Abstract
LAY ABSTRACT
Autism is known to be highly heritable, and has been associated with abnormalities in the development of several brain structures, including the cerebellum. Previous research has hinted that a gene controlling the development of posterior brain regions such as the cerebellum, may influence risk for autism. This gene is called Homeobox Domain A1 (HOXA1), and the variant within HOXA1 that has been most studied in relation to autism (A218G) falls within a gene region that is important for HOXA1 protein functioning. Although we know that autism appear to influence the dynamics of brain development, and that cerebellar anatomy continues to change over the lifespan – we do not know if A218G genotype influences cerebellar development over time. We studied this issue in typically developing controls who had a total of 296 repeat structural brain scans taken between ages 5 and 23 years of age. The volume of multiple cerebellar components was measures by hand in each scan, and we related developmental changes in these volumes to A218G genotype. We found that, in a part of the cerebellum implicated in autism, A218G genotype modified the rate of cerebellar growth. This suggests for the first time that the putative ASD risk gene HOXA1 has the capacity to modify the longitudinal development of cerebellar systems implicated in ASD neurobiology.
SCIENTIFIC ABSTRACT
Homeobox-A-1 (HOXA1) has been proposed as a candidate gene for autism spectrum disorder (ASD) as it regulates embryological patterning of hind-brain structures implicated in autism neurobiology. In line with this notion, a non-synonymous single nucleotide polymorphism within a highly conserved domain of HOXA1 - A218G (rs10951154) - has been linked to both ASD risk, and cross-sectional differences in superior posterior lobar cerebellar anatomy in late adulthood. Despite evidence for early onset and developmentally dynamic cerebellar involvement in ASD, little is known of the relationship between A218G genotype and maturation of the cerebellum over early development. We addressed this issue using 296 longitudinally acquired structural magnetic resonance imaging brain scans from 116 healthy individuals between 5 and 23 years of age. Mixed models were used to compare the relationship between age and semi-automated measures of cerebellar volume in A-homozygotes (AA) and carriers of the G allele (Gcar). Total cerebellar volume increased between ages 5 and 23 years in both groups. However, this was accelerated in the Gcar relative to the AA group (Genotype-by-age interaction term, p=0.03), and driven by genotype-dependent differences in the rate of bilateral superior posterior lobar volume change with age (p=0.002). Resultantly, although superior posterior lobar volume did not differ significantly between genotype groups at age 5 (p=0.9), by age 23 it was 12% greater in Gcar than AA (p=0.002). Our results suggest that common genetic variation within this putative ASD risk gene has the capacity to modify the development of cerebellar systems implicated in ASD neurobiology.
Keywords: Autism, HOXA1, Cerebellum, Gene, Brain, MRI
INTRODUCTION
In recent years there has been increasing interest in using combinations of neuroimaging and genetics to delineate the biological pathways through which specific genetic variants may modify risk for heritable neurodevelopmental conditions (Hariri & Weinberger, 2003; Meyer-Lindenberg, 2010). To date however, relatively few studies have applied this approach to study putative risk genes for Autism Spectrum Disorder (ASD) (Raznahan et al., 2010; Raznahan et al., 2009; Wassink et al., 2007). Here, we use such an “Imaging Genetics” approach to relate genetic variation within a candidate risk gene for ASD to longitudinal measures of regional structural cerebellar maturation within a large sample of typically developing controls.
Interest in regional cerebellar maturation as a potential marker of genetic risk for ASD stems from several observations. First, reduced cerebellar purkinje cell count has become one of the most consistent findings in ASD post-mortem studies (Palmen, van, Hof, & Schmitz, 2004), which suggests that disruption of cerebellar development in ASD may be evident very early in life. Second, there are multiple structural magnetic resonance imaging (sMRI) reports of abnormal cerebellar anatomy in children and adults with ASD (Stanfield et al., 2008). Such reports have examined diverse anatomical indices including volume and mid-sagittal cerebellar surface area. The best-replicated finding in these studies is abnormality within lobule VI of the cerebellum (Mitchell et al., 2009; Stanfield et al., 2007; Webb et al., 2009) – arguing that a regional approach to measurement of cerebellar anatomy is desirable in ASD neurobiology. Furthermore, several cross-sectional reports suggest that cerebellar abnormalities in ASD may vary as a function of age (Cleavinger et al., 2008; Courchesne et al., 2001; Stanfield et al., 2007) – indicating that candidate cerebellar markers of ASD risk would be best studied using longitudinal study designs. Finally, the notion that genetic risk factors may shape cerebellar alterations in a developmentally dynamic manner within ASD is supported by evidence from twin and family neuroimaging studies demonstrating that the magnitude of genetic contribution may vary with age (Posthuma et al., 2000; Wallace et al., 2006). Despite the above lines of evidence, the relationship between specific putative ASD risk alleles and developmental changes in regional cerebellar anatomy remains unexamined.
The strongest candidate gene for such a longitudinal imaging genetics approach is Homeobox-A-1 (HOXA1), which encodes a protein of known importance for hind-brain patterning in vertebrates. Homeobox-A-1 (HOXA1) is one of a family of evolutionarily conserved genes that govern morphogenesis and cellular differentiation of the central nervous system through their function as transcription factors (Akin & Nazarali, 2005). The initial impetus for examining HOXA1 as a candidate gene for ASD came from overlap in the hind-brain abnormalities seen in post-mortem studies of people with idiopathic ASD (Bauman & Kemper, 2005), hind-brain alterations in both animal HOXA1 knockout models, and post-mortem studies of people with ASD in the context of teratogenic syndromes caused by molecules known to impact HOXA1 signaling (e.g. sodium valproate (Ingram et al., 2000; Rodier, Ingram, Tisdale, Nelson, & Romano, 1996)). Subsequently, case-series of humans with rare HOXA1 mutations found that learning disability and ASD were central to the resulting syndrome, which was accompanied by radiological evidence of hind-brain malformation (Tischfield et al., 2005).
To date, three independent groups have linked a common non-synonymous mutation with an evolutionarily conserved HOXA1 exon to risk for ASD (Conciatori et al., 2004; Ingram et al., 2000) or related traits (Chakrabarti et al., 2009) [although a different allele was implicated as imparting risk in each study, and several negative association studies also exist (Collins, Schroer, Bird, & Michaelis, 2003; Devlin et al., 2002; Romano et al., 2003; Sen et al., 2007; Talebizadeh et al., 2002)]. This single nucleotide polymorphism (SNP) (A218G, rs10951154) results in a His->Arg amino acid sequence change in a highly conserved polyhistidine tail HOXA1 protein domain. Although there are no published studies that directly test A218G functionality, sequence variation within this polyhistidine domain is known to result in increased cell death and impaired neuronal differentiation – suggesting that allelic variation at A218G may impact HOXA1 functioning and brain development (Paraguison et al., 2005; Paraguison et al., 2007). Give this, it is notable that allelic variation at the A218G SNP has been shown to modulate head circumference in people with ASD diagnoses (Conciatori et al., 2004), and in typically developing children (Muscarella et al., 2007). The lack of a relationship between A218G genotype and head circumference in healthy adults (Muscarella et al., 2007) suggests that the relationship between this SNP and brain anatomy may change with development. This possibility has never been explored, but is supported by a recent observation that HOXA1 expression in human brain tissue varies dramatically as a function of age (Siegmund et al., 2007).
Despite extensive evidence that HOXA1 plays a role in hind-brain development, only one study has related HOXA1 A218G genotype to sMRI measures of hind-brain anatomy. Canu et al (Canu et al., 2009) found that carriage of the G allele was associated with regional reductions of cerebellar volume within the anterior and superior posterior lobes in two independent cohorts of healthy adults (Canu et al., 2009). Because this was a cross-sectional study limited to adults, it remains unknown if A218G influences cerebellar anatomy in childhood, and whether the relationship between A218G genotype and anatomy changes with age. As outlined above, these questions are especially relevant to the potential role of HOXA1 in ASD neurobiology given meta-analytic evidence that differences in cerebellar anatomy between people with ASD and TDCs vary with age (Stanfield et al., 2007). Therefore, in order to more fully assess the potential for HOXA1 to influence structural cerebellar phenotypes in ASD, we examined the relationship between A218G genotype and longitudinal measures of regional cerebellar anatomy in a large sample of typically developing children, adolescents and young adults.
METHODS
Sample
Please see Table 1 for full details of participant demographics and genotype composition. We included 296 structural magnetic imaging brain scans that had been acquired longitudinally on 116 healthy unrelated Caucasian children and adolescents aged between ages 5 and 23 years or age. Participants were recruited through local advertisement. The absence of neurological or psychiatric illness was established through completion of a screening questionnaire (Child Behavior Checklist) and a structured diagnostic interview administered by a child psychiatrist (Giedd et al., 1999). Participants were of mixed handedness (handedness established using Physical and Neurological Examination of Soft Signs).
TABLE 1.
PARTICIPANT CHARACTERISTICS
| CHARACTERISTIC | GENOTYPE GROUP | |
|---|---|---|
| AA | Gcar | |
| Number of individuals | 93 | 23 |
| Male, No | 50 | 13 |
| Handedness, No | ||
| L | 17 | 5 |
| R | 76 | 18 |
| IQ, mean (SD) | 113 (11.8) | 109 (14.6) |
| VIQ | 112 (12.1) | 109 (14.8) |
| PIQ | 112 (12.5) | 105 (14.9) |
| SES | 41 (18.5) | 49 (20.9) |
| Total number of scans | 243 | 53 |
| Number of scans, No | ||
| 1 scan | 21 | 7 |
| 2 scans | 22 | 6 |
| ≥3 scans | 50 | 10 |
| Age at each scan in years, mean (SD) | ||
| 1st scan | 10.9 (3.6) | 11.9 (4) |
| 2nd scan | 12.9 (3.4) | 14.7 (4.6) |
| 3rd scan | 15.5 (3.4) | 15.9 (4.1) |
| 4th scan | 18.1 (2.5) | 19.3 (1.8) |
| Age distribution of scans, years | ||
| Mean (SD) | 13.4 (4.3) | 14.0 (4.5) |
| Range | 5.6–23.5) | 5.3–23.5 |
All participants had a full-scale intelligence quotient (FSIQ) of greater than 80 as determined using age-appropriate Wechsler Intelligence Scales including WISC-R, WAIS-R and WASI (Shaw et al., 2006)). Socioeconomic status (SES) was quantified using Hollingshead scales (Hollingshead, 1975). The institutional review board of the National Institutes of Health approved the research protocol employed in this study and written informed consent and assent to participate in the study were obtained from parents and children respectively.
Genotyping
For each participant, DNA was extracted from previously prepared lymphoblastoid cell lines using standard methods (Qiagen, MD, USA). It has been established that conversion of cells into lymphoblastoid lines does not cause errors into SNP genotyping (Herbeck et al., 2009). Genotyping was performed by Prevention Genetics (Marshfield, WI, USA), using sub-microlitre allele-specific polymerase-chain-reactions (Hawkins, Khripin, Valdes, & Weaver, 2002). DNA sequencing of positive controls was conducted to ensure correct assignment of genotypes. Allele frequencies were A 0.89 and G 0.11 - in keeping with reference CEPH data for populations of European descent. Participants were grouped as G-carriers (20%) or A-homozygotes (80%). Genotype frequencies did not deviate from Hardy-Weinberg equilibrium.
Neuroimaging
Of all 116 participants with at least one brain sMRI scan, 60% had 2 or more, and 15% had 3 or more scans. Scans were acquired at approximately 2-year intervals. All sMRI scans were T-1 weighted images with contiguous 1.5 mm axial slices and 2.0 mm coronal slices, obtained on the same 1.5-T General Electric (Milwaukee, WI) Signa scanner using a 3D spoiled gradient recalled echo sequence with the following parameters: Echo time, 5ms; Repetition time, 24ms; flip angle 45 (DEG); acquisition matrix, 256 × 192; number of excitations, 1; and field of view, 24 cm. Head placement was standardized as described previously.
All measures of cerebellar anatomy were derived using the semi-automated method described in Pierson et al (Pierson et al., 2002). Briefly, 31 pre-specified cerebellar landmarks were defined in each scan. Each landmark in the cerebellar co-ordinate space was averaged over all scans in which landmarks had been defined. This created a reference landmark standard to which all subsequent scans could be warped prior to segmentation. Segmentation was achieved using an artificial neural network (ANN) approach (Magnotta et al., 2002), which was “trained” on 30 different cerebella that had been manually segmented into the following structures: Corpus Medullare (Cerebellar white matter and output nuclei), Cerbellar Vermis, Anterior Lobe (comprising lobules I, II, III, IV and V), Superior Posterior Lobe (comprising lobules VI, and VIIA-crus I) and Inferior Posterior Lobe (comprising lobules VIIA-crus II, VIIB, VIII, IX and X). Each cerebellum to be segmented was submitted to the ANN for labeling, and the output then hand-edited to conform with reference tracing standards. Final overlap and intra-class correlation coefficients between semi-automated and manual approaches, as established in the original methodological publication for this method were favorable. For all sub-regions, overlap values exceeded 82%, and ICCs were greater than 0.9. This technique was implemented within our study cohort with high inter- and intra-rater reliabilities (all intra-class correlation coefficients over 0.7). As was reported elsewhere (Mackie et al., 2007), for cerebellar lobes, initial intra-rater and interr-ater intraclass correlations (ICCs) were greater than 0.80 for all 5 tracers. For the vermis, initial intra-rater ICCs were greater than 0.80, and inter-rater ICCs were greater than 0.68. Following the initial phase of measuring, two tracers continued measuring cerebellar lobes over a four-year span. Longitudinal lobar (left and right anterior, superior posterior and inferior posterior) intra-rater ICCs over this period ranged from 0.69 (superior vermis) to 0.99. All of these ICCs were approximately or at least 0.70, which is a qualitatively acceptable (good) level of reliability (based on Fleiss (1986) who proposes a standard of 0.40 to 0.75 as fair to good reliability, and above 0.75 as excellent). Concurrent lobar inter-rater ICCs were excellent and ranged from 0.91 to 0.99, and longitudinal lobar inter-rater ICCs were good to excellent and ranged from 0.72 to 0.96. ICCs were calculated using the “one -way random” option in SPSS. In addition to calculating ICCs, measurements were graphed to examine tracer drift and systematic biases within tracers. In the current sample, one tracer completed all of the vermis measurements that followed the initial phase of measuring. Longitudinal ICCs for the vermis were 0.92 for the total vermis, and ranged from 0.70 to 0.98 for the three vermal sub-regions (anterior, superior and inferior).
Statistical Analysis
Longitudinal mixed-models were used to estimate the effects of genotype on the rate of linear volumetric change over time. We used mixed-models as these permit the inclusion of multiple measurements per person at different ages, and irregular intervals between measurements, thereby increasing statistical power (Pinheiro & DM, 2000). A nested random effects term that modeled within family and within person dependence of observations was included. All models were run with and without sex, handedness, IQ and SES status as both main effects and in interaction with genotype terms. These terms were not included in the final model because (i) their inclusion did not add significantly to the predictive power of the model (as determined using a likelihood ratio test), (ii) their addition did not alter the findings. Linear age terms were used for all cerebellar measures as preliminary analyses established that higher-order age terms could not significantly predict volumetric measures within the smaller genotype sub-group. This is likely to reflect sample-size limitations because curvilinear volume changes in cerebellar volume have been identified in larger samples (Tiemeier et al., 2010). Use of linear age protected against the risk of finding trajectory differences between genotype groups as an artifact of differences in group-size impacting the ability to identify significant higher-order age effects. We also protected against artifactual effects of differences in genotype group-sizes, by confirming that all significant findings in the whole sample held when re-examined within a smaller subsample where each G-carrier were longitudinally matched 1:1 with an AA homozygotes.
We analyzed the relationship between HOXA1 genotype and cerebellar maturation in a staged manner, beginning with total cerebellar volume change as our developmental phenotype of interest. Next we distinguished vermal and non-vermal (i.e cereballar hemispheres) structures given evidence for a difference in their embryonic origins (Yachnis & Rorke, 1999). Finally, we separately analyzed genetic influences on each of three vermal sub-components - Anterior Lobe (encompassing lobules I, II, III, IV and V), Superior Posterior Lobe (encompassing lobules VI and Crus I of lobule VIIA) and Inferior Posterior Lobe (encompassing Crus II of lobule VIIA and lobules VIIB, VIII, IX and X). Non-vermal cerebellar volume was divided into the same three lobar compartments and the corpus medullare (central cerebellar white matter and output nuclei).
RESULTS
As has been previously shown in another longitudinal study of cerebellar development (Tiemeier et al., 2010), total cerebellar volume increased between ages 5 and 23 years. However, this was accelerated in G-carriers relative to A-homozygotes (Genotype-by-age interaction term, p=0.03). The interaction was driven by non-vermal rather than rather than vermal structures (Genotype-by-age interaction terms p=0.01 and p=0.3 respectively). Analysis of non-vermal subdivisions revealed that allelic effects on cerebellar anatomy were localized to the Superior Posterior Lobe (p=0.002) only (Anterior Lobe p=0.2, Inferior Posterior Lobe p=0.5, Corpus Medullare p=0.1). Genotypic effects on bilateral Superior Posterior Lobe volumes held in both hemispheres (Left p=0.01, Right p= 0.002). Resultantly, although bilateral Superior Posterior Lobe volume did not differ significantly between genotype groups at age 5 (p=0.9), by age 23 it was 12% greater in G-carriers than A-homozygotes (p=0.002). There were no statistically significant relationships between A218G genotype and vermal volume – either as a main effect (p=0.11), or in interaction with age (p=0.32).
DISCUSSION
Our results suggest that there are significant differences in bilateral Superior Posterior Lobe growth according to A218G genotype. As a result, although there were no statistically significant differences in the volume of this cerebellar sub-region between genotype groups in childhood, a highly significant volume increase in G-carriers relative to A-homozygotes emerged by the third decade of life. Cross-sectional analyses in two independent groups of healthy adults in the sixth decade of life have also linked A218G genotype to Superior Posterior lobar volume, although G-carriers had volumetric reductions compared to A-homozygotes (Canu et al., 2009). Different “directions” of genotype effects in our current study and that of Canu et al (Canu et al., 2009) could be due to differences in the age-ranges studied. The upper limit of the age-range examined in our study was 23 years, whereas the sample studies by Canu et al began at age 40 years, and extended to beyond 60 years of age. Therefore, it is possible that the relationship between A218G genotype and Superior Posterior Lobe anatomy changes between the third and sixth decades of life, such that exaggerated growth in G carriers relative to A homozygotes during the early decades of life when cerebellar volume is known to increase in age (Tiemeier et al., 2010), gives way to exaggerated volume loss in G carriers relative to A homozygotes in later life, when cerebellar volume is known to reduce with age (Raz, Dupuis, Briggs, McGavran, & Acker, 1998). This hypothesis is in keeping with existing evidence for developmentally dynamic relationships between A218G genotype and another anatomical index of relevance to the central nervous system – head circumference (Conciatori et al., 2004; Muscarella et al., 2007).
Although the molecular mechanism and cellular pathways through which allelic variation at the A218G SNP might modify cerebellar development remain unclear, our findings argue for a close assessment of the potential functional consequences of this non-synonymous SNP. While the canonical understanding that HOXA1 expression is restricted to hind-brain regions including and caudal to rhombomere 4 argues against the possibility that a functional HOXA1 polymorphim might modify cerebellar structures (which derive from rhombomere 1), recent evidence for more rostral HOXA1 expression suggests several mechanisms through which genetically-determined variation in HOXA1 functioning could impact cerebellar development (McClintock et al., 2003; Wang & Zoghbi, 2001). In addition to arguing that HOXA1 may play a role in normal human cerebellar development, our findings raise the possibility that the tentative links that have been made between functional variation within HOXA1 signaling pathways and ASD risk (Rasalam et al., 2005) may be mediated by the capacity of HOXA1 to modify development of the cerebellum. This argument is strengthened by the fact that our study localizes A218G genotype influences on cerebellar maturation to the superior posterior cerebellar lobe, which includes the cerebellar lobule that has most consistently shown abnormalities in ASD - lobule VI (Courchesne et al., 1994; Hashimoto et al., 1995; Kaufmann et al., 2003; Stanfield et al., 2007; Webb et al., 2009).
An important next step would be to determine if our findings can be replicated in independent samples of typically developing individuals. To more directly examine if HOXA1 regulation of cerebellar development is relevant to ASD pathogenesis, it will also be necessary to compare the relationship between HOXA1 genotype and cerebellar anatomy in people with ASD and typically developing controls. Disruption of normative relationships between HOXA1 genotype and cerebellar development in people with ASD could either reflect the action of primary risk factors for ASD, or the secondary consequences of having ASD. Theoretically, these two possibilities would be best distinguished by studying fully discordant monozygotic twin pairs, although this is not practically possible in ASD due to very high monozygotic concordance rates (Folstein & Rutter, 1977). Moreover, sample size restrictions of ASD twin studies would make it very hard to longitudinally model developmental changes in cerebellar anatomy. An alternative approach would be to study gene-brain anatomy in a longitudinal sample of probands with ASD and their unaffected siblngs. This paradigm has been used with some success in psychiatric conditions other than ASD (Raznahan et al., 2011), but comes with its own compilations. These include the inability of sibling studies to distinguish shared genetic and environmental risks for ASD; the fact genetic imaging findings in unaffected siblings of probands with ASD may not only reflect the action of primary ASD risk factors in the absence of active disease, but could also reflect causes of resilience in healthy siblings that are absent in affected probands; and the inadequacy of sib-designs for defining markers of genetic risk in non-polygenic scenarios of ASD causation (Frazer, Murray, Schork, & Topol, 2009)(Frazer et al., 2009).
Our study has several limitations. Firstly, our sample of typically developing controls is restricted to people of European descent, and while this reduced the likelihood of population stratification, it limits the generalizability of out findings. Secondly, our parcellation of the cerebellum did not distinguish white and gray matter, which could show different relationships with HOXA1 genotype. Thirdly, our sample size was such that we were not able to model the curvilinear changes in cerebellar volume with age that have been described in larger longitudinal studies (Tiemeier et al., 2010).
Despite these limitations, our study represents the largest, and only longitudinal, imaging-genetic investigation of HOXA1 to date. Our findings suggest that this putative ASD risk gene has the capacity to modify the development of cerebellar systems implicated in ASD neurobiology.
Figure 1. HOXA1 genotype modulates cerebellar development.
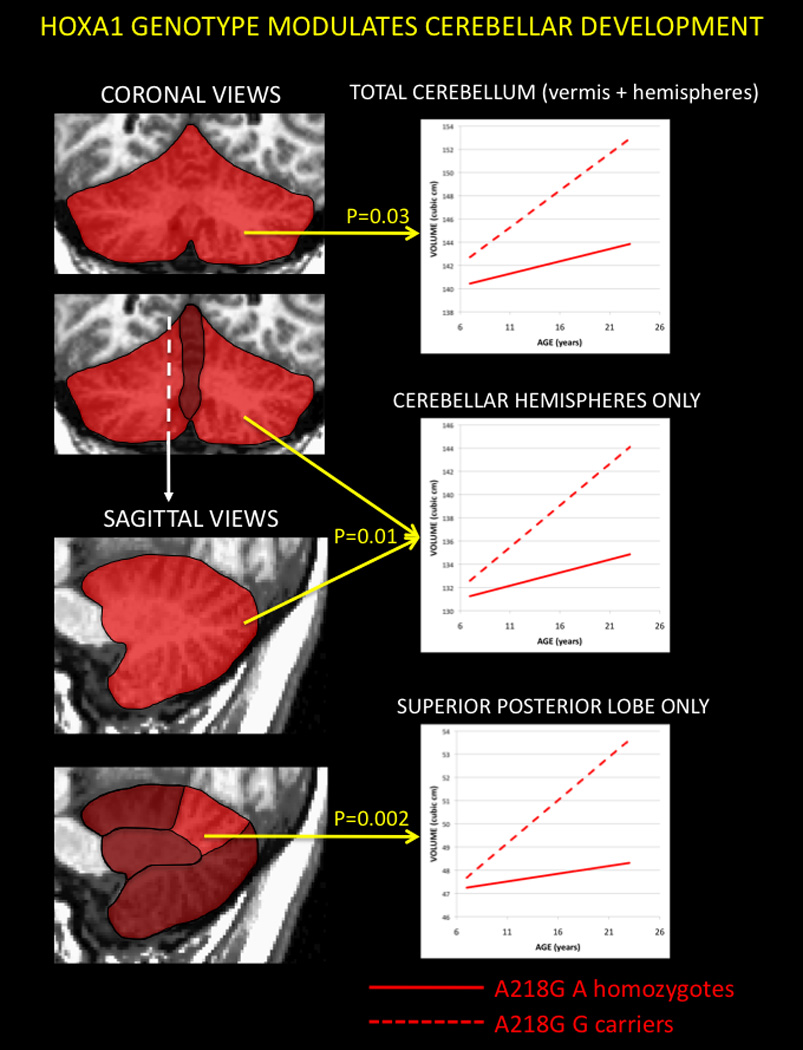
Relationship Between Genotype at HOXA1 A218G Polymorphism and Developmental Changes in Cerebellar Volume. Geneotypic influences on cerebellar anatomy are presented in a staged manner. Shaded coronal and then sagittal views of the cerebellum are used to highlight successive localization of significant HOXA1 modulation of cerebellar maturation, moving from total cerebellar volume, through non-vermal cerebellar volume to posterior superior lobe volume. P values for the interaction between age and A218G genotype are provided by each line plot.
ACKNOWLEDGMENTS
AR was funded by the UK Medical Research Council Fellowship (G0701370) while carrying out aspects of this work. AR would like to thanks Profs Patrick Bolton, Declan Murphy and Gareth Barker for their helpful comments on earlier versions of this manuscript.
REFERENCES
- Akin ZN, Nazarali AJ. Hox genes and their candidate downstream targets in the developing central nervous system. Cellular and Molecular Neurobiology. 2005;25(3–4):697–741. doi: 10.1007/s10571-005-3971-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman ML, Kemper TL. Neuroanatomic observations of the brain in autism: A review and future directions. International Journal of Developmental Neuroscience. 2005;23(2–3):183–187. doi: 10.1016/j.ijdevneu.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Canu E, Boccardi M, Ghidoni R, Benussi L, Duchesne S, Testa C, et al. HOXA1 A218G polymorphism is associated with smaller cerebellar volume in healthy humans. Journal of Neuroimaging. 2009;19(4):353–358. doi: 10.1111/j.1552-6569.2008.00326.x. [DOI] [PubMed] [Google Scholar]
- Chakrabarti B, Dudbridge F, Kent L, Wheelwright S, Hill-Cawthorne G, Allison C, et al. Genes related to sex steroids, neural growth, and social-emotional behavior are associated with autistic traits, empathy, and Asperger syndrome. Autism Res. 2009;2(3):157–177. doi: 10.1002/aur.80. [DOI] [PubMed] [Google Scholar]
- Cleavinger HB, Bigler ED, Johnson JL, Lu J, McMahon W, Lainhart JE. Quantitative magnetic resonance image analysis of the cerebellum in macrocephalic and normocephalic children and adults with autism. Journal of the International Neuropsychological Society. 2008;14(3):401–413. doi: 10.1017/S1355617708080594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JS, Schroer RJ, Bird J, Michaelis RC. The HOXA1 A218G polymorphism and autism: lack of association in white and black patients from the South Carolina Autism Project. Journal of Autism & Developmental Disorders. 2003;33(3):343–348. doi: 10.1023/a:1024414803151. [DOI] [PubMed] [Google Scholar]
- Conciatori M, Stodgell CJ, Hyman SL, O'Bara M, Militerni R, Bravaccio C, et al. Association between the HOXA1 A218G Polymorphism and Increased Head Circumference in Patients with Autism. [References] Biological Psychiatry. 2004;55(4):413–419. doi: 10.1016/j.biopsych.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Courchesne E, Karns CM, Davis HR, Ziccardi R, Carper RA, Tigue ZD, et al. Unusual brain growth patterns in early life in patients with autistic disorder: an MRI study. Neurology. 2001;57(2):245–254. doi: 10.1212/wnl.57.2.245. [DOI] [PubMed] [Google Scholar]
- Courchesne E, Saitoh O, Yeung-Courchesne R, Press GA, Lincoln AJ, Haas RH, et al. Abnormality of cerebellar vermian lobules VI and VII in patients with infantile autism: identification of hypoplastic and hyperplastic subgroups with MR imaging. AJR Am.J.Roentgenol. 1994;162(1):123–130. doi: 10.2214/ajr.162.1.8273650. [DOI] [PubMed] [Google Scholar]
- Devlin B, Bennett P, Cook EH, Jr, Dawson G, Gonen D, Grigorenko EL, et al. No evidence for linkage of liability to autism to HOXA1 in a sample from the CPEA network. American Journal of Medical Genetics. 2002;114(6):667–672. doi: 10.1002/ajmg.10603. [DOI] [PubMed] [Google Scholar]
- Folstein S, Rutter M. Genetic influences and infantile autism. Nature. 1977;265(5596):726–728. doi: 10.1038/265726a0. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10(4):241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Blumenthal J, Jeffries NO, Castellanos FX, Liu H, Zijdenbos A, et al. Brain development during childhood and adolescence: a longitudinal MRI study. Nature Neuroscience. 1999;2(10):861–863. doi: 10.1038/13158. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Weinberger DR. Imaging genomics. British Medical Bulletin. 2003;65(1):259–270. doi: 10.1093/bmb/65.1.259. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Tayama M, Murakawa K, Yoshimoto T, Miyazaki M, Harada M, et al. Development of the brainstem and cerebellum in autistic patients. Journal of Autism & Developmental Disorders. 1995;25(1):1–18. doi: 10.1007/BF02178163. [DOI] [PubMed] [Google Scholar]
- Hawkins JR, Khripin Y, Valdes AM, Weaver TA. Miniaturized sealed-tube allele-specific PCR. Hum Mutat. 2002;19(5):543–553. doi: 10.1002/humu.10060. [DOI] [PubMed] [Google Scholar]
- Herbeck JT, Gottlieb GS, Wong K, Detels R, Phair JP, Rinaldo CR, et al. Fidelity of SNP array genotyping using Epstein Barr virus-transformed B-lymphocyte cell lines: implications for genome-wide association studies. PLoS One. 2009;4(9):e6915. doi: 10.1371/journal.pone.0006915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingshead AB. Four-factor index for social status. New Haven: Yale UP; 1975. [Google Scholar]
- Ingram JL, Stodgell CJ, Hyman SL, Figlewicz DA, Weitkamp LR, Rodier PM. Discovery of allelic variants of HOXA1 and HOXB1: genetic susceptibility to autism spectrum disorders. Teratology. 2000;62(6):393–405. doi: 10.1002/1096-9926(200012)62:6<393::AID-TERA6>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Cooper KL, Mostofsky SH, Capone GT, Kates WR, Newschaffer CJ, et al. Specificity of cerebellar vermian abnormalities in autism: a quantitative magnetic resonance imaging study. Journal of Child Neurology. 2003;18(7):463–470. doi: 10.1177/08830738030180070501. [DOI] [PubMed] [Google Scholar]
- Magnotta VA, Harris G, Andreasen NC, O'Leary DS, Yuh WTC, Heckel D. Structural MR image processing using the BRAINS2 toolbox. Computerized Medical Imaging & Graphics. 2002;26(4):251–264. doi: 10.1016/s0895-6111(02)00011-3. [DOI] [PubMed] [Google Scholar]
- Mark M, Lufkin T, Vonesch JL, Ruberte E, Olivo JC, Dolle P, et al. Two rhombomeres are altered in Hoxa-1 mutant mice. Development. 1993;119(2):319–338. doi: 10.1242/dev.119.2.319. [DOI] [PubMed] [Google Scholar]
- McClintock JM, Jozefowicz C, Assimacopoulos S, Grove EA, Louvi A, Prince VE. Conserved expression of Hoxa1 in neurons at the ventral forebrain/midbrain boundary of vertebrates. Development Genes and Evolution. 2003;213(8):399–406. doi: 10.1007/s00427-003-0335-7. [DOI] [PubMed] [Google Scholar]
- McIntosh AM, Baig BJ, Hall J, Job D, Whalley HC, Lymer GK, et al. Relationship of catechol-O-methyltransferase variants to brain structure and function in a population at high risk of psychosis. Biological Psychiatry. 2007;61(10):1127–1134. doi: 10.1016/j.biopsych.2006.05.020. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A. Intermediate or brainless phenotypes for psychiatric research? Psychological Medicine. 2010;40(7):1057–1062. doi: 10.1017/s0033291709991929. [DOI] [PubMed] [Google Scholar]
- Mitchell SR, Reiss AL, Tatusko DH, Ikuta I, Kazmerski DB, Botti JA, et al. Neuroanatomic alterations and social and communication deficits in monozygotic twins discordant for autism disorder. American Journal of Psychiatry. 2009;166(8):917–925. doi: 10.1176/appi.ajp.2009.08101538. [DOI] [PubMed] [Google Scholar]
- Muscarella LA, Guarnieri V, Sacco R, Militerni R, Bravaccio C, Trillo S, et al. HOXA1 gene variants influence head growth rates in humans. Am.J.Med.Genet.B Neuropsychiatr.Genet. 2007;144B(3):388–390. doi: 10.1002/ajmg.b.30469. [DOI] [PubMed] [Google Scholar]
- Palmen SJ, van EH, Hof PR, Schmitz C. Neuropathological findings in autism. [Review] [117 refs] Brain. 2004;127(Pt 12):2572–2583. doi: 10.1093/brain/awh287. [DOI] [PubMed] [Google Scholar]
- Paraguison RC, Higaki K, Sakamoto Y, Hashimoto O, Miyake N, Matsumoto H, et al. Polyhistidine tract expansions in HOXA1 result in intranuclear aggregation and increased cell death. Biochemical and Biophysical Research Communications. 2005;336(4):1033–1039. doi: 10.1016/j.bbrc.2005.08.212. [DOI] [PubMed] [Google Scholar]
- Paraguison RC, Higaki K, Yamamoto K, Matsumoto H, Sasaki T, Kato N, et al. Enhanced autophagic cell death in expanded polyhistidine variants of HOXA1 reduces PBX1-coupled transcriptional activity and inhibits neuronal differentiation. Journal of Neuroscience Research. 2007;85(3):479–487. doi: 10.1002/jnr.21137. [DOI] [PubMed] [Google Scholar]
- Pierson R, Corson PW, Sears LL, Alicata D, Magnotta V, Oleary D, et al. Manual and semiautomated measurement of cerebellar subregions on MR images. Neuroimage. 2002;17(1):61–76. doi: 10.1006/nimg.2002.1207. [DOI] [PubMed] [Google Scholar]
- Pinheiro J, DM B. Mixed-effects models in S and S-PLUS. New York: Springer; 2000. [Google Scholar]
- Posthuma D, De Geus EJ, Neale MC, Hulshoff Pol HE, Baare WEC, Kahn RS, et al. Multivariate genetic analysis of brain structure in an extended twin design. Behavior Genetics. 2000;30(4):311–319. doi: 10.1023/a:1026501501434. [DOI] [PubMed] [Google Scholar]
- Rasalam AD, Hailey H, Williams JH, Moore SJ, Turnpenny PD, Lloyd DJ, et al. Characteristics of fetal anticonvulsant syndrome associated autistic disorder. Developmental Medicine and Child Neurology. 2005;47(8):551–555. doi: 10.1017/s0012162205001076. [DOI] [PubMed] [Google Scholar]
- Raz N, Dupuis JH, Briggs SD, McGavran C, Acker JD. Differential effects of age and sex on the cerebellar hemispheres and the vermis: a prospective MR study. AJNR. American Journal of Neuroradiology. 1998;19(1):65–71. [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Greenstein D, Lee Y, Long R, Clasen L, Gochman P, et al. Catechol-o-methyl transferase (COMT) val158met polymorphism and adolescent cortical development in patients with childhood-onset schizophrenia, their non-psychotic siblings, and healthy controls. Neuroimage. 2011;57(4):1517–1523. doi: 10.1016/j.neuroimage.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Lee Y, Long R, Greenstein D, Clasen L, Addington A, et al. Common functional polymorphisms of DISC1 and cortical maturation in typically developing children and adolescents. Molecular Psychiatry. 2010 doi: 10.1038/mp.2010.72. 2010/07/16 ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Toro R, Proitsi P, Powell J, Paus T, Bolton P, et al. A functional polymorphism of the brain derived neurotrophic factor gene and cortical anatomy in autism spectrum disorder. Journal of Neurodevelopmental Disorders. 2009;1(3):215–223. doi: 10.1007/s11689-009-9012-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier PM, Ingram JL, Tisdale B, Nelson S, Romano J. Embryological origin for autism: developmental anomalies of the cranial nerve motor nuclei. Journal of Comparative Neurology. 1996;370(2):247–261. doi: 10.1002/(SICI)1096-9861(19960624)370:2<247::AID-CNE8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Romano V, Cali F, Mirisola M, Gambino G, D AR, Di RP, et al. Lack of association of HOXA1 and HOXB1 mutations and autism in Sicilian (Italian) patients. Molecular Psychiatry. 2003;8(8):716–717. doi: 10.1038/sj.mp.4001285. [DOI] [PubMed] [Google Scholar]
- Sen B, Sinha S, Ahmed S, Ghosh S, Gangopadhyay PK, Usha R. Lack of association of HOXA1 and HOXB1 variants with autism in the Indian population. Psychiatric Genetics. 2007;17(1):1. doi: 10.1097/YPG.0b013e328010de0d. [DOI] [PubMed] [Google Scholar]
- Shaw P, Greenstein D, Lerch J, Clasen L, Lenroot R, Gogtay N, et al. Intellectual ability and cortical development in children and adolescents. Nature. 2006;440(7084):676–679. doi: 10.1038/nature04513. [DOI] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One. 2007;2(9):e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield AC, McIntosh AM, Spencer MD, Philip R, Gaur S, Lawrie SM. Towards a neuroanatomy of autism: A systematic review and meta-analysis of structural magnetic resonance imaging studies. Eur.Psychiatry. 2007 doi: 10.1016/j.eurpsy.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Stanfield AC, McIntosh AM, Spencer MD, Philip R, Gaur S, Lawrie SM. Towards a neuroanatomy of autism: a systematic review and meta-analysis of structural magnetic resonance imaging studies. Eur Psychiatry. 2008;23(4):289–299. doi: 10.1016/j.eurpsy.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Talebizadeh Z, Bittel DC, Miles JH, Takahashi N, Wang CH, Kibiryeva N, et al. No association between HOXA1 and HOXB1 genes and autism spectrum disorders (ASD) Journal of Medical Genetics. 2002;39(11):e70. doi: 10.1136/jmg.39.11.e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemeier H, Lenroot RK, Greenstein DK, Tran L, Pierson R, Giedd JN. Cerebellum development during childhood and adolescence: a longitudinal morphometric MRI study. Neuroimage. 2010;49(1):63–70. doi: 10.1016/j.neuroimage.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield MA, Bosley TM, Salih MA, Alorainy IA, Sener EC, Nester MJ, et al. Homozygous HOXA1 mutations disrupt human brainstem, inner ear, cardiovascular and cognitive development. Nature Genetics. 2005;37(10):1035–1037. doi: 10.1038/ng1636. [DOI] [PubMed] [Google Scholar]
- Trainor PA, Krumlauf R. Patterning the cranial neural crest: hindbrain segmentation and Hox gene plasticity. Nat Rev Neurosci. 2000;1(2):116–124. doi: 10.1038/35039056. [DOI] [PubMed] [Google Scholar]
- Wallace GL, Eric Schmitt J, Lenroot R, Viding E, Ordaz S, Rosenthal MA, et al. A pediatric twin study of brain morphometry. J Child Psychol Psychiatry. 2006;47(10):987–993. doi: 10.1111/j.1469-7610.2006.01676.x. [DOI] [PubMed] [Google Scholar]
- Wang VY, Zoghbi HY. Genetic regulation of cerebellar development. Nat Rev Neurosci. 2001;2(7):484–491. doi: 10.1038/35081558. [DOI] [PubMed] [Google Scholar]
- Wassink TH, Hazlett HC, Epping EA, Arndt S, Dager SR, Schellenberg GD, et al. Cerebral cortical gray matter overgrowth and functional variation of the serotonin transporter gene in autism. Archives of General Psychiatry. 2007;64(6):709–717. doi: 10.1001/archpsyc.64.6.709. [DOI] [PubMed] [Google Scholar]
- Webb SJ, Sparks BF, Friedman SD, Shaw DW, Giedd J, Dawson G, et al. Cerebellar vermal volumes and behavioral correlates in children with autism spectrum disorder. Psychiatry Research. 2009;172(1):61–67. doi: 10.1016/j.pscychresns.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachnis AT, Rorke LB. Cerebellar and brainstem development: an overview in relation to Joubert syndrome. Journal of Child Neurology. 1999;14(9):570–573. doi: 10.1177/088307389901400904. [DOI] [PubMed] [Google Scholar]


