Abstract
Background
Recently, there have been a number of studies on the association between XRCC1 polymorphisms and childhood acute lymphoblastic leukemia (ALL) risk. However, the results of previous reports are inconsistent. Thus, we performed a meta-analysis to clarify the effects of XRCC1 variants on childhood ALL risk.
Methods
A meta-analysis was performed to examine the association between XRCC1 polymorphisms (Arg399Gln, Arg194Trp, and Arg280His) and childhood ALL risk. We critically reviewed 7 studies with a total of 880 cases and 1311 controls for Arg399Gln polymorphism, 3 studies with a total of 345 cases and 554 controls for Arg280His polymorphism, and 6 studies with a total of 783 cases and 1180 controls for Arg194Trp polymorphism, respectively. Odds ratio (OR) and its 95% confidence interval (CI) were used.
Results
Significant association between XRCC1 Arg399Gln polymorphism and childhood ALL risk was observed in total population analyses (ORadditive model = 1.501, 95% CI 1.112–2.026, POR = 0.008; ORdominant model = 1.316, 95% CI = 1.104–1.569, POR = 0.002) and Asian subgroup analyses (ORadditive model = 2.338, 95%CI = 1.254–4.359, POR = 0.008; ORdominant model = 2.108, 95%CI = 1.498–2.967, POR = 0.000). No association was detected in Caucasians, Metizo and mixed populations. Ethnicity was considered as a significant source of heterogeneity in the meta-regression model. For the other two XRCC1 polymorphisms, no association with childhood ALL risk was found.
Conclusions
The meta-analysis results suggested that XRCC1 Arg399Gln polymorphism might be associated with elevated childhood ALL risk among Asian population.
Introduction
While survival rates for childhood acute lymphoblastic leukemia (ALL) have been improved significantly over the past 50 years, ALL is still the most common pediatric cancer in developed countries with high incidence and mortality [1].
As a complex and multifactorial process, leukemogenesis is still not fully understood. It is widely known that dysregulated immune response to infection may be a cause of childhood ALL [2]. Although the role of environmental exposure is still currently undefined, it is likely that the environmental carcinogenesis exposure is influenced by co-inheritance of multiple low-risk variants, such as single nucleotide polymorphisms (SNPs) in susceptible genes [3]. These variants can be identified by comparing the frequency of polymorphic genotypes in cases and controls.
To date, the candidates for childhood ALL susceptibility genes have been categorized into those coding for carcinogen metabolism enzymes, folate metabolism enzymes, DNA repair proteins, and others [4]. The DNA repair system plays an important role in maintaining the genome integrity and stability through the reversal of DNA damage. If accumulated mutations are occurred in corresponding DNA repair genes, their reversal capacity could be damaged, substantially increasing the risk of cancer. SNPs in common DNA repair genes have been identified and demonstrated to be linked to several sporadic carcinogenesis [5], [6].
X-ray repair cross-complementing group 1 (XRCC1), located on chromosome 19q13.2–13.3, with 33 kilobases in length, is one of the most important proteins in base excision repair (BER) [7]. BER is also the predominant DNA damage repair pathway for the processing of small base lesions derived from oxidation and alkylation damage [8]. There have been more than 300 validated SNPs in the XRCC1 gene reported in the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP). Nevertheless, only three genetic changes have been extensively studied including Arg194Trp on exon 6 (rs1799782 in dbSNP, C/T), Arg280His on exon 9 (rs25489 in dbSNP, G/A), and Arg399Gln on exon 10 (rs25487 in dbSNP, G/A). There have been a number of studies on the association between XRCC1 polymorphisms and childhood ALL risk [9]–[15]. However, these inconsistent results fail to clarify this complicated genetic relationship because of the small sample size and its low statistical power. To reliably demonstrate the effect of XRCC1 variants (Arg399Gln, Arg280His, and Arg194Trp) on childhood ALL risk, we conduct a meta-analysis of all eligible studies to resolve this pivotal issue.
Materials and Methods
Study identification and selection
Computer searches of PubMed, EMBASE, Medline, Google Scholar and Cochrane Library were performed by two authors independently using the following key words (“childhood acute lymphoblastic leukemia” or ”childhood ALL”)and(“XRCC1” or “X-ray repair cross-complementation group 1”), covering all papers published before November 30, 2011. All eligible articles were retrieved and their references were searched simultaneously to find other relevant articles. Inclusion criteria was defined as follows: (1) case-control studies evaluating the association between XRCC1 polymorphisms and childhood ALL risk; (2) studies based on unrelated individuals; (3) sufficient published data available to estimate an odds ratio (OR) with 95% confidence interval (CI). We excluded studies that were not full-length publications articles or letters in peer-reviewed English journals. When the same patient population was included in different articles, the one with the largest population of participants or the most recent one was selected.
Data extraction
The following information was extracted from each study by two authors independently: first author, publication year, ethnicity, area, mean age of the study subjects, gender component, matching criteria, genotyping method, numbers of cases and controls, and genotype frequency of cases and controls. The two authors achieved a consensus at last.
Statistical analysis
The strength of XRCC1 polymorphisms and childhood ALL risk was assessed by odds ratios (ORs) with the corresponding 95% CI for each study. The OR and its 95% CI in each comparison were assessed in additive (aa versus AA; a was for the minor allele and A was for the major allele), dominant (aa+Aa versus AA), and recessive (aa versus Aa+AA) genetic models. Heterogeneity among studies was tested by chi-square-based Q test, and I 2 statistics was calculated to quantify the proportion of the total variation due to heterogeneity [16]. The pooled ORs were calculated by a fixed-effects model (the Mantel-Haenszel method) when the P value>0.05 for the Q test which indicated a lack of heterogeneity among the studies [17]. Otherwise, a random-effects model (DerSimonian-Laird method) was used [18]. The source of heterogeneity was explored in a meta-regression model. The significance of pooled ORs was determined by Z test (P<0.05 was considered statistically significant). The potential publication bias was examined visually in a funnel plot of log[OR] against its standard error (SE), and the degree of asymmetry was tested by Egger's test (P<0.05 was considered a significant publication bias) [19]. In the control populations, Hardy–Weinberg equilibrium (HWE) was tested. In addition, subgroup analysis for ethnicity (Asian, Caucasian, Mestizo, and Mixed population) was conducted, and influence analysis was performed by omitting each study to find potential outliers [20]. The two authors inputted the data in the statistic software programs STATA version 11.0 to perform the statistical analysis independently and got the same results.
Results
Extraction process and study characteristics
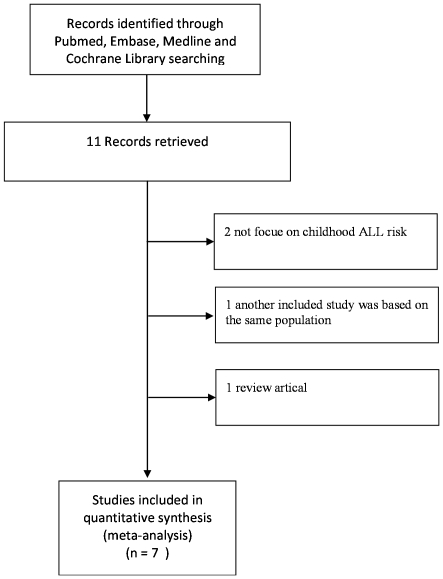
With our search criterion, a total of eleven full-text articles [4], [9]–[15], [21]–[23] were preliminarily identified for further detailed evaluation (Fig. 1). Two studies that not focused on childhood ALL risk were excluded after title review. One study [23] was excluded as another included study [13] was based on the same population, and one was a systematic review. At last, seven case-control studies [9]–[15] were selected, including a total of 880 cases and 1311 controls originally. A list of characteristics of these included studies was provided in Table 1. There were 7 studies with a total of 880 cases and 1311 controls for Arg399Gln polymorphism, 3 studies with a total of 345 cases and 554 controls for Arg280His polymorphism, and 6 studies with a total of 783 cases and 1180 controls for Arg194Trp polymorphism. Genotype distributions in the controls of all studies were in agreement with HWE except one [13].
Figure 1. Flowchart of selection of studies for inclusion in meta-analysis.
Table 1. General characteristics of studies included in the meta-analysis.
| Author | Year | Country | Ethnicity | Source of controls | Matching criteria | Case | Control | SNP studied | HWEa |
| Joseph | 2005 | India | Asian | Hospital-based | sex, age, ethnicity | 117 | 117 | 194, 280, 399 | Yes |
| Pakakasama | 2007 | Thailand | Asian | Hospital-based | ethnicity | 108 | 317 | 194, 280, 399 | Yes |
| Batar | 2009 | Turkey | Caucasian | Hospital-based | sex, age, ethnicity | 70 | 75 | 194, 399 | Yes |
| Meza-Espinoza | 2009 | Mexico | Mestizo | Hospital-based | ethnicity | 120 | 120 | 194, 280, 399 | Yes |
| Tumer | 2010 | Turkey | Caucasian | Hospital-based | ethnicity | 167 | 190 | 194, 399 | Yesb |
| Canalle | 2011 | Brizal | Mixed | Hospital-based | - | 201 | 361 | 194,399 | Yes |
| Stanczyk | 2011 | Poland | Caucasian | Hospital-based | ethnicity | 97 | 131 | 399 | Yes |
HWE Hardy–Weinberg equilibrium.
Genotype distributions of Arg194Trp in the controls were significantly deviated from HWE 399 Arg399Gln, 194 Arg194Trp, 280 Arg280His.
Meta-analysis results
Table 2 listed the main results of the meta-analysis
Table 2. Results of the meta-analysis on XRCC1 polymorphisms and childhood ALL risk.
| Polymorphism | Analysis | Case/Control | Additive model(aa vs AA) | Dominant model(Aa+aa vs AA) | Recessive model (aa vs AA+Aa) | |||
| OR[95% CI] | P/P het | OR[95% CI] | P/P het | OR[95% CI] | P/P het | |||
| Arg399Gln | Overall | 880/1311 | 1.501[1.112, 2.026] | 0.008/0.384 | 1.316[1.104, 1.569] | 0.002/0.016 | 1.324[0.998, 1.757] | 0.052/0.628 |
| Asian | 225/434 | 2.338[1.254, 4.359] | 0.008/0.903 | 2.108[1.498, 2.967] | 0.000/0.821 | 1.698[0.937, 3.077] | 0.081/0.706 | |
| Caucasian | 334/396 | 1.289[0.830, 2.000] | 0.258/0.168 | 1.275[0.945, 1.722] | 0.112/0.381 | 1.138[0.760, 1.704] | 0.530/0.228 | |
| Mestizo | 120/120 | 1.711[0.653, 4.479] | 0.274/− | 1.306[0.787, 2.169] | 0.302/− | 1.556[0.612, 3.954] | 0.353/− | |
| Mixed | 201/361 | 1.227[0.629, 2.397] | 0.548/− | 0.845[0.597, 1.194] | 0.339/− | 1.358[0.707, 2.606] | 0.358/− | |
| Arg280His | Overall | 345/554 | 1.251[0.371, 4.220] | 0.709/0.675 | 1.125[0.805, 1.574] | 0.490/0.534 | 1.203[0.357, 4.055] | 0.765/0.693 |
| Asian | 225/434 | 1.053[0.251, 4.426] | 0.944/0.425 | 1.172[0.774, 1.775] | 0.453/0.284 | 0.989[0.234, 4.175] | 0.988/0.461 | |
| Mestizo | 120/120 | 2.023[0.180, 22.720] | 0.568/− | 1.043[0.590, 1.843] | 0.885/− | 2.017[0.180, 22.545] | 0.569/− | |
| Arg194Trp | Overall | 783/1180 | 0.806[0.451, 1.438] | 0.465/0.041 | 1.056[0.850, 1.312] | 0.625/0.059 | 0.797[0.445, 1.426] | 0.444/0.070 |
| Asian | 225/434 | 0.710[0.319, 1.578] | 0.400/0.014 | 0.966[0.684, 1.363] | 0.843/0.007 | 0.714[0.319, 1.600] | 0.414/0.028 | |
| Caucasian | 237/265 | 0.585[0.145, 2.351] | 0.450/0.056 | 1.240[0.781, 1.969] | 0.361/0.166 | 0.551[0.136, 2.233] | 0.404/0.061 | |
| Mestizo | 120/120 | 2.150[0.520, 8.885] | 0.290/− | 1.265[0.730, 2.190] | 0.402/− | 2.053[0.501, 8.405] | 0.317/− | |
| Mixed | 201/361 | 0.443[0.049, 4.000] | 0.469/− | 0.929[0.586, 1.474] | 0.755/− | 0.446[0.050, 4.020] | 0.472/− | |
OR Odds ratio; 95% CI 95% confidence interval.
a:minor allele; A:major allele.
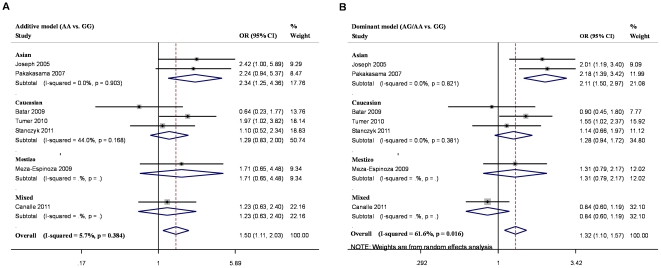
The results of the associations between XRCC1 Arg399Gln polymorphism and childhood ALL risk, and the heterogeneity test were shown in Table 2. When all the eligible studies were pooled into the meta-analysis, elevated childhood ALL risk was revealed in additive model (OR = 1.501, 95% CI 1.112–2.026, POR = 0.008, P = 0.384 for heterogeneity)(Fig. 2a). We also found a significant association with childhood ALL risk in the dominant model (OR = 1.316, 95% CI = 1.104–1.569, POR = 0.002, P = 0.016 for heterogeneity) (Fig. 2b). No significant association was found in the recessive model (OR = 1.324, 95% CI = 0.998–1.757, P = 0.628 for heterogeneity). In subgroup analysis by ethnicity, the results revealed significant associations between the XRCC1 Arg399Gln polymorphism and childhood ALL in Asian population (Additive model: OR = 2.338, 95%CI = 1.254–4.359, POR = 0.008, P = 0.903 for heterogeneity; Dominant model: OR = 2.108, 95%CI = 1.498–2.967, POR = 0.000 P = 0.821 for heterogeneity). We did not observe any significant association in any genetic model among other subgroups. Moreover, meta-regression analysis revealed that ethnicity was a significant source of between-study heterogeneity (P = 0.007).
Figure 2. Meta-analysis of XRCC1 Arg399Gln polymorphism in childhood ALL. a Additive model, b Dominant model.
There was no statistical difference in all contrasts of genotypes for Arg280His polymorphism (Additive model: OR = 1.251, 95%CI 0.371–4.220, POR = 0.709; Dominant model: OR = 1.125, 95% CI 0.805–1.574, POR = 0.490; Recessive model: OR = 1.203, 95%CI 0.357–4.055, POR = 0.765) and Arg194Trp polymorphism(Additive model: OR = 0.806, 95% CI 0.451–1.438, POR = 0.465; Dominant model OR = 1.056, 95% CI 0.850–1.312, POR = 0.625; Recessive model = 0.797, 95% CI 0.445–1.426, P OR = 0.444). Subgroup analysis based on ethnicity also showed no significant association between the two SNPs and childhood ALL risk.
Tests of heterogeneity
We have found heterogeneities in four studies: Arg399Gln polymorphism dominant model (p = 0.016, I 2 = 61.6%); Arg194Trp polymorphism additive model (p = 0.041, I 2 = 56.9%), dominant model (p = 0.059, I 2 = 53%), recessive model (p = 0.070, I 2 = 50.9%) (Table 2). A random-effects model was employed in these studies.
Sensitivity analysis
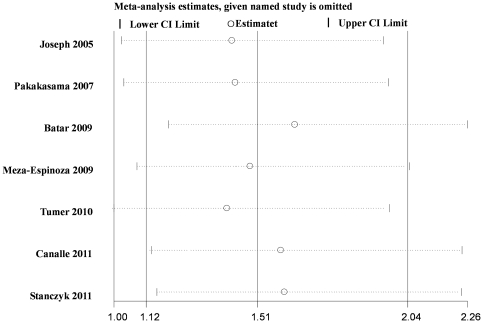
Influence analysis was performed to assess the influence of each individual study on the pooled OR by sequential omission of individual studies. The results suggested that no individual study significantly affected the pooled ORs (Fig. 3).
Figure 3. Influence analysis for AA versus GG in the overall meta-analysis.
This figure shows the influence of individual studies on the summary OR. The middle vertical axis indicates the overall OR and the two vertical axes indicate its 95% CI. Open circles indicate the pooled OR when the left study is omitted in this meta-analysis. The two ends of the dotted lines represent the 95% CI.
Publication bias
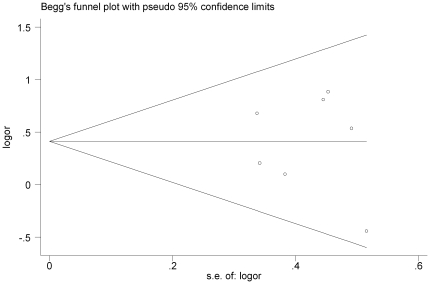
Publication bias was examined by Funnel plot and Egger's regression test. The shapes of the funnel plot did not indicate any evidence of obvious asymmetry in additive model (Fig. 4) and the Egger's test suggested the absence of publication bias (P = 0.810).
Figure 4. Funnel plot of XRCC1 Arg399Gln polymorphism and childhood ALL risk for publication bias.
Discussion
XRCC1 plays an important role in the DNA damage repair pathway for the processing of small base lesions, which has been thought of as the predominant DNA-damage repair pathway for the processing of small base lesions derived from oxidation and alkylation damage [7]. It is widely accepted that alterations in DNA repair genes play roles in the process associated with the etiology of cancers. In some of the previous studies, it has been reported that carriers of the variant allele were at higher risk of lung cancer [24], breast cancer [25], and prostate cancer among Asians [26], whereas the result was controversial in gastric cancer [27], as well as in bladder caner [28]. In this meta-analysis, we focused on XRCC1 genetic polymorphisms and provide the most comprehensive assessment of its association with childhood ALL risk. By critically reviewing 7 studies on XRCC1 Arg399Gln polymorphism (a total of 880 cases and 1311 controls), 3 studies on XRCC1 Arg280His polymorphism (a total of 345 cases and 554 controls), and 6 studies on Arg194Trp polymorphism (a total of 783 cases and 1180 controls), we performed a meta-analysis to indicate that the polymorphisms in XRCC1 Arg399Gln was significantly associated with risk of childhood ALL. However, we did not observe associations of XRCC1 Arg280His polymorphism and Arg194Trp polymorphism with childhood ALL risk.
This study showed that the mean frequency of the XRCC1 variant 399Gln allele was 28.99% (Table S1). Previous investigations found that the frequency distribution of 399Gln allele significantly varied in different ethnicities. Among Asian population, 22%∼28% had at least one copy of the variant allele XRCC1 399Gln [9]–[10], [29]–[30], while among Caucasian, Turkish, and other ethnic populations, the frequencies were 28%∼43% [11], [13], [31]–[33]. This may lead to XRCC1 Arg399Gln polymorphism genotype distribution disequilibrium when all ethnic populations were pooled together. Ethnicity was significantly associated with childhood ALL risk and it was considered as a significant source of heterogeneity in the meta-regression model. It was essential to conduct a subgroup analysis based on ethnicities. In this meta-analysis, all subjects were classified into four ethnic groups (Caucasian, Asian, Metizo and mixed populations). No association was detected in Caucasians, Metizo and mixed populations, while increased risk was found in Asian population carrying variant 399Gln allele homozygote, and it should be further investigated in large scale Asian populations.
Despite lots of investigations in many aspects of childhood ALL, little attention has been paid to its pathogenesis, particularly with respect to genetic susceptibility. Several reports have demonstrated the association between some DNA repair gene variants and childhood ALL, so the possible relationship between polymorphisms of DNA repair genes and childhood ALL may be helpful in understanding the pathogenesis of childhood ALL and the prevention of this disease. Our results suggest that the risk of childhood ALL may be associated with DNA repair mechanisms. XRCC1 polymorphisms may be used as an important predictive factor, and ethnic background might have an impact on the results in the studies of its polymorphisms in childhood ALL. Analysis of these polymorphisms, particularly XRCC1 codon 399 Arg/Gln may help in identifying individuals at risk of developing ALL and providing an essential information source for future improvement of ALL treatment.
Although our result is suggestive, there are still some limitations in this meta-analysis. First, heterogeneity among the studies, resulting from different sources of controls, matching criteria of age and gender or some other factors, may influence the results of this analysis. The matching criteria of the control group, such as age, gender, and environment exposures, are different between studies. Furthermore, childhood ALL is heterogeneous considering its underlying cellular and molecular biology, so subtypes of B- or T-cell precursor ALL may not be suspected to share a common etiology. To date none of the studies have examined the relationship between XRCC1 variants and risk by subtype. Second, specific environmental and lifestyle factors may alter those associations between gene polymorphisms and cancer risk. Evidence supports that there is no association between Gln/Gln genotype of Arg399Gln and bladder cancer risk in total population, but it is associated with a decreased risk of bladder cancer among ever smokers [34], [35]. The relationship between XRCC1 gene polymorphism and childhood ALL risk was analyzed without consideration of gene–gene and gene-environment interactions because of the lack of sufficient data, which should be further investigated. Third, the Funnel plot did not reveal any evidence of obvious publication bias, while there is still a possibility that our meta-analysis was biased toward a positive result since negative findings were likely to be unreported.
In summary, it is a worthy and meaningful enterprise to search for polymorphic variants influencing the risk of childhood ALL. This meta-analysis suggests that XRCC1 399Gln might be a susceptibility allele for childhood ALL. However, we could not observe any association of XRCC1 Arg194Trp and Arg280His with childhood ALL. We have searched as many publications as we could by means of various searching approaches, comprehensively assessed publication biases and pinpointed the potential sources of heterogeneity via subgroup and sensitivity analysis. However, these results may be biased by the relatively small number of subjects, and therefore need to be validated by larger studies and subsequent update of the current meta-analysis. In order to understand the mechanisms underlying childhood ALL better, future research should be considered and investigated.
Supporting Information
Frequencies of XRCC1 Arg194Trp, Arg280His and Arg399Gln allele among control population in different studies.
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was financially supported by Young Scholar Grant from National Natural Science Foundation of China (Grant Number: 30800488) and Excellent Young Scholarship from Shanghai Health Bureau(XYQ2011007). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schafer ES, Hunger SP. Optimal therapy for acute lymphoblastic leukemia in adolescents and young adults. Nat Rev Clin Oncol. 2011;8:417–424. doi: 10.1038/nrclinonc.2011.77. [DOI] [PubMed] [Google Scholar]
- 2.Cardwell CR, McKinney PA, Patterson CC, Murray LJ. Infections in early life and childhood leukaemia risk: a UK case-control study of general practitioner records. Br J Cancer. 2008;99:1529–1533. doi: 10.1038/sj.bjc.6604696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehta PA, Davies SM, Kumar A, Devidas M, Lee S, et al. Perforin polymorphism A91V and susceptibility to B-precursor childhood acute lymphoblastic leukemia: a report from the Children's Oncology Group. Leukemia. 2006;20:1539–1541. doi: 10.1038/sj.leu.2404299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vijayakrishnan J, Houlston RS. Candidate gene association studies and risk of childhood acute lymphoblastic leukemia: a systematic review and meta-analysis. Haematologica. 2010;95:1405–1414. doi: 10.3324/haematol.2010.022095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts MR, Shields PG, Ambrosone CB, Nie J, Marian C, et al. Single-nucleotide polymorphisms in DNA repair genes and association with breast cancer risk in the web study. Carcinogenesis. 2011. 2011;32:1223–1230. doi: 10.1093/carcin/bgr096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shiraishi K, Kohno T, Tanai C, Goto Y, Kuchiba A, et al. Association of DNA repair gene polymorphisms with response to platinum-based doublet chemotherapy in patients with non-small-cell lung cancer. J Clin Oncol. 2010;28:4945–4952. doi: 10.1200/JCO.2010.30.5334. [DOI] [PubMed] [Google Scholar]
- 7.Chou WC, Wang HC, Wong FH, Ding SL, Wu PE, et al. Chk2-dependent phosphorylation of XRCC1 in the DNA damage response promotes base excision repair. EMBO J. 2008;27:3140–3150. doi: 10.1038/emboj.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, et al. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci U S A. 2007;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joseph T, Kusumakumary P, Chacko P, Abraham A, Pillai MR. DNA repair gene XRCC1 polymorphisms in childhood acute lymphoblastic leukemia. Cancer Lett. 2005;217:17–24. doi: 10.1016/j.canlet.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 10.Pakakasama S, Sirirat T, Kanchanachumpol S, Udomsubpayakul U, Mahasirimongkol S, et al. Genetic polymorphisms and haplotypes of DNA repair genes in childhood acute lymphoblastic leukemia. Pediatr Blood Cancer. 2007;48:16–20. doi: 10.1002/pbc.20742. [DOI] [PubMed] [Google Scholar]
- 11.Batar B, Güven M, Bariş S, Celkan T, Yildiz I. DNA repair gene XPD and XRCC1 polymorphisms and the risk of childhood acute lymphoblastic leukemia. Leuk Res. 2009;33:759–763. doi: 10.1016/j.leukres.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Meza-Espinoza JP, Peralta-Leal V, Gutierrez-Angulo M, Macias-Gomez N, Ayala-Madrigal ML, et al. XRCC1 polymorphisms and haplotypes in Mexican patients with acute lymphoblastic leukemia. Genet Mol Res. 2009;8:1451–1458. doi: 10.4238/vol8-4gmr687. [DOI] [PubMed] [Google Scholar]
- 13.Tumer TB, Yilmaz D, Tanrikut C, Sahin G, Ulusoy G, et al. DNA repair XRCC1 Arg399Gln polymorphism alone, and in combination with CYP2E1 polymorphisms significantly contribute to the risk of development of childhood acute lymphoblastic leukemia. Leuk Res. 2010;34:1275–1281. doi: 10.1016/j.leukres.2010.02.035. [DOI] [PubMed] [Google Scholar]
- 14.Canalle R, Silveira VS, Scrideli CA, Queiroz RG, Lopes LF, et al. Impact of thymidylate synthase promoter and DNA repair gene polymorphisms on susceptibility to childhood acute lymphoblastic leukemia. Leuk Lymphoma. 2011;52:1118–1126. doi: 10.3109/10428194.2011.559672. [DOI] [PubMed] [Google Scholar]
- 15.Stanczyk M, Sliwinski T, Cuchra M, Zubowska M, Bielecka-Kowalska A, et al. The association of polymorphisms in DNA base excision repair genes XRCC1, OGG1 and MUTYH with the risk of childhood acute lymphoblastic leukemia. Mol Biol Rep. 2011;30:445–451. doi: 10.1007/s11033-010-0127-x. [DOI] [PubMed] [Google Scholar]
- 16.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. [Google Scholar]
- 17.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 18.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 19.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tobias A. Assessing the influence of a single study in the meta-analysis estimate. Stata Tecnol Bull. 1999;8:15–17. [Google Scholar]
- 21.Krajinovic M, Labuda D, Mathonnet G, Labuda M, Moghrabi A, et al. Polymorphisms in genes encoding drugs and xenobiotic metabolizing enzymes, DNA repair enzymes, and response to treatment of childhood acute lymphoblastic leukemia. Clin Cancer Res. 2002;8:802–810. [PubMed] [Google Scholar]
- 22.da Silva Silveira V, Canalle R, Scrideli CA, Queiroz RG, Bettiol H, et al. Polymorphisms of xenobiotic metabolizing enzymes and DNA repair genes and outcome in childhood acute lymphoblastic leukemia. Leuk Res. 2009;33:898–901. doi: 10.1016/j.leukres.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Tumer TB, Sahin G, Arinç E. Association between polymorphisms of EPHX1 and XRCC1 genes and the risk of childhood acute lymphoblastic leukemia. Arch Toxicol. 2012;86:431–439. doi: 10.1007/s00204-011-0760-8. [DOI] [PubMed] [Google Scholar]
- 24.Kiyohara C, Takayama K, Nakanishi Y. Association of genetic polymorphisms in the base excision repair pathway with lung cancer risk: a meta-analysis. Lung Cancer. 2006;54:267–283. doi: 10.1016/j.lungcan.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Saadat M, Ansari-Lari M. Polymorphism of XRCC1 (at codon 399) and susceptibility to breast cancer, a meta-analysis of the literatures. Breast Cancer Res Treat. 2009;115:137–144. doi: 10.1007/s10549-008-0051-0. [DOI] [PubMed] [Google Scholar]
- 26.Geng J, Zhang Q, Zhu C, Wang J, Chen L. XRCC1 genetic polymorphism Arg399Gln and prostate cancer risk: a meta-analysis. Urology. 2009;72:4648–4653. doi: 10.1016/j.urology.2009.02.046. [DOI] [PubMed] [Google Scholar]
- 27.Xue H, Ni P, Lin B, Xu H, Huang G. X-ray repair cross-complementing group 1 (XRCC1) genetic polymorphisms and gastric cancer risk: A HuGE review and meta-analysis. Am J Epidemiol. 2011;173:363–375. doi: 10.1093/aje/kwq378. [DOI] [PubMed] [Google Scholar]
- 28.Wang C, Sun Y, Han R. XRCC1 genetic polymorphisms and bladder cancer susceptibility: a meta-analysis. Urology. 2008;72:869–872. doi: 10.1016/j.urology.2007.12.059. [DOI] [PubMed] [Google Scholar]
- 29.Xing D, Qi J, Miao X, Lu W, Tan W, et al. Polymorphisms of DNA repair Genes XRCC1 and XPD and their associations with risk of esophageal squamous cell carcinoma in a Chinese population. Int J Cancer. 2002;100:600–605. doi: 10.1002/ijc.10528. [DOI] [PubMed] [Google Scholar]
- 30.Lee SG, Kim B, Choi J, Kim C, Lee I, et al. Genetic polymorphisms of XRCC1 and risk of gastric cancer. Cancer Lett. 2002;187:53–60. doi: 10.1016/s0304-3835(02)00381-6. [DOI] [PubMed] [Google Scholar]
- 31.Vural P, Değirmencioğlu S, Doğru Abbasoğlu S, Saral NY, Akgül C, et al. Genetic polymorphisms in DNA repair gene APE1, XRCC1 and XPD and the risk of preeclampsia. Eur J Obstet Gynecol Reprod Biol. 2009;146:160–164. doi: 10.1016/j.ejogrb.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 32.Harth V, Schafer M, Abel J, Maintz L, Neuhaus T, et al. Head and neck squamous-cell cancer and its association with polymorphic enzymes of xenobiotic metabolism and repair. J Toxicol Environ Health A. 2008;71:887–897. doi: 10.1080/15287390801988160. [DOI] [PubMed] [Google Scholar]
- 33.Coppedè F, Migheli F, Lo Gerfo A, Fabbrizi MR, Carlesi C, et al. Association study between XRCC1 gene polymorphisms and sporadic amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2009;25:1–3. doi: 10.3109/17482960903220297. [DOI] [PubMed] [Google Scholar]
- 34.Wang C, Sun Y, Han R. XRCC1 genetic polymorphisms and bladder cancer susceptibility: a meta-analysis. Urology. 2008;72:869–872. doi: 10.1016/j.urology.2007.12.059. [DOI] [PubMed] [Google Scholar]
- 35.Lao T, Gu W, Huang Q. A meta-analysis on XRCC1 R399Q and R194W polymorphisms, smoking and bladder cancer risk. Mutagenesis. 2008;23:523–532. doi: 10.1093/mutage/gen046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Frequencies of XRCC1 Arg194Trp, Arg280His and Arg399Gln allele among control population in different studies.
(DOC)