004;106:139–143. [Google Scholar]
- 13.Xenoulis PG, Gray PL, Brightsmith D, Palculict B, Hoppes S, et al. Molecular characterization of the cloacal microbiota of wild and captive parrots. Vet Microbiol. 2010;146:320–325. doi: 10.1016/j.vetmic.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 14.Godoy-Vitorino F, Ley RE, Gao Z, Pei Z, Ortiz-Zuazaga H, et al. Bacterial community in the crop of the hoatzin, a neotropical folivorous flying bird. Appl Environ Microb. 2008;74:5905–5912. doi: 10.1128/AEM.00574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godoy-Vitorino F, Goldfarb KC, Brodie EL, Garcia-Amado MA, Michelangeli F, et al. Developmental microbial ecology of the crop of the folivorous hoatzin. ISME J. 2010;4:611–620. doi: 10.1038/ismej.2009.147. [DOI] [PubMed] [Google Scholar]
- 16.Godoy-Vitorino F, Goldfarb KC, Karaoz U, Leal S, Garcia-Amado MA, et al. Comparative analyses of foregut and hindgut bacterial communities in hoatzins and cows. ISME J. 2012;6:531–541. doi: 10.1038/ismej.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shawkey MD, Pillai SR, Hill GE, Siefferman LM, Roberts SR. Bacteria as an agent for change in structural plumage color: correlational and experimental evidence. Am Nat. 2007;169:S112–S121. doi: 10.1086/510100. [DOI] [PubMed] [Google Scholar]
- 18.Burt EH, Schroeder MR, Smith LA, Sroka JE, McGraw KJ. Colourful parrot feathers resist bacterial degradation. Biol Letters. 2011;7:214–216. doi: 10.1098/rsbl.2010.0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, et al. Metagenomic Analysis of the Human Distal Gut Microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 21.Ohkuma M. Symbioses of flagellates and prokaryotes in the gut of lower termites. Trends Microbiol. 2008;16:345–352. doi: 10.1016/j.tim.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Torok VA, Ophel-Keller K, Loo M, Hughes RJ. Application of methods for identifying broiler chicken gut bacterial species linked with increased energy metabolism. Appl Environ Microb. 2008;74:783–791. doi: 10.1128/AEM.01384-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Backhed F, Ding H, Wang T, Hooper LV, Koh GY, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meinl W, Sczensy S, Brigelius-Flohe R, Blaut M, Glatt H. Impact of gut microbiota on intestinal and hepatic levels of phase 2 xenobiotic-metabolizing enzymes in the rat. Drug Metab Dispos. 2008;37:1179–1186. doi: 10.1124/dmd.108.025916. [DOI] [PubMed] [Google Scholar]
- 26.Bjorkholm B, Bok CM, Lundin A, Rafter J, Hibberd ML, et al. Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS ONE. 2009;4:e6958. doi: 10.1371/journal.pone.0006958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Wielen PWJJ, Keuzenkamp DA, Lipman LJA, van Knapen F, Biesterveld S. Spatial and temportal variation of the intestinal bacterial community in commercially raised broiler chickens during growth. Microb Ecol. 2002;44:286–293. doi: 10.1007/s00248-002-2015-y. [DOI] [PubMed] [Google Scholar]
- 28.Khachatryan ZA, Ktsoyan ZA, Manukyan GP, Kelly D, Ghazaryan KA, et al. Predominant role of host genetics in controlling the composition of gut microbiota. PLoS ONE. 2008;3:e3064. doi: 10.1371/journal.pone.0003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brisbin JT, Gong J, Sharif S. Interactions between commensal bacteria and the gut-associated immune system of the chicken. Anim Health Res Rev. 2008;9:101–110. doi: 10.1017/S146625230800145X. [DOI] [PubMed] [Google Scholar]
- 31.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hudson JA, Luckey TD. Bacteria induced morphologic changes. P Soc Exp Biol Med. 1964;116:628–631. doi: 10.3181/00379727-116-29324. [DOI] [PubMed] [Google Scholar]
- 34.Hooijkaas H, Benner R, Pleasants JR, Wostmann BS. Isotypes and specificities of immunoglobulins produced by germ-free mice fed chemically defined ultrafiltered antigen-free diet. Eur J Immunol. 1984;14:1127–1130. doi: 10.1002/eji.1830141212. [DOI] [PubMed] [Google Scholar]
- 35.Brangenberg N, McInnes C, Connolly JH, Rogers LE. Absence of Salmonella and Campylobacter species in fecal and cloacal swab samples from kakapo (Strigops habroptilus) on Codfish Island, New Zealand. J Avian Med Surg. 2003;17:203–205. [Google Scholar]
- 36.Gartrell BD, Alley MR, Mack H, Donald J, McInnes K, et al. Erysipelas in the critically endangered kakapo (Strigops habroptilus). Avian Pathol. 2005;34:383–387. doi: 10.1080/03079450500268583. [DOI] [PubMed] [Google Scholar]
- 37.Janiga M, Sedlarova A, Rigg R, Novotna M. Patterns of prevalence among bacterial communities of alpine accentors (Prunella collaris) in the Tatra Mountains. J Ornithol. 2007;148:135–143. [Google Scholar]
- 38.Eason D, Moorhouse R. Hand-rearing kakapo (Strigops habroptilus), 1997–2005. Notornis. 2006;53:116–125. [Google Scholar]
- 39.Morton ES. Avian arboreal folivores: why not? In: Montgomery GG, editor. The ecology of arboreal folivores. Washington, D.C.: Smithsonian Institution Press; 1978. pp. 123–130. [Google Scholar]
- 40.Grajal A, Strahl SD, Parra R, Dominguez MG, Neher A. Foregut fermentation in the hoatzin, a neotropical leaf-eating bird. Science. 1989;245:1236–1238. doi: 10.1126/science.245.4923.1236. [DOI] [PubMed] [Google Scholar]
- 41.Clench MH, Mathias JR. The avian cecum: a review. Wilson Bull. 1995;107:93–121. [Google Scholar]
- 42.Boom R, Sol CJA, Salimans MMM, Jansen CL, Wertheim van Dillen PME, et al. Rapid and simple method for purification of nucleic acid. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kribe ERK, Olsen I. Leptotrichia species in human infections. Anaerobe. 2008;14:131–137. doi: 10.1016/j.anaerobe.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 44.O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16 S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hopkins MJ, Sharp R, Macfarlane GT. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16 S rRNA abundance, and community cellular fatty acid profiles. Gut. 2001;48:198–205. doi: 10.1136/gut.48.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajilic-Stojanovic M, Heilig HGHJ, Molenaar D, Kajander K, Surakka A, et al. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol. 2009;11:1736–1751. doi: 10.1111/j.1462-2920.2009.01900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaahtovuo J, Toivanen P, Eerola E. Study of murine faecal microflora by cellular fatty acid analysis; effect of age and mouse strain. Antonie Leeuwenhoek. 2001;80:35–42. doi: 10.1023/a:1012058107731. [DOI] [PubMed] [Google Scholar]
- 49.Hiura T, Hashidoko Y, Kobayashi Y, Tahara S. Effective degradation of tannic acid by immobilized rumen microbes of a sika deer (Cervus nippon yesoensis) in winter. Anim Feed Sci Tech. 2010;155:1–8. [Google Scholar]
- 50.Shoham Y, Lamed R, Bayer EA. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 1999;7:275–280. doi: 10.1016/s0966-842x(99)01533-4. [DOI] [PubMed] [Google Scholar]
- 51.Warnick TA, Methe BA, Leschine SB. Clostridium phytofermentans sp. nov., a cellulolytic mesophile from forest soil. Int J Syst Evol Micr. 2002;52:1155–1160. doi: 10.1099/00207713-52-4-1155. [DOI] [PubMed] [Google Scholar]
- 52.Varel VH, Pond WG. Characteristics of a New Cellulolytic Clostridium sp. Isolated from Pig Intestinal Tract. Appl Environ Microb. 1992;58:1645–1649. doi: 10.1128/aem.58.5.1645-1649.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sabathe F, Belaich A, Soucaille P. Characterization of the cellulolytic complex (cellulosome) of Clostridium acetobutylicum. FEMS Microbiol Lett. 2002;217:15–22. doi: 10.1111/j.1574-6968.2002.tb11450.x. [DOI] [PubMed] [Google Scholar]
- 54.Laure R, Yu Z, Parisi VA, Egan AR, Morrison M. Novel microbial diversity adherent to plant biomass in the herbivore gastrointestinal tract, as revealed by ribosomal intergenic spacer analysis and rrs gene sequencin. Environ Microbiol. 2005;7:530–543. doi: 10.1111/j.1462-2920.2005.00721.x. [DOI] [PubMed] [Google Scholar]
- 55.Kim YS, Milner JA. Dietary modulation of colon cancer risk. J Nutr. 2007;137:2576–2579. doi: 10.1093/jn/137.11.2576S. [DOI] [PubMed] [Google Scholar]
- 56.Thomas F, Hehemann J, Rebuffet E, Czjzek M, Michel G. Environmental and gut Bacteroidetes: the food connection. Front Microbiol. 2011;2 doi: 10.3389/fmicb.2011.00093. doi: 10.3389/fmicb.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dongowski G, Lorenz A, Anger H. Degradation of pectins with different degrees of esterigication by Bacteroides thetaiotaomicron isolated from human gut flora. Appl Environ Microb. 2000;66:1321–1327. doi: 10.1128/aem.66.4.1321-1327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chassard C, Goumy V, Leclerc M, Del'homme C, Bernalier-Donadille A. Characterization of the xylan-degrading microbial community from human faeces. FEMS Microbiol Ecol. 2007;61:121–131. doi: 10.1111/j.1574-6941.2007.00314.x. [DOI] [PubMed] [Google Scholar]
- 59.Martens EC, Koropatkin NM, Smith TJ, Gordon JI. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem. 2009;284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bolam DN, Sonnenburg JL. Mechanistic insight into polysaccharide use within the intestinal microbiota. Gut Microbes. 2011;2:86–90. doi: 10.4161/gmic.2.2.15232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horrocks M, Salter J, Braggins J, Nichol S, Moorhouse R, et al. Plant microfossil analysis of coprolites of the critically endangered kakapo (Strigops habroptilus) parrot from New Zealand. Rev Palaeobot Palyno. 2008;149:229–245. [Google Scholar]
- 62.Finegold SM, Attebery HR, Sutter VL. Effect of diet on human fecal flora: comparison of Japanese and American diets. Am J Clin Nutr. 1974;27:1456–1469. doi: 10.1093/ajcn/27.12.1456. [DOI] [PubMed] [Google Scholar]
- 63.Hehemann J, Correc G, Barbeyron T, Helbert W, Czjzek M, et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–914. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 64.Martinez I, Kim J, Duffy PR, Schlegel VL, Walter J. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS ONE. 2010;5:e15046. doi: 10.1371/journal.pone.0015046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shanks OC, Kelty CA, Archibeque S, Jenkins M, Newton RJ, et al. Community structures of fecal cacteria in cattle from different animal feeding operation. Appl Environ Microb. 2011;77:2992–3001. doi: 10.1128/AEM.02988-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreno J, Briones V, Merino S, Ballesteros C, Sanz JJ, et al. Beneficial effects of cloacal bacteria on growth and fledging size in Nestling Pied Flycatchers (Fidecula hypoleuca) in Spain. Auk. 2003;120:784–790. [Google Scholar]
- 67.Blanco G, Lemus JA, Grande J. Faecal bacteria associated with different diets of wintering red kites: influence of livestock carcass dumps in microflora alteration and pathogen acquisition. J Appl Ecol. 2006;43:990–998. [Google Scholar]
- 68.Klomp JE, Murphy MT, Smith SB, McKay JE, Ferrera I, et al. Cloacal microbial communities of female spotted towhees Pipilo maculatus: microgeographic variation and individual sources of variability. J Avian Biol. 2008;39:530–538. [Google Scholar]
- 69.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 71.Ley RE, Hamady M, Lozupone C, Turnbaugh P, Ramey RR, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tokura M, Chagan I, Ushida K, Kojima Y. Phylogenetic study of methanogens associated with rumen ciliates. Curr Microbiol. 1999;39:123–128. doi: 10.1007/s002849900432. [DOI] [PubMed] [Google Scholar]
- 73.Tajima K, Nagamine T, Matsui H, Nakamura M, Aminov RI. Phylogenetic analysis of archaeal 16 S rRNA libraries from the rumen suggests the existence of a novel group of archaea not associated with known methanogens. FEMS Microbiol Lett. 2001;200:67–72. doi: 10.1111/j.1574-6968.2001.tb10694.x. [DOI] [PubMed] [Google Scholar]
- 74.Irbis C, Ushida K. Detection of methanogens and proteobacteria from a single cell of rumen ciliate protozoa. J Gen Appl Microbiol. 2004;50:203–212. doi: 10.2323/jgam.50.203. [DOI] [PubMed] [Google Scholar]
- 75.Shin EC, Choi BR, Lim WJ, Hong SY, An CL, et al. Phylogenetic analysis of archaea in three fractions of cow rumen based on the 16 S rDNA sequence. Anaerobe. 2004;10:313–319. doi: 10.1016/j.anaerobe.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 76.Yu Z, Garcia-Gonzalez R, Schanbacher FL, Morrison M. Evaluations of different hypervariable regions of archaeal 16 S rRNA genes in profiling of methanogens by archaea-specific PCR and denaturing gradient gel electrophoresis. Appl Environ Microb. 2008;74:889–893. doi: 10.1128/AEM.00684-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Russel JB, Rychlik JL. Factors that alter rumen microbial ecology. Science. 2001;292:1119–1122. doi: 10.1126/science.1058830. [DOI] [PubMed] [Google Scholar]
- 78.Oliver WRB. New Zealand Birds. Wellington: A.H. & A.W. Reed; 1955. 641 [Google Scholar]
- 79.Christensen H, Foster G, Christense JP, Pennycott T, Olsen JE, et al. Phylogenetic analysis by 16 S rDNA gene sequence comparison of avian taxa of Bisgaard and characterization and description of two new taxa of Pasteurellaceae. J Appl Microbiol. 2003;95:354–363. doi: 10.1046/j.1365-2672.2003.01986.x. [DOI] [PubMed] [Google Scholar]
- 80.Gregersen RH, Neubauer C, Christensen H, Korczak B, Bojesen AM, et al. Characterization of Pasteurellaceae-like bacteria isolated from clinically affected psittacine birds. J Appl Microbiol. 2010;108:1235–1243. doi: 10.1111/j.1365-2672.2009.04518.x. [DOI] [PubMed] [Google Scholar]
- 81.Sadeghi HMM, Najafabadi AJ, Abedi D, Dehkordi AJ. Identification of an isolate of Pseudomonas aeroginosa desposited in PTCC as a PHA producer strains: comparison of three different bacterial genomic DNA extraction methods. J Biol Sci. 2008;8:826–830. [Google Scholar]
- 82.Peterson A, Bisgaard M, Christensen H. Real-time PCR detection of Enterococcus faecalis associated with amyloid arthropathy. Lett Appl Microbiol. 2010;51:61–64. doi: 10.1111/j.1472-765X.2010.02861.x. [DOI] [PubMed] [Google Scholar]
- 83.Costa R, Gomes NCM, Milling A, Smalla K. An optimized protocol for simultaneous extraction of DNA and RNA from soils. Braz J Microbiol. 2004;35:230–234. [Google Scholar]
- 84.Webster NS, Negri AP, Munro MMHG, Battershill CN. Diverse microbial communities inhabit Antarctic sponges. Environ Microbiol. 2004;6:288–300. doi: 10.1111/j.1462-2920.2004.00570.x. [DOI] [PubMed] [Google Scholar]
- 85.Wintzingerode FV, Gobel UB, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 86.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, et al. 2010. Geneious. 5.5 ed. Available from http://www.geneious.com.
- 87.Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. New screening software shows that most recent large 16 S rRNA gene clone libraries contain chimeras. Appl Environ Microb. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ludwig W, Strunk O, Westram R, Richter L, Meier H, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb. 2009;75:7537–7540. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Spring S, Lins U, Amann R, Schleifer K, Ferreira LCS, et al. Phylogenetic affiliation and ultrastructure of uncultured magnetic bacteria with unusually large magnetosomes. Arch Microbiol. 1998;169:136–147. doi: 10.1007/s002030050553. [DOI] [PubMed] [Google Scholar]
- 92.Polz MF, Cavanaugh CM. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microb. 1998;64:3472–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.DeLong EF. Archaea in costal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ishii K, Fukui M. Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Appl Environ Microb. 2001;67:3753–3755. doi: 10.1128/AEM.67.8.3753-3755.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16 S rRNA. Appl Environ Microb. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]