Abstract
The centromere is a key region for cell division where the kinetochore assembles, recognizes and attaches to microtubules so that each sister chromatid can segregate to each daughter cell. The centromeric chromatin is a unique rigid chromatin state promoted by the presence of the histone H3 variant CENP-A, in which epigenetic histone modifications of both heterochromatin or euchromatin states and associated protein elements are present. Although DNA sequence is not regarded as important for the establishment of centromere chromatin, it has become clear that this structure is formed as a result of a highly regulated epigenetic event that leads to the recruitment and stability of kinetochore proteins. We describe an integrative model for epigenetic processes that conform regional chromatin interactions indispensable for the recruitment and stability of kinetochore proteins. If alterations of these chromatin regions occur, chromosomal instability is promoted, although segregation may still take place.
Keywords: Chromosome Instability, DNA Methylation, Histone Modifications, Kinetochore
Introduction
The centromere is an essential locus that is required for the accurate segregation of genetic material during mitosis and meiosis. It serves as a platform upon which the kinetochore assembles; thus, it is a vital structure for mitotic spindle attachment that is required to guide chromosomal movements during cell division. Centromeres are vital in this task and serve a conserved role in many organisms; however, there is a surprising variability in the structure’s sequence and organization among eukaryotes. Eukaryotic centromeres are characterized by the presence of a histone H3 variant known as centromeric protein A (CENP-A) in mammals.1 Centromeres are located near or within repetitive DNA sequences, but sequence specificity has only been found in budding yeast. The budding yeast centromere is determined by a 125 bp DNA element that is assembled into a single Cse4 nucleosome, which captures a single microtubule.2 Other organisms lack this sequence specificity in such a way that even centromeric DNA within the same organism varies among chromosomes.
In humans, centromeres are defined by AT-rich repeats called α satellites, which are based on a 171 bp monomer that is tandemly arranged into higher order arrays that extend from 0.2 to 5 Mb.3 In human chromosomes, CENP-A is located on the α satellite DNA. However, its binding does not appear to be sequence-specific, as CENP-A is confined to just a portion of a given multi-mega base array. It also does not bind to scattered monomeric α satellite DNA or at inactive centromeres of naturally occurring dicentric human centromeres that contain two regions of α satellite DNA.4,5 Neocentromeres are ectopic centromeres formed de novo typically at regions of non-repetitive DNA and may be formed locally at sequences near the centromere or hundreds of kilobases away from a deleted centromere in gene-poor regions with some repetitive sequence.6
The deposition of CENP-A to the centromere is mediated by the histone chaperone HJURP (Holliday junction recognition protein). In particular, HJURP’s short N-terminal domain, is responsible for specific and stoichiometric binding to the CENP-A/H4 complex.7,8 The expression of HJURP chaperone is tightly regulated since perturbation of its expression leads to mitotic defects.9
Alpha satellite monomers contain a 17 bp motif, known as the CENP-B box that is recognized by centromere protein B (CENP-B).10 CENP-B is important during de novo centromere assembly and for the proper phasing of centromeric nucleosomes, with the exception of human chromosome Y, which lacks CENP-B boxes at the α satellite and does not associate with CENP-B, although all other centromere proteins are recruited to this site.11,12 Interestingly, in α satellites devoid of CENP-B boxes or those that contain mutated CENP-B boxes, euchromatic DNA and Y alphoid DNA do not form artificial chromosomes.4,13 These findings suggest that CENP-B is essential for centromere formation and that α satellites are the preferred sequence for de novo CENP-A assembly. However, not all α satellite sequences can form de novo centromeres.14
To date, it has become increasingly clear that the chromatin environment has a relevant impact on centromere determination and establishment. Nevertheless, the necessary genomic and chromatin elements that establish and maintain the centromere are still unknown. Moreover, it has been suggested that DNA sequence alone is not always sufficient for centromere establishment or function, which supports theories postulating the involvement of epigenetic or chromatin based mechanisms.
In this review, we focus on centromere chromatin structure and its relationship with epigenetic regulation. We will also discuss centromere epigenetics as a cause of chromosomal instability (CIN) (for further review on centromere epigenetics, see references 15 and 16).
Centromere chromatin and epigenetics
One obstacle in the study of the chromatin environment at normal human centromeres is the nature of the repetitive sequence and shared sequence regions at non-homologous centromeres, making centromeres difficult to evaluate using molecular approaches for long-range chromatin organization analysis. It is known that centromeric chromatin in humans and flies are arranged as CENP-A nucleosomes that are interspersed with H3K4me2 nucleosomes.17 CENP-A is a histone H3 variant found only at functional centromeres over which the kinetochore will eventually assemble18,19; it represents an epigenetic mark necessary for centromere activation. Recently, high-resolution structural data for a CENP-A/H4 heterotetramer have been reported, showing significant structural differences between CENP-A/H4 and the canonical H3-containing nucleosomes. Also, the crystal structure of the human CENP-A has been reported, showing specific differences with the H3 canonical histone, in particular the loop 1 contains two extra aminoacid residues (Arg 80 and Gly 81), which may stabilize centromere chromatin containing CENP-A.20
CENP-A-mediated differences at centromeric chromatin between CATD (CENP-A centromere targeting domain) and H4 beginning at interphase alter the global physical properties of the nucleosome, thus converting the nucleosome into a more rigid structure. This finding supports the existence of a CENP-A-driven self-assembly mechanism that mediates the maintenance of centromere identity.21,22 These differences are essential for centromeric incorporation of CENP-A nucleosomes and reveal the contribution of the histone analog to a specialized chromatin structure at the centromere that differs from typical heterochromatin and euchromatin.
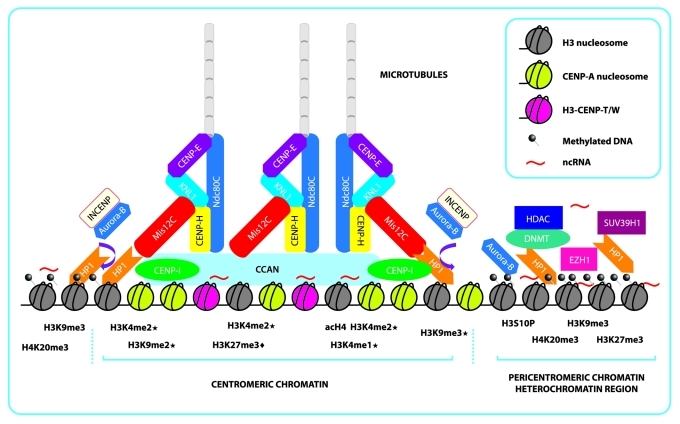
Chicken centromeres also contain CENP-A nucleosomes that are interspersed with H3K9me3 nucleosomes, although H3K4me2 is present in lower amounts.23 In plants such as maize, centromeres are also enriched with H3K9me2 and H3K9me3, while exhibiting low enrichment levels of H3K4me2. Maize centromeric H3 domains are interspersed with H3K27me1,24,25 and high resolution studies of these centromeres also revealed the presence of active genes within the region that are associated with H3K4me2 and acH4 enrichment, suggesting that centromeres are organized as euchromatic pockets surrounded by regions of heterochromatin enriched with H3K9me2 (Fig. 1).26 Also, the H3K4me2 histone mark is an essential part of the chromatin environment of vertebrate kinetochore required for long-term maintenance and function.27
Figure 1. Schematic representation of centromeric and pericentromeric chromatin and the formation of an epigenetic complex that further shapes the kinetochore. Centromeric chromatin is shaped mainly by the presence of rigid CENP-A nucleosomes, interspersed H3-CENP-T/W complex and interspersed histone H3 nucleosomes, that present the repressive marks, H3K9me2/me3 and H3K27me1/3 (“♦” indicates marks only found in plants species and “★” indicates marks reported in more than one species), active marks scattered throughout the region, H3k4me2 and acH4, and ncRNAs that form the foundation. This foundation then recruits the CCAN that will further link the other complexes that recruit the microtubules needed for chromosome segregation. In mammals, many of the CENP proteins that form the inner centromere are recruited by DNA interactions and histone mark-dependent proteins because there is evidence that Mis12C depends on HP1 for its incorporation into the kinetochore. Aurora-B/INCENP is recruited from methylated pericentromeric chromatin in order for Mis12C to interact with NDC80C (represented by the brown arrow). Aurora-B/INCENP dimer is removed from the kinetochore by phosphorylation of H3S10 Pericentromeric chromatin is mainly constitutive heterochromatin composed of repetitive satellite DNA that is heavily methylated and enriched with repressive marks, principally H3K9me3 and scattered H3K27me3, dependent on SUV39H1 and EZH1, which serve to recruit HP1, HDAC, and DNMT. Pericentromeric chromatin is further stabilized and regulated by ncRNAs generated from these satellites regions
All together, these results suggest that centromeric chromatin varies among different species. In spite of these dissimilarities in histone modifications, some findings could be obscured due to resolution limitations caused by the nature of these regions.
Centromere inactivation (centromeres without CENP-A incorporation) is considered an epigenetic phenomenon. Therefore, inactive centromeres may adopt a chromatin configuration that is not compatible with centromere maintenance.28 The chromatin environment in centromeres is different from the usual active and inactive chromatin configuration. It has been suggested by studying artificial chromosomes with integrated α satellite and TetO sequences constructed to tether chromatin-modifying proteins, both transcriptional activators and repressors, which disrupt centromere function on the artificial chromosomes. Thus, purely euchromatic and heterochromatic environments are incompatible with CENP-A assembly and maintenance.29 It has also been suggested that neither large domains of euchromatic nor heterochromatic chromatin are required for the formation of functional neocentromeres.30 A closer analysis using a transcriptional repressor, KAP1, targeted to the synthetic centromere of the artificial chromosome resulted in the depletion of CENP-C and CENP-H followed by depletion of CENP-A, thus providing more evidence for a hierarchy of centromere disassembly in which CENP-A is one of the last proteins to be removed from a centromere that is being inactivated.31 However, it remains unclear whether the changes in chromatin promote CENP disassembly or if CENP removal allows heterochromatin to replace centromere chromatin. In this respect, it has been shown that histone H3-containing nucleosomes readily replace sites of CENP-A occupancy when this protein is depleted, but when overexpressed, CENP-A can replace sites of histone H3 assembly.17 Considering that H3K9me3 is a marker that is thought to function as a limiting factor or antagonist of CENP-A chromatin, the overall ratio of chromatin containing CENP-A to heterochromatin may be more important than just the simple presence or absence of a particular modification.32,33
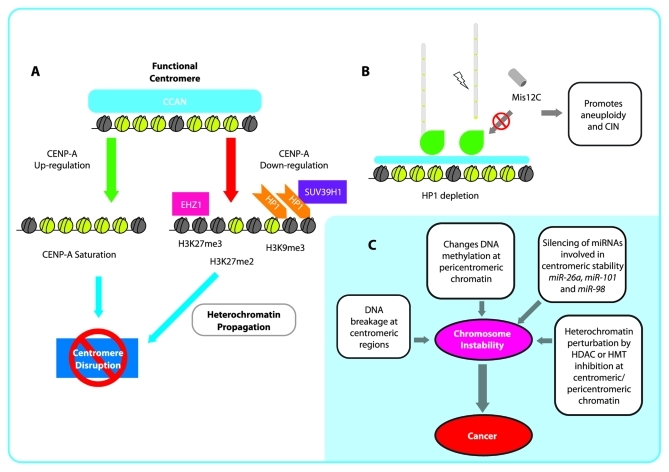
H3K27 methylation is another epigenetic mark that defines heterochromatin, and in some species of plants, it has been associated with centromeres in which H3K27me2/me3 enrichment promotes centromere inactivation of two centromeres of a tricentric chromosome in wheat (Fig. 1).34 Thus, the modification of histone marks in either a heterochromatic or euchromatic state abrogates the capacity of the centromere from generating a kinetochore. Therefore, modifications cause the loss of function to the region during mitosis, which supports the significance of an epigenetic model in the establishment and regulation of the unique centromeric chromatin (Fig. 2a).
Figure 2. Epigenetic disruption of the centromere and implications for chromosomal instability (CIN). (A): Schematic representation of centromere disruption pathways by CENP-A upregulation and heterochromatization. B: HP1 depletion causes that localization of Misc12C to the kinetochore is reduced, which may promote microtubule misincorporation and kinetochore unsteadiness generating aneuploidy and CIN. C: Chromosomal instability generated by different mechanisms at centromeric and pericentromeric regions as a plausible early cause of cancer.
The kinetochore: A macromolecular protein complex
In eukaryotes, accurate chromosome segregation requires each chromosome to interact appropriately with microtubules from the mitotic spindle, which provides the structural framework upon which chromosome segregation occurs. This interaction is mediated by a macromolecular complex known as the kinetochore, which is a structure composed of more than 90 proteins (Fig. 1).35 The kinetochore must facilitate the interaction between centromeric chromatin and dynamic microtubules to ensure the bi-orientation of chromosomes on the metaphase plate and the segregation of sister chromatids at anaphase.36
During S phase, CENP-A is equally segregated between sister chromatids, but new CENP-A is not incorporated into centromeric chromatin until telophase and G1.18 The significance of this abnormal timing remains elusive, but it has been suggested that this timing could represent a defense mechanism against misincorporation and the subsequent assembly of the kinetochore structures at non-centromeric sites caused by temporarily separating the incorporation of CENP-A from replication and the incorporation of other histones.37 In some species, such as Drosophila, CENP-A alone is sufficient to ensure kinetochore formation; however, this is not true in human cells, and CENP-A alone is not sufficient for complete kinetochore assembly in mitosis.38 Additional proteins are also required for the correct assembly. To date, many approaches have been attempted to identify core centromere components in mammals, and 15 proteins were identified and defined as the constitutive centromere associated network (CCAN).1,35,39,40 It has been suggested based on several functional analyses that these proteins play a key structural role in forming a stable foundation for dynamic kinetochore assembly and for providing a proper environment for new CENP-A incorporation.1,35 It was also suggested that CCAN might also function to directly control microtubule dynamics.39
Recently, the CENP-T/W complex has been shown to interact stably with histone H3-containing nucleosomes.39 Although the precise molecular organization of the histone H3-CENP-T/W nucleosome population is not known, but it has been suggested that they are interspersed closely with CENP-A nucleosomes (Fig. 1). The functional consequence of these assembly events would be an expansion of the histone H3-CENP-T/W compartment within post-replicative centromeric chromatin. The dynamic behavior of proteins within this compartment kinetically parallels the active establishment of the kinetochore complex. This suggests that the CENP-T/W complex plays a functional role in kinetochore formation following DNA replication.41
It has been shown that CCAN proteins remain associated with centromeric chromatin through the entire cell cycle. In conjunction with CENP-A, they may form a stable environment for the assembly of the mitotic kinetochore structure. DNA binding activity or direct interaction with CENP-A has been observed for several CCAN proteins.10,35,42
The outer kinetochore plate and fibrous corona assemble upon entry into mitosis and contain proteins required for interactions with microtubules. These proteins include those with direct microtubule binding activity, such as the KL1, Mis12, Ndc80 complex (Ndc80C) (together forming the KMN network),35 CENP-E, and the Ska1 complex,43 all of which are transient factors that modulate this interaction or monitor attachment status.15
Recent experiments on microtubule tension in human cells treated with taxol helped map the internal architecture of the kinetochore in the presence and absence of tension across kinetochore pairs. These studies identified surprising changes in the organization of the layer structure, where the absence of tension leads to a reduced distance between inner kinetochore proteins, such as CENP-C, and the microtubule interacting complex Ndc80. However, the localization of inner kinetochore proteins with respect to each other remained unchanged.44 The reduction of tension across kinetochores also caused striking rearrangements of components of the KMN network, suggesting that some kinetochore-proteins complexes are more dependent than others on forces exerted by microtubule interactions.
Although the kinetochore appears as a stable structure during mitosis, it has recently been suggested that kinetochore proteins are highly dynamic through the cell cycle. Whereas the inner kinetochore proteins of the CCAN are present at the centromere in a constitutive manner throughout the cycle, some outer kinetochore proteins, such as the Mis12 complex (Mis12C) and KLN1, are recruited in G2.45 Because this recruitment is suggested to prime centromere assembly, the assembly of remaining components occurs at prophase and prometaphase to generate the structure capable of binding to microtubules.
From the more than 90 proteins that contribute to kinetochore assembly, four groups have recently been suggested to prime centromere assembly given their known functions: linkers, scaffolds, chaperones and structural stabilizers.15 However, a striking feature of the vertebrate kinetochore is the massive reorganization that takes place during mitosis. In a time period of less than one hour, the kinetochore recruits more than 40 mitotic components in a hierarchical manner and then subsequently disassembles these proteins to return to an interphase state. It has been proposed that this process may be controlled by the presence of the nuclear envelope, which restricts proteins from the nucleus such that they are unable to associate with the kinetochore until nuclear envelope break down occurs.46 However, recent work has demonstrated that at least some proteins are present within the nucleus at times when they are not localized at the kinetochores, suggesting that the formation of the kinetochore during mitosis is not necessarily blocked from assembly by the nuclear envelope.45 Furthermore, post-translational modifications may regulate kinetochore formation. Recent work has demonstrated that the ubiquitin-like protein SUMO modifies CENP-I. When CENP-I is extensively SUMOylated, it is targeted for degradation. Thus, for this protein to become incorporated into the kinetochore during mitosis, the removal of the SUMO group by the SUMO protease SENP6 is required.47
Heterochromatin protein 1: A kinetochore partner
Heterochromatin protein 1 (HP1) was first discovered in Drosophila as a dominant suppressor of position-effect variegation (PEV) and was later found to participate in the formation of compact heterochromatin in an array of pericentric heterochromatin.48 Although initial studies demonstrated the role of HP1 in the formation of heterochromatin, especially in centromeric and pericentromeric regions, it has become increasingly evident that HP1 has multiple functions and is also present in actively transcribed euchromatic regions.49 HP1 also plays a role in centromeric sister chromatid cohesion,50 telomere maintenance,51 and DNA repair.52 In humans, these functions are performed in a specific manner by each of the three subtypes of HP1 that have been identified: HP1α, Hp1β and HP1γ.53,54
HP1 binds to histone H3 that has been methylated at lysine 9 by SUV39H1, and, in turn, it recruits SUV39H1 to the DNA, which further propagates methylation along the chromatin.55 This relationship between HP1 and SUV39H1 is conserved in their Saccharomyces pombe homologs, Swi6 and Clr4,56 suggesting evolutionary conservation of this mechanism of heterochromatin formation. It has been demonstrated that Swi6 and other factors are required for the establishment of de novo centromeres, but not for their maintenance.57
The function of HP1 is highly important in the establishment, propagation and maintenance of constitutive heterochromatin,58 especially at the pericentromeric region that has been demonstrated to be enriched in the H3K9me3 and H4K20me3 marks, hypoacetylated histones H3 and H4, and highly methylated regions along the satellites repeats.59,60 Due to its juxtaposition next to centromeric chromatin, it has been suggested that the organization and stability of the pericentromeric region is crucial to ensuring correct chromosomal segregation during mitosis; therefore, this region is important for genome stability.59,61 Increasing evidence has shown that the KMN network in humans and flies is a binding partner of HP1, where HP1 may participate in recruiting and directing Mis12C to the centromere during interphase (Fig. 1).62-64 It is also suggested that the recruitment of Mis12 protein is performed by HP1.63 Each HP1 subunit consists of a chromodomain, which binds to methylated H3K9, a hinge implicated in the regulation of the protein/DNA/RNA interactions, and a chromoshadow domain, which is responsible for dimerization and for the interactions with binding partners containing the defined motifs comprise of the PXVXL consensus.65 These properties of the HP1 proteins give rise to its function as an adaptor that enables other proteins to interact with chromatin. Recently, in vitro analyses have suggested that Mis12C dimerizes with HP1, but that its interaction with the PVIHL motif located at residues 209–213 of protein NSL1 is largely responsible for binding to the PXVXL consensus.62,66
Moreover, these results give rise to the following question. If H3K9me3 functions as a boundary marker or an antagonizing marker for CENP-A,33 why does a protein that is recruited to the centromeric chromatin in a Suv39h-dependent manner play a major role in directing the Mis12C complex to the kinetochore? Is HP1 involvement regulated in a cycle-dependent manner? Even though enrichment of H3K9me2 and H3K9me3 has been found in the centromere, other marks, such as H3K4me2 and H3K27me3, have been found to be interspersed among centromeres, as previously discussed. In vivo microscopic analyses have demonstrated that human HP1α and HP1β localization has a specific role at different times during the cell cycle. Thus, the localization of human HP1α and HP1β to centromeric heterochromatin at interphase and metaphase is exchanged. Specifically, while in metaphase, HP1β (which is preferentially found at centromeric chromatin) is replaced by HP1α (which is typically located at pericentric and telomeric chromatin). These exchanges are mediated by differences in HP1 chromoshadow domain sequence.67 Recently, in vitro protein interaction analyses demonstrated that the DSN1-NSL1 dimer is a crucial binding partner for HP1, Ndc80C and KNL1C. Therefore, HP1α and Ndc80C are competitive binders of Mis12C, suggesting that they have either identical or overlapping binding sites.66 Hence, for Ndc80C to localize to the kinetochore, it is necessary to displace most of the HP1α from Mis12C. It is clear that this exchange must occur rapidly and in a coordinated fashion during mitosis if chromosome segregation is to occur.68 The disruption or abrogation of HP1 is believed to lead to the formation of some tumors, and it may also be possible that the absence of HP1 may lead to the loss of incorporation of Mis12C into the kinetochore. Therefore, centromere structure and kinetochore relaxation further promote CIN69 (Fig. 2b).
It has been proposed that INCENP localizes to the Aurora-B/AIM-1 complex in heterochromatin, where its kinase activity is required for the dissociation of HP1 from chromosome arms in mitotic cells. This process is mediated by the phosphorylation of H3S10. It has also been shown that Aurora-B/AIM1 regulates the localization of SUV39H.70 These results indicate that Aurora-B/AIM1 is necessary for the regulated histone modifications involved in the binding of HP1 to centromere chromatin during mitosis. However, it is not sufficient by itself to completely regulate the localization of HP1 subtypes during mitosis, implying that other mechanisms are necessary for the event. Moreover, strong evidence suggests that the HP1α localized at the mitotic centromeric heterochromatin contributes to the stability of sister chromatid cohesion or activation of the kinetochore checkpoint. Reduced HP1α at the metaphase centromere may be a cause of chromosomal instability in cancer cells.71,72 Centromeres and kinetochore formation seem to be directly or indirectly regulated by epigenetic mechanisms in most eukaryotes. In particular, this hypothesis is supported by the fact that several kinetochore components are related to epigenetic factors. This finding suggests the deregulation epigenetic components at the kinetochore complex could lead to chromosome defects and the development of chromosomal instability.
Chromosomal instability: Epigenetics and centromere involvement
Two major models of genetic instability have been described. The first model is associated with microsatellite instability (MIN), and the second describes CIN. Microsatellites are repeated sequences of DNA that vary in length among individuals but over the course of an individual’s lifetime, the lengths remain constant. Abnormally long or short microsatellites of DNA are referred as MIN. This phenomenon may be associated with diseases such as cancer. Approximately 15% of colorectal cancers present a MIN phenotype.73 Meanwhile, CIN can develop in two principal ways. One is related to abnormalities in chromosome number, which mainly occur due to the gain or loss of the whole chromosome (W-CIN); the other is associated with an abnormal organization of the chromosome (S-CIN). This faulty organization is characterized by structural changes of the chromosomes by gain, loss or translocation of chromosome fragments, which are mainly caused by breakage. This phenomenon is associated with mitotic errors that allow chromosome missegregation, which can lead to oncogenesis.74 In particular, labile regions of DNA, known as chromosomal fragile sites, are heritable and contain specific loci that are especially prone to breakage and rearrangement.75 These sites lead to rearrangements of large genomic regions by the insertions, deletions or translocations deriving in S-CIN. Thus, the CIN phenomenon promotes the expression of altered oncogenes, the loss of tumor suppressor genes and the deletion of several other genes, such as those encoding microRNAs.76
In cancer, CIN is associated with poor prognosis in solid tumors and results in phenotypic variations that promote drug resistance.77 CIN is a likely cause of tumor cell heterogeneity.77 One of the main hypotheses is that these tumors rapidly acquire multidrug resistance, leading to lower rates of disease-free survival.
It was previously believed that genomic instability develops from strictly genetic mechanisms. However, there is now some evidence that epigenetic processes are also involved. The perturbation of the epigenetic balance may lead to alterations in gene expression and CIN, resulting in cellular transformation and cancer development (Fig. 2B,C).78 DNA methylation, one of the primary epigenetic processes, is performed by the addition of a methyl group to the cytosine base of DNA to form 5-methyl-cytosine (for more information, see references 79–80). DNA methylation has been linked to the silencing of imprinting genes, X-chromosome inactivation and repetitive elements, leading to chromosome stability.81 A growing number of human diseases linked to epigenetic defects are currently being studied. In particular, DNA methylation in cancer gained attention with studies reporting that there is local hypermethylation, mainly at CpG islands, and global hypomethylation in cancer.82,83 This phenomenon is present in the majority of cancers, suggesting that it plays an important role in oncogenic transformation.83-85 Recently, it has been suggested that epigenetic instability represents a theoretical alternative to genetic instability in cancer.86 Several cancers exhibit high degrees of DNA methylation. The difference in the methylation degree gave rise to a tumor sub-classification called a CpG island methylator phenotype, or CIMP. This classification represents a clinically and etiologically distinct group of tumors that is characterized by epigenetic instability.87
It has been reported that some colon cancers that demonstrate genetic instability do not exhibit MIN nor CIN, but do present the CIMP phenotype, suggesting the importance of epigenetic deregulation in cancer.88 The CIMP-positive tumors are clinically distinct from those in the rest of the patient population. These differences could help improve the understanding of the tumor’s origins. Several human genetic disorders have been linked to epigenetic deregulation, such as Prader-Willi, Angelman and Fragile X syndrome,89 but only one human genetic disease is currently known to arise from a germline mutation, namely the immunodeficiency, centromeric region instability and facial anomalies syndrome (ICF).90 ICF is an autosomal recessive disease that involves spontaneous CIN and immunodeficiency. The molecular basis for this disease is related to the mutation of DNA methyltransferase 3B (DNMT3B). This disease is extremely rare and is characterized by profound immunodeficiency due to the absence of or significant reduction in the expression of at least two immunoglobulin isotypes.91
Diseases can have many causes, from a single nucleotide modification to structural changes at the chromosome level, genetic damage, chromosomal rearrangements, mutations, or germinal and somatic deficiencies in genes associated with DNA repair.92 However, the study of epigenetic components has become more relevant in the last two decades because of its implications in multiple cellular processes, such as transcriptional regulation, differentiation, and genomic protection against viral infections. Deregulation of any of these processes is associated with the development of syndromes and diseases such as cancer.93
An epigenetic component implied in CIN is CENP-A. This protein has been reported to be overexpressed in primary colorectal cancer.94 In particular, diminished levels of pRb have been associated with the CENP-A overexpression and the induction of hypodiploid aneuploidy. Bioinformatics analysis at the 5′ upstream sequence of the human CENPA gene revealed a potential E2F motif. This observation could explain the increase of CENP-A transcript in pRb-depleted cells.95,96 A novel hypothesis is that CENP-A overexpression might cause spreading along the centromere heterochromatin through chromosome arms and interfere with the correct kinetochore complex assembly, this being a cause of genomic instability.96
It has also been reported that the overexpression and mislocalization of the CENP-A chaperone HJURP has been observed in lung cancer cell lines. These observations were associated with CIN and immortality of cancer cells.9 In clinical trials, the overexpression of HJURP was associated with an increased sensitivity to radiotherapy but with a decreased survival in patients with breast cancers.97
DNA methylation is another epigenetic process associated with neoplastic disorders in many reports.98 Global hypomethylation and local hypermethylation are broadly represented in cancer, and it is suggested that they might promote CIN as a result of gene expression deregulation.83,86,98,99 The gain or loss of histone marks is associated with gene silencing at a local level,100 and chromatin rearrangements at a global level; both have a profound effect on the local function of the cell and can promote certain diseases. Examples of histone modifying proteins include the deregulation of histone methyltransferases, such as EZH2, and the downregulation of HP1, either by the loss of H3K9me3 or gene mutations. Additionally, the inhibition or reduction of HDAC levels at the centromeric region promotes the accumulation of H3K9ac and H3K14ac, which is expected to cause a loss of chromosomal segregation due to the acetylation-dependent inhibition of H3K9me3 (Fig. 2c).69,101,102
Therefore, it is clear that aberrant changes that modify chromatin structure are important for chromosome stability. Particularly, the co-existence of epigenetic components, such as H3K9me3 and HP1, which are highly enriched at pericentromeric chromatin regions and satellites, may strengthen the hypothesis that HP1 is not only a component that helps establish heterochromatin (thereby making it a protein that is associated with genetic silencing) but is also an important scaffold protein that is involved in kinetochore assembly. If HP1 is disrupted, studies suggest CIN is promoted; therefore, HP1 disruption may lead to cancer.51,62
Non-coding RNA: Covering the centromere
Non-coding RNA (ncRNA) has become an increasingly studied field of research in epigenetic studies, is believed to be involved in chromatin regulation at the level of the centromeres and the kinetochore.103 There are many types of ncRNAs including the following: small interfering RNAs (siRNAs), microRNAs (miRNAs) and long ncRNAs. The expression of these RNAs has a direct effect on chromosomal architecture. Moreover, the transcription of siRNA from the satellite regions that form and stabilize pericentromeric and centromeric DNA has been reported, and this transcription is conserved in species such as Drosophila, mice and humans. Although their function is not yet clear, it is possible that they are involved in the establishment and regulation of chromatin structure in pericentromeric and centromeric regions.
Fission yeast centromeres resemble those of human in their organization and epigenetic nature, but provide a simplified model for the study of complex regional centromeres. The discovery that ncRNA, specifically iRNA, directs chromatin modifying activities to outer repeats of fission yeast centromeres, become a key precedent concerning heterochromatin formation.104 The modulation of such heterochromatin formation is engaged by RNA-induced silencing complexes (RISCs) which downregulate homologous gene expression.105 Its effect in transcriptional gene silencing has been intensively studied in Schizosaccharomyces pombe, where members of the RNAi pathway such as: Dicer, Argonaute (ago1), Chp1 and the RNA-dependent RNA polymerase (Rdp1) plays an important role. Evidence suggest that at sites of active RNAi, chromatin-based activities drives the formation of self-enforcing loop coupling siRNA biogenesis to promote H3K9me2 expansion, where RNAPII transcription of centromeric repeats together with chromodomain proteins bound to H3K9me2 mediate recruitment of silencing factors.106 Interestingly, in fission yeast, RNAPII transcribe centromeric pre-siRNAs at heterochromatin, which act in transcriptional gene silencing. These transcripts require a particular subunit of RNAPII, Rbp7, for initiation of centromeric siRNA precursor transcription that will drive centromeric chromatin silencing.107
For efficient production of centromere repeat homologous to be attained, siRNA is followed by the loading of RNA-induced transcriptional silencing effector complexes (RITS). By means of a component of RITS complex, Chp1, that contain a chromodomain that binds to H3K9me2 modification, RITS associates with heterochromatin repeats.108 RITS, along with its encapsulated ss-siRNA, might be targeted to homologous chromatin via siRNA-nacent transcript complementarity. The siRNA response is amplified by the RNA-dependent RNA polymerase complex (RDRC), which promotes further dsRNA, and, thus, more siRNA synthesis.108
Recently, a surveillance mechanism was proposed, where small RNA degradation products are generated independently of Dicer or RDRC activities that becomes loaded to Ago1. Such Ago1-priRNAs complexes engage homologous centromeric transcripts and recruit Clr4 to promote basal H3K9me2 levels that are sufficient to induce RNAi-mediated heterochromatin establishment.109 However, similar Dicer-independent Ago mediated small regulatory RNA have been characterized in zebrafish and mice.110,111
Moreover, centromeric ncRNA transcript might have different functions rather than heterochromatin propagation and silencing. There is evidence that suggest that transcripts homologous to centromere-associated DNAs are detected in various organisms, since then centromeric transcripts have been found to associate with kinetochore proteins.112-114 In fission yeast, it has been reported that Hrp1, an ATP-dependent remodeling factor (orthologous to S. cerevisiae chd1), affects CENP-A deposition.115 In S. pombe, Hrp1 facilitates the assembly of CENP-A analogous to siRNA derived of outer repeats transcripts drive heterochromatin formation. Thus, Hrp1 facilitates the assembly of CENP-A chromatin, and becomes essential when MIs6 or CENP-A function is impaired. Also, Hrp1 acts at a subset of gene promoters to dissemble histone H3-containing nucleosomes close to the transcription start sites, allowing the deposition of CENP-A-nucleosome at the promoter of some genes in the centromere.116 Such remodeling resembles the transcription-coupled replacement of H3.1, H2A with H3.3 and H2A.Z in metazoans.117 The fact that Hrp1 promotes H3-nucleosome eviction suggests that similar remodeling processes may occur at RNAPII promoters within centromeres. Therefore, it is possible that transcription within centromeres occurs merely as a consequence of having RNAPII promoters that might contribute to promote CENP-A deposition.116 It is also possible that the discrete transcripts of 0.5 kb detected at centromere repeats could be processed into a specific class of small RNAs that have a roll in CENP-A chromatin formation and kinetochore assembly analogous to the siRNA derived from outer repeats transcripts drive heterochromatin formation. Interestingly, in S. cerevisiae it has been shown that CENP-A also tends to associate with some RNAPII promoters where RNAPII binding is enriched; however, not clear association with CENP-A depositions has been observed.118
In humans, recent analyses have found that the human chromatin remodeling factor FACT, whose function is implicated in transcription, interacts with affinity purified CENP-A chromatin.1 Moreover, depletion of FACT was found to impair incorporation of newly synthesized CENP-A in chicken cells.40 Taken together, these results might give evidence of possible transcripts regulated by FACT acting at centromere chromatin.
Hence, if centromeric chromatin in fission yeast and other organisms contains RNPII transcripts that are implicated in many processes at these regions, it is natural to think that such transcripts can be modulated through cell cycle and development. Such an example in development has been recently explored in mammals, where pericentromeric chromatin flanks the centromere; these regions are important in the stability of the centromere and in kinetochore formation. It has been observed that pericentromeric regions in mice, specifically the major satellite region, present high peaks of transcription from the zygote genome followed by rapid downregulation, which coincides with the organization of the chromocenters. Particularly, the paternal genome forward strand DNA was predominantly expressed, suggesting that this paternal bias might reflect the asymmetry in histone marks between maternal and paternal pericentric domains.119 This evidence strongly supports the idea that pericentric satellites have an important functional role during embryo development.
In cancer, it has been reported that the ncRNAs transcripts are expressed in repetitive satellite regions in some solid tumors, suggesting that their deregulation could be involved in the carcinogenesis process.120
Alternatively, extensive studies of miRNAs have revealed their importance in the regulation of multiple gene targets, including key epigenetic components. Interestingly, the overexpression of EZH2 deregulates multiple miRNAs, especially miR-26a, miR-101 and miR-98, in many types of cancers, such as lung, gastric and nasopharyngeal cancer and glioblastoma. Given that ncRNAs are shown to play a key role in establishing the characteristic heterochromatin epigenetic patterns necessary for centromere regions, they may function as structural components.120
Conclusion and final remarks
Centromeric chromatin regions are highly structured and regulated during the cell cycle, leading to the formation of one of the most complex macromolecular machineries that function to maintain genetic information for progeny. It is clear that this huge assembly of proteins must be regulated at many levels and is not dependent on DNA sequence. Centromeres must be an epigenetically regulated region that is needed to generate a unique rigid chromatin state, as defined by the CENP-A nucleosome surrounded by a constitutive heterochromatin region. The conjunction of the centromeric chromatin and euchromatin and heterochromatin is indispensable for the recruitment and stability of kinetochore proteins. If alterations of these chromatin regions occur, chromosomal instability is promoted, although segregation may still take place. This disruption could occur in the early stages of cancer development, supporting the fact that epigenetic mechanisms might be one of the first steps of carcinogenesis. However, there are still many open questions regarding centromere epigenetics and how it regulates kinetochore assembly as well as how the communication between centromeric and pericentromeric chromatin is established.
Acknowledgments
We are particularly indebted with Dr. María Eugenia Gonsebatt Bonaparte and Nicolas Alcaraz Millman for critical reading of the manuscript. This work was supported by the Consejo Nacional de Ciencia y Tecnología (grant number 83959) and by the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica, Universidad Nacional Autónoma de México (grant number IN213311). In loving memory of Carlos Hesselbart, “Carlitos.”
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/18504
References
- 1.Foltz DR, Jansen L, Black B, Yates J, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–69. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 2.Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci USA. 2007;104:14706–11. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ugarković DI. Centromere-competent DNA: structure and evolution. Prog Mol Subcell Biol. 2009;48:53–76. doi: 10.1007/978-3-642-00182-6_3. [DOI] [PubMed] [Google Scholar]
- 4.Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, et al. Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol. 1998;16:431–9. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 5.Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–4. doi: 10.1016/S0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- 6.Ketel C, Wang H, McClellan M, Bouchonville K, Selmecki A, Lahav T, et al. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 2009;5:e1000400. doi: 10.1371/journal.pgen.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci USA. 2010;107:1349–54. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–6. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato T, Sato N, Hayama S, Yamabuki T, Ito T, Miyamoto M, et al. Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 2007;67:8544–53. doi: 10.1158/0008-5472.CAN-07-1307. [DOI] [PubMed] [Google Scholar]
- 10.Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol. 1989;109:1963–73. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choo KH. Domain organization at the centromere and neocentromere. Dev Cell. 2001;1:165–77. doi: 10.1016/S1534-5807(01)00028-4. [DOI] [PubMed] [Google Scholar]
- 12.Earnshaw WC, Ratrie HR, Stetten G. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- 13.Grimes BR, Babcock J, Rudd M, Chadwick B, Willard H. Assembly and characterization of heterochromatin and euchromatin on human artificial chromosomes. Genome Biol. 2004;5:R89. doi: 10.1186/gb-2004-5-11-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrington JJ, Van BG, Mays R, Gustashaw K, Willard H. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet. 1997;15:345–55. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 15.Gascoigne KE, Cheeseman I. Kinetochore assembly: if you build it, they will come. Curr Opin Cell Biol. 2011;23:102–8. doi: 10.1016/j.ceb.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeitlin SG. Centromere: the wild west of the post-genome age. Epigenetics. 2010;5:34–40. doi: 10.4161/epi.5.1.10629. [DOI] [PubMed] [Google Scholar]
- 17.Blower MD, Sullivan B, Karpen G. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–30. doi: 10.1016/S1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jansen LE, Black B, Foltz D, Cleveland D. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stimpson KM, Sullivan B. Histone H3K4 methylation keeps centromeres open for business. EMBO J. 2011;30:233–4. doi: 10.1038/emboj.2010.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–5. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 21.Black BE, Brock M, Bédard S, Woods V, Cleveland D. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci USA. 2007;104:5008–13. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sekulic N, Bassett E, Rogers D, Black B. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–51. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen B, Fukugawa T, et al. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci USA. 2010;107:10484–9. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin W, Lamb J, Zhang W, Kolano B, Birchler J, Jiang J. Histone modifications associated with both A and B chromosomes of maize. Chromosome Res. 2008;16:1203–14. doi: 10.1007/s10577-008-1269-8. [DOI] [PubMed] [Google Scholar]
- 25.Shi J, Dawe R. Partitioning of the maize epigenome by the number of methyl groups on histone H3 lysines 9 and 27. Genetics. 2006;173:1571–83. doi: 10.1534/genetics.106.056853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan H, Jin W, Nagaki K, Tian S, Ouyang S, Buell C, et al. Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell. 2005;17:3227–38. doi: 10.1105/tpc.105.037945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergmann JH, Rodríguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, et al. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–40. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higgins AW, Gustashaw K, Willard H. Engineered human dicentric chromosomes show centromere plasticity. Chromosome Res. 2005;13:745–62. doi: 10.1007/s10577-005-1009-2. [DOI] [PubMed] [Google Scholar]
- 29.Nakano M, Cardinale S, Noskov V, Gassmann R, Vangarelli P, Kandels-Lewis S, et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–22. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alonso A, Fritz B, Hasson D, Abrusan G, Cheung F, Yoda K, et al. Co-localization CENP-C and CENP-H to discontinuous domains of CENP-A chromatin at human neocentromeres. Genome Biol. 2007;8:R148. doi: 10.1186/gb-2007-8-7-r148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardinale S, Bergmann J, Kelly D, Nakano M, Valdivia M, Kimura H, et al. Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol Biol Cell. 2009;20:4194–204. doi: 10.1091/mbc.E09-06-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam AL, Boivin C, Bonney C, Rudd M, Sullivan B. Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc Natl Acad Sci USA. 2006;103:4186–91. doi: 10.1073/pnas.0507947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okamoto Y, Nakano M, Ohzeki J-I, Larionov V, Masumoto H. A minimal CENP-A core is required for nucleation and maintenance of a functional human centromere. EMBO J. 2007;26:1279–91. doi: 10.1038/sj.emboj.7601584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, Friebe B, Gill B, Jiang J. Centromere inactivation and epigenetic modifications of a plant chromosome with three functional centromeres. Chromosoma. 2010;119:553–63. doi: 10.1007/s00412-010-0278-5. [DOI] [PubMed] [Google Scholar]
- 35.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 36.McEwen BF, Dong Y. Contrasting models for kinetochore microtubule attachment in mammalian cells. Cell Mol Life Sci. 2010;67:2163–72. doi: 10.1007/s00018-010-0322-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukugawa T, Cheeseman I. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–22. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heun P, Erhardt S, Blower M, Weiss S, Skora A, Karpen G. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–15. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hori T, Amano M, Suzuki A, Backer C, Welburn J, Dong Y, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–52. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 40.Okada M, Okawa K, Isobe T, Fukagawa T. CENP-H-containing complex facilitates centromere deposition of CENP-A in cooperation with FACT and CHD1. Mol Biol Cell. 2009;20:3986–95. doi: 10.1091/mbc.E09-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prendergast L, van Vuuren C, Kaczmarczyk A, Doering V, Hellwig D, Quinn N, et al. Premitotic assembly of human CENPs -T and -W switches centromeric chromatin to a mitotic state. PLoS Biol. 2011;9:e1001082. doi: 10.1371/journal.pbio.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carroll CW, Milks K, Straight A. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–55. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wood KW, Sakowicz R, Goldstein L, Cleveland D. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–66. doi: 10.1016/S0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- 44.Wan X, O'Quinn R, Pierce H, Joglekar A, Gall W, DeLuca J, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–84. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheeseman IM, Hori T, Fukagawa T, Desai A. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol Biol Cell. 2008;19:587–94. doi: 10.1091/mbc.E07-10-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26:1853–64. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukhopadhyay D, Dasso M. The fate of metaphase kinetochores is weighed in the balance of SUMOylation during S phase. Cell Cycle. 2010;9:3194–201. doi: 10.4161/cc.9.16.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wallrath LL, Elgin S. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 1995;9:1263–77. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- 49.de Wit E, Greil F, Steensel BV. High-resolution mapping reveals links of HP1 with active and inactive chromatin components. PLoS Genet. 2007;3:e38. doi: 10.1371/journal.pgen.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 51.Inoue A, Hyle J, Lechner M, Lahti J. Perturbation of HP1 localization and chromatin binding ability causes defects in sister-chromatid cohesion. Mutat Res. 2008;657:48–55. doi: 10.1016/j.mrgentox.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 52.Ayoub N, Jeyasekharan A, Venkitaraman A. Mobilization and recruitment of HP1: a bimodal response to DNA breakage. Cell Cycle. 2009;8:2945–50. doi: 10.4161/cc.8.18.9486. [DOI] [PubMed] [Google Scholar]
- 53.Singh PB, Miller J, Pearce J, Kothary R, Burton R, Paro R, et al. A sequence motif found in a Drosophila heterochromatin protein is conserved in animals and plants. Nucleic Acids Res. 1991;19:789–94. doi: 10.1093/nar/19.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ye Q, Worman H. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem. 1996;271:14653–6. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- 55.Jenuwein T. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 2001;11:266–73. doi: 10.1016/S0962-8924(01)02001-3. [DOI] [PubMed] [Google Scholar]
- 56.Bannister AJ, Zegerman P, Partridge J, Miska E, Thomas J, Allshire R. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–4. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 57.Folco HD, Pidoux A, Urano T, Allshire R. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–7. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Probst AV, Almouzni G. Heterochromatin establishment in the context of genome-wide epigenetic reprogramming. Trends Genet. 2011;27:177–85. doi: 10.1016/j.tig.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 59.Cowell IG, Aucott R, Mahadevaiah S, Burgoyne P, Huskisson N, Bongiorni S, et al. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma. 2002;111:22–36. doi: 10.1007/s00412-002-0182-8. [DOI] [PubMed] [Google Scholar]
- 60.Heit R, Underhill D, Chan G, Hendzel M. Epigenetic regulation of centromere formation and kinetochore function. Biochem Cell Biol. 2006;84:605–18. doi: 10.1139/o06-080. [DOI] [PubMed] [Google Scholar]
- 61.Vos LJ, Famulski J, Chan G. How to build a centromere: from centromeric and pericentromeric chromatin to kinetochore assembly. Biochem Cell Biol. 2006;84:619–39. doi: 10.1139/o06-078. [DOI] [PubMed] [Google Scholar]
- 62.Kiyomitsu T, Iwasaki O, Obuse C, Yanagida M. Inner centromere formation requires hMis14, a trident kinetochore protein that specifically recruits HP1 to human chromosomes. J Cell Biol. 2010;188:791–807. doi: 10.1083/jcb.200908096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Obuse C, Iwasaki O, Kiyomitsu T, Goshima G, Toyoda Y, Yanagida M. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat Cell Biol. 2004;6:1135–41. doi: 10.1038/ncb1187. [DOI] [PubMed] [Google Scholar]
- 64.Przewloka MR, Zhang W, Costa P, Archambault V, D’Avino P, Lilley KS, et al. Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS ONE. 2007;2:e478. doi: 10.1371/journal.pone.0000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lomberk G, Wallrath L, Urrutia R. The Heterochromatin Protein 1 family. Genome Biol. 2006;7:228. doi: 10.1186/gb-2006-7-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Petrovic A, Pasqualato S, Dube P, Krenn V, Santaguida S, Cittaro D, et al. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J Cell Biol. 2010;190:835–52. doi: 10.1083/jcb.201002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hayakawa T, Haraguchi T, Masumoto H, Hiraoka Y. Cell cycle behavior of human HP1 subtypes: distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase. J Cell Sci. 2003;116:3327–38. doi: 10.1242/jcs.00635. [DOI] [PubMed] [Google Scholar]
- 68.Thomsen R, Christensen D, Rosborg S, Linnet T, Blechingberg J, Nielsen A. Analysis of HP1a regulation in human breast cancer cells. Mol Carcinog. 2011;50:601–13. doi: 10.1002/mc.20755. [DOI] [PubMed] [Google Scholar]
- 69.Terada Y. Aurora-B/AIM-1 regulates the dynamic behavior of HP1alpha at the G2-M transition. Mol Biol Cell. 2006;17:3232–41. doi: 10.1091/mbc.E05-09-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kirschmann DA, Lininger R, Gardner L, Selfor E, Odero V, Ainsztein A, et al. Down-regulation of HP1Hsalpha expression is associated with the metastatic phenotype in breast cancer. Cancer Res. 2000;60:3359–63. [PubMed] [Google Scholar]
- 71.Bernard P, Maure J, Partridge J, Genier S, Javerzat J, Allshire R. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–42. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 72.Rao CV, Yamada H, Yao Y, Dai W. Enhanced genomic instabilities caused by deregulated microtubule dynamics and chromosome segregation: a perspective from genetic studies in mice. Carcinogenesis. 2009;30:1469–74. doi: 10.1093/carcin/bgp081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thoma CR, Toso A, Meraldi P, Krek W. Mechanisms of aneuploidy and its suppression by tumour suppressor proteins. Swiss Med Wkly. 2011;141:w13170. doi: 10.4414/smw.2011.13170. [DOI] [PubMed] [Google Scholar]
- 74.Laganà A, Russo F, Sismeiro C, Giugno R, Pulvirenti A, Ferro A. Variability in the incidence of miRNAs and genes in fragile sites and the role of repeats and CpG islands in the distribution of genetic material. PLoS ONE. 2010;5:e11166. doi: 10.1371/journal.pone.0011166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Calin GA, Sevignani C, Dumitru C, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee AJ, Endesfelder D, Rowan A, Walther A, Birkbak N, Futreal P, et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 2011;71:1858–70. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carter SL, Eklund A, Kohane I, Harris L, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–8. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 78.Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu Rev Pharmacol Toxicol. 2005;45:629–56. doi: 10.1146/annurev.pharmtox.45.120403.095832. [DOI] [PubMed] [Google Scholar]
- 79.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–68. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 80.Watanabe Y, Maekawa M. Methylation of DNA in cancer. Adv Clin Chem. 2010;52:145–67. doi: 10.1016/S0065-2423(10)52006-7. [DOI] [PubMed] [Google Scholar]
- 81.Jones PA, Wolkowicz MJ, Rideout WM, 3rd, Gonzales FA, Marziasz CM, Coetzee GA, et al. De novo methylation of the MyoD1 CpG island during the establishment of immortal cell lines. Proc Natl Acad Sci USA. 1990;87:6117–21. doi: 10.1073/pnas.87.16.6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hansen KD, Timp W, Bravo H, Sabaunciyan S, Langmead B, McDonald OG, et al. Incraesed methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–75. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kanai Y. Genome-wide DNA methylation profiles in precancerous conditions andcancers. Cancer Sci. 2010;101:36–45. doi: 10.1111/j.1349-7006.2009.01383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Issa JP. Colon cancer: it's CIN or CIMP. Clin Cancer Res. 2008;14:5939–40. doi: 10.1158/1078-0432.CCR-08-1596. [DOI] [PubMed] [Google Scholar]
- 85.Ndlovu MN, Denis H, Fuks F. Exposing the DNA methylome iceberg. Trends Biochem Sci. 2011;36:381–7. doi: 10.1016/j.tibs.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 86.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–93. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 87.Georgiades IB, Curtis L, Morris R, Bird C, Wyllie A. Heterogeneity studies identify a subset of sporadic colorectal cancers without evidence for chromosomal or microsatellite instability. Oncogene. 1999;18:7933–40. doi: 10.1038/sj.onc.1203368. [DOI] [PubMed] [Google Scholar]
- 88.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 89.Toyota M, Suzuki H. Epigenetic drivers of genetic alterations. Adv Genet. 2010;70:309–23. doi: 10.1016/B978-0-12-380866-0.60011-3. [DOI] [PubMed] [Google Scholar]
- 90.Jin B, Tao Q, Peng J, Soo H, Wu W, Ying J, et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum Mol Genet. 2008;17:690–709. doi: 10.1093/hmg/ddm341. [DOI] [PubMed] [Google Scholar]
- 91.Ehrlich M. The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease. Clin Immunol. 2003;109:17–28. doi: 10.1016/S1521-6616(03)00201-8. [DOI] [PubMed] [Google Scholar]
- 92.Sawan C, Vaissière T, Murr R, Herceg Z. Epigenetic drivers and genetic passengers on the road to cancer. Mutat Res. 2008;642:1–13. doi: 10.1016/j.mrfmmm.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 93.Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana M, Bibikova M, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 94.Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, et al. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–6. [PubMed] [Google Scholar]
- 95.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 96.Amato A, Schillaci T, Lentini L, Di Leonardo A. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol Cancer. 2009;8:119. doi: 10.1186/1476-4598-8-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hu Z, Huang G, Sadanandam A, Gu S, Lenburg ME, Pai M, et al. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res. 2010;12:R18. doi: 10.1186/bcr2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Toyota M, Ahuja N, Ohe-Toyota M, Herman J, Baylin S, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–6. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Soto-Reyes E, Recillas-Targa F. Epigenetic regulation of the human p53 gene promoter by the CTCF transcription factor in transformed cell lines. Oncogene. 2010;29:2217–27. doi: 10.1038/onc.2009.509. [DOI] [PubMed] [Google Scholar]
- 100.Simon JA, Lange C. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–9. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 101.Jagani Z, Wiederschain D, Loo A, He D, Mosher R, Fordjour P, et al. The Polycomb group protein Bmi-1 is essential for the growth of multiple myeloma cells. Cancer Res. 2010;70:5528–38. doi: 10.1158/0008-5472.CAN-09-4229. [DOI] [PubMed] [Google Scholar]
- 102.Volpe TA, Kidner C, Hall I, Teng G, Grewal S, Martienssen R. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–7. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 103.Pezer Z, Ugarkovic D. Role of non-coding RNA and heterochromatin in aneuploidy and cancer. Semin Cancer Biol. 2008;18:123–30. doi: 10.1016/j.semcancer.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 104.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–7. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 105.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SIS. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–7. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 107.Djupedal I, Portoso M, Spahr H, Bonilla C, Gustafsson CM, Allshire RC, et al. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19:2301–6. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 109.Shanker S, Job G, George OL, Creamer KM, Shaban A, Partridge JF. Continuous requirement for the Clr4 complex but not RNAi for centromeric heterochromatin assembly in fission yeast harboring a disrupted RITS complex. PLoS Genet. 2010;6:e1001174. doi: 10.1371/journal.pgen.1001174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cheloufi S, Dos Santos CO, Chong MMW, Hannon GJ. A dicer- independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–9. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–8. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Topp CN, Zhong CX, Dawe RK. Centromere-encoded RNAs are integral components of the maize kinethochore. Proc Natl Acad Sci USA. 2004;101:15986–91. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146–60. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Carone DM, Longo M, Ferreri GC, Hall L, Harris M, Shook N, et al. A new Class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma. 2009;118:113–25. doi: 10.1007/s00412-008-0181-5. [DOI] [PubMed] [Google Scholar]
- 115.Walfridsson J, Bjerling P, Thalen M, Yoo EJ, Park SD, Ekwall K. The CHD remodeling factor Hrp1 stimulates CENP-A loading to centromeres. Nucleic Acids Res. 2005;33:2868–79. doi: 10.1093/nar/gki579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Choi ES, Strålfors A, Castillo AG, Durand-Dubief M, Ekwall K, Allshire RC. Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J Biol Chem. 2011;286:23600–7. doi: 10.1074/jbc.M111.228510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, et al. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet. 2009;41:941–5. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lefrançois P, Euskirchen GM, Auerbach RK, Rozowsky J, Gibson T, Yellman CM, et al. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics. 2009;10:37. doi: 10.1186/1471-2164-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Probst AV, Okamoto I, Casanova M, El Marjou F, Le Baccon P, Almouzni G. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev Cell. 2010;19:625–38. doi: 10.1016/j.devcel.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 120.Doi A, Park I-H, Wen B, Murakami P, Aryee M, Irizarry R, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–3. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]