Abstract
Lymphoid tyrosine phosphatase (LYP) and C-terminal Src kinase (CSK) are negative regulators of signaling mediated through the T cell antigen receptor (TCR) and are thought to act in a cooperative manner when forming a complex. Here, we studied the spatio-temporal dynamics of the LYP/CSK complex in T cells. We demonstrate that dissociation of this complex is necessary for recruitment of LYP to the plasma membrane, where it down-modulates TCR signaling. Development of a potent and selective chemical probe of LYP confirmed that LYP inhibits T cell activation when removed from CSK. Our findings may explain the reduced TCR-mediated signaling associated with a single nucleotide polymorphism, which confers increased risk for certain autoimmune diseases, including type 1 diabetes and rheumatoid arthritis, and results in expression of a LYP allele that is unable to bind CSK. Our compound also represents a starting point for the development of a LYP-based treatment of autoimmunity.
A dynamic balance between tyrosine phosphorylation and dephosphorylation of signaling molecules is crucial for maintaining the homeostasis of the immune system1. In T cells, engagement of the TCR by cognate antigen leads to mobilization of the CD4/CD8-associated Src family kinase LCK, which through autophosphorylation of Y394 in its activation loop adopts an active conformation2. Activated LCK phosphorylates tyrosine residues in the immunoreceptor tyrosine-based activation motifs (ITAMs) of the TCR-associated CD3 and ζ-chains. Tyrosine phosphorylated ITAMs serve as docking sites for the tandem Src homology 2 (SH2) domains of ζ-associated protein of 70 kDa (ZAP70), which through its tyrosine kinase activity propagates the signals, eventually leading to downstream responses such as activation of transcription factors (e.g. nuclear factor of activated T cells (NFAT) and activator protein 1 (AP1)), cell growth, proliferation, and production of cytokines2.
TCR-induced responses are transient, and different mechanisms are involved in signal termination. The most TCR-proximal mechanisms for down-regulation include receptor internalization/degradation, phosphorylation of LCK on its negative regulatory residue Y505 by CSK, and dephosphorylation of the positive regulatory residue Y394 in LCK and/or the ITAMs of the CD3 and ζ-chains by a number of protein tyrosine phosphatases (PTPs), including LYP, SHP1, PTPH1, PTP-MEG1, and perhaps CD45 and PTP-PEST. Although these PTPs have overlapping substrate specificities, subtle differences between their actions do exist, e.g. due to different subcellular localization and recruitment in response to TCR stimulation1. In addition, recent findings indicate that even a minor alteration in the LYP sequence can substantially affect TCR signaling. The C1858T single-nucleotide polymorphism (SNP) in PTPN22, the gene encoding LYP, confers an increased risk for various autoimmune disorders (reviewed in ref. 3). The SNP results in alteration of amino acid 620 from arginine in the ‘normal’ allele (LYP*R620) to tryptophan in the disease-associated allele (LYP*W620). Residue 620 is located in the first of four proline-rich motifs (named P1–P4) that are found on the C-terminal part of LYP. Interestingly, R620 of the major allele is crucial for the interaction between LYP and the Src homology 3 (SH3) domain of CSK4–6, rendering the minor allele LYP*W620 incapable of binding CSK7,8. Experiments with primary T cells have indicated that LYP*W620 is a gain-of-function mutant that has approximately 50% higher catalytic activity and acts as a more potent inhibitor of TCR signaling8–10.
Since the risk allele LYP*W620 cannot bind CSK and is a stronger inhibitor of TCR signaling, we hypothesized that the interaction between CSK and the major allele LYP*R620 could interfere with the catalytic activity of the latter. Here, we show that the major allele of LYP down-modulates TCR-induced signaling mainly when dissociated from CSK. We also describe the development of a specific chemical probe for LYP and use this probe to strengthen our model. Our findings may explain why the risk allele LYP*W620 is a more potent inhibitor of TCR signaling and suggest a positive regulatory role for the pool of CSK molecules that interact with LYP.
RESULTS
The LYP/CSK complex dissociates after TCR stimulation
The disease-associated R620W mutation in LYP alters the surface properties in the P1 motif such that it can no longer bind CSK (Supplementary Results, Supplementary Fig. 1)7,8. However, binding properties in other parts of LYP seem unaffected by the mutation. For instance, both LYP variants display comparable interactions with PSTPIP (also known as CD2 binding protein), which, through its coiled-coiled domain, binds equally well to the P4 motif in LYP*R620 and LYP*W620 (Supplementary Fig. 1)11. These findings, combined with the increased risk of autoimmunity associated with LYP*W620, prompted us to study the LYP*R620/CSK interaction more closely (hereafter referred to as the LYP/CSK interaction).
In resting human T cells (LYP*R620 homozygous), endogenous LYP and CSK interacted as evidenced by co-immunoprecipitation experiments (Fig. 1a). Based on similar experiments, we estimate that approximately 50% (47.9 ± 6.0, average ± SEM, n=5) of total cellular LYP bound to CSK while only 6% (5.8 ± 2.6, average ± SEM, n=5) of total cellular CSK interacted with LYP, suggesting a significant molar excess of CSK over LYP in T cells. Phosphoprotein associated with glycolipid-enriched membranes (PAG) is exclusively found in lipid rafts and is important for recruiting CSK to raft-associated LCK1. In resting T cells PAG co-precipitated with CSK but not with LYP (Fig. 1a and Supplementary Fig. 2a), suggesting that LYP-bound CSK does not interact with PAG in lipid rafts. As expected, TCR-stimulation resulted in transient phosphorylation of LCK-Y394 and the ITAMs of the ζ-chains, both of which are LYP substrates (Fig. 1b). Interestingly, the LYP/CSK complex dissociated after TCR-stimulation, and the timing of the dissociation paralleled the decline of the TCR proximal signals (Fig. 1b). Similar observations were made with Jurkat TAg T cells (Supplementary Fig. 2b), which are also homozygous for LYP*R620. Since LYP*W620, which cannot bind CSK, is a more potent inhibitor of TCR-induced responses, these findings led us to hypothesize that the CSK interaction with LYP*R620 may exert an inhibitory effect on this LYP variant by preventing it from interacting with its endogenous substrates. To test this, we overexpressed the CSK-SH3 domain in Jurkat TAg T cells, which normally express LYP but not the other known CSK-SH3 binding partner, PTP-PEST12. As shown in Supplementary Figure 2c, the CSK-SH3 domain disrupted the LYP/CSK complex. Consistent with our hypothesis, TCR proximal responses were lower in Jurkat TAg T cells overexpressing the CSK-SH3 domain (Supplementary Fig. 2d). A similar reduction in TCR-proximal signaling as well as in the production of interleukin-2 (IL-2) and interferon γ (IFNγ) cytokines were observed when the CSK-SH3 domain was overexpressed in primary human T cells homozygous for LYP*R620 (Fig. 1c and Supplementary Fig. 2e). Although PTP-PEST is expressed in primary T cells, significant effects on TCR-mediated signaling due to the interaction between CSK-SH3 and PTP-PEST are less likely, the interaction between PTP-PEST and CSK is 10-fold less strong than the corresponding interaction between PEP (the mouse version of LYP) and CSK13. Finally, by using an NFAT-AP1 reporter assay in Jurkat TAg T cells, we also compared the inhibitory effect of the CSK-SH3 domain with that of full-length CSK, a well-established inhibitor of TCR signaling (Supplementary Fig. 2f). Overexpression of the CSK-SH3 domain reduced the TCR-induced responses almost as potently as overexpressed full-length CSK. However, while the inhibitory effect of full-length CSK is largely due to its kinase activity, which ensures phosphorylation of the negative regulatory Y505 in LCK14,15, the inhibitory effect mediated by the CSK-SH3 domain is clearly different and adds a new level of complexity to the role of CSK in T cell signaling.
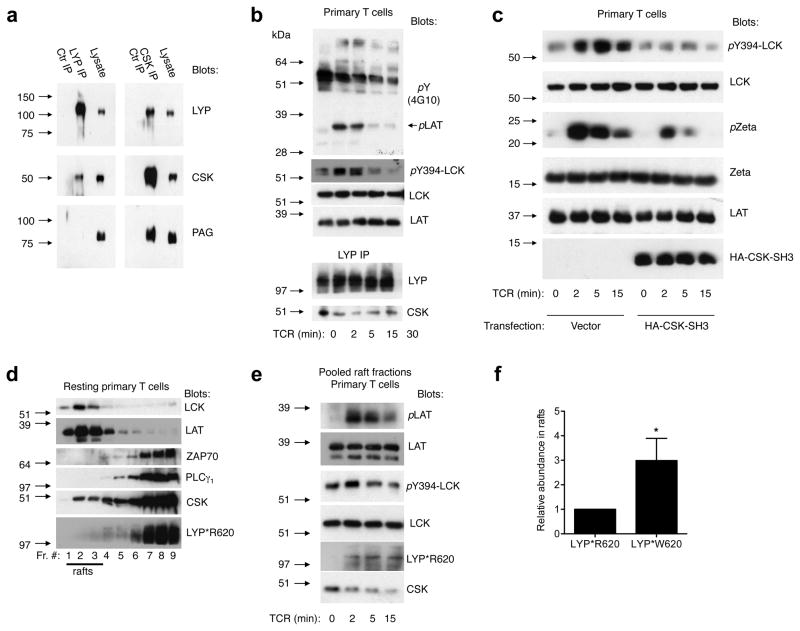
Figure 1. LYP recruited to lipid rafts down-modulates TCR-signaling independently of CSK.
(a) Lysates prepared from resting human T cells (LYP*R620 homozygous) were subjected to immunoprecipitation with the indicated antibodies or isotype controls, followed by immunoblotting with the given antibodies. (b) TCR time course (CD3 cross-linking) with human T cells (LYP*R620 homozygous), thereafter prepared lysates and LYP immunoprecipitates were analyzed with the indicated antibodies. (c) Human T cells (LYP*R620 homozygous) were transfected with empty vector or a plasmid encoding HA-tagged CSK-SH3, followed by TCR time course (CD3 cross-linking) and immunoblotting. (d) Homogenized resting human T cells (LYP*R620 homozygous) were subjected to sucrose gradient centrifugation and subsequent immunoblotting of the harvested fractions with the indicated antibodies. (e) Human T cells (LYP*R620 homozygous) were TCR-stimulated (CD3 cross-linking) for the indicated time periods. After homogenization, sucrose gradient centrifugation was performed. The harvested lipid raft fractions were pooled and analyzed with immunoblotting. (f) Jurkat TAg T cells were transfected with plasmids encoding either HA-tagged LYP*R620 or LYP*W620. Only experiments with comparable LYP*R620 and LYP*W620 expression levels were included. After cell lysis and sucrose gradient centrifugation, harvested lipid raft fractions were pooled and subjected to immunoblotting. Relative amounts of HA-reactivity (i.e. HA-LYP*R620 vs. HA-LYP*620W, LCK reactivity used as reference) in the pooled raft fractions were determined by densitometry. Data from four independent experiments are presented (average± SD, *p < 0.05). All experiments presented in Fig. 1 are representative of three or more experiments with similar results.
To better understand this complexity, we conducted a more elaborate study of the spatiotemporal regulation of LYP and CSK following TCR stimulation. The most TCR-proximal substrate for LYP in T cells is LCK phosphorylated on Y394. Purification of lipid rafts from human T cells (LYP*R620 homozygous) using the relatively mild detergent Brij-58 revealed that most LCK molecules partitioned into lipid rafts, and notably, a significant and transient induction in LCK-Y394 phosphorylation was observed (Supplementary Fig. 2g). In resting T cells (LYP*R620 homozygous), a fraction of the CSK molecules (typically 5–10%) was found in lipid rafts while no LYP*R620 could be detected (Fig. 1d). However, in response to TCR stimulation, LYP*R620 was recruited to lipid rafts (Fig. 1e) with kinetics paralleling the dissociation of the LYP/CSK complex (Fig. 1b) and the decline in the LCK-Y394 phosphorylation signal (Fig. 1b/e). Noteworthy, the LYP*R620 recruitment to rafts occurred independently of CSK since the latter quickly dissociated from rafts after TCR stimulation (Fig. 1e). We then investigated how the R620W mutation affected the ability of LYP to partition into lipid rafts. Since these experiments require large numbers of T cells and would necessitate fresh samples from several donors carrying the polymorphism, we used Jurkat TAg T cells, which, similar to primary T cells, displayed recruitment of endogenous LYP*R620 to lipid rafts in response to TCR stimulation (Supplementary Fig. 2h). Small amounts of LYP*R620 could also be detected in rafts in Jurkat TAg T cells under resting conditions (Supplementary Fig. 2h). Similarly, both ectopically expressed LYP*R620 and LYP*W620 partitioned into lipid rafts in resting Jurkat TAg T cells (Supplementary Fig. 2i), but LYP*W620 did so approximately three times more efficiently (3.0 ± 0.9, average ± standard deviation, n = 4, two-tailed paired t-test: p = 0.022, Fig. 1f and Supplementary Fig. 2j). Because overexpression of either LYP*R620 or LYP*W620 blocked TCR signaling, it was not possible to study TCR-induced recruitment of transfected LYP to rafts. Collectively, these results suggest that dissociation of the LYP/CSK complex is necessary for the recruitment of LYP to lipid rafts, where it down-modulates TCR-mediated signaling. In addition, they also suggest that augmented raft partitioning of the LYP*W620 variant likely contributes to its increased inhibitory activity.
Screening for specific LYP inhibitors
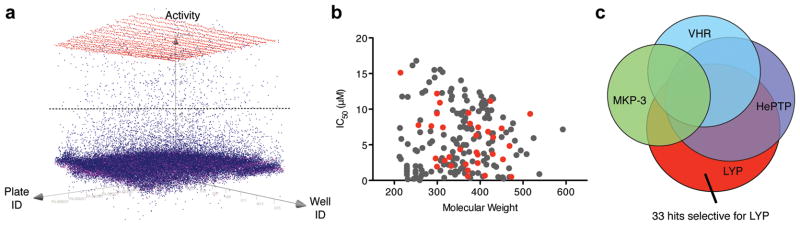
In order to corroborate that LYP down-modulates TCR-induced signaling only after it dissociates from CSK, we set out to determine whether the decrease in signaling depends on the LYP catalytic activity. Because existing inhibitors were not potent and/or selective enough, we developed a novel, specific inhibitor of LYP. We screened 50,000 drug-like molecules using a 384-well format phosphatase assay16, which used 3-O-methylfluorescein phosphate (OMFP) as substrate, and was robust with an average Z′ value of 0.86 and a signal-to-background ratio of 9.3 (Fig. 2a and Supplementary Table 1). 712 compounds inhibited the catalytic activity of LYP by ≥50%, of which 190 compounds exhibited dose-dependent inhibition with IC50 values <0.005 mg/mL. Corresponding molar IC50 values ranged from 0.047 to 16.8 μM. Plotting IC50 values versus molecular weights showed no correlation between inhibitory activity and molecule size (Fig. 2b). In fact, some of the most potent inhibitors had relatively low molecular weights. While the majority of these compounds exhibited no selectivity against a panel of PTPs, compounds 1–33 were selective for LYP, with IC50 values ranging from 0.5 to 15 μM (Fig. 2b/c and Supplementary Table 2). Clustering the selective inhibitors by chemical similarity using a Tanimoto distance17 of 0.5 resulted in 18 different clusters and singletons, indicating that these molecules cover a diverse range of chemical space.
Figure 2. HTS and counter screens for LYP inhibitors.
(a) Representation of activity versus assay plate versus well. The horizontal line indicates the 50% activity cut-off for hit determination. The color code means red, positive control; magenta, negative control; blue, compounds. (b) Scatter plot of 190 confirmed (grey + red) and 33 selective (red) LYP inhibitors shows the distribution of IC50 values versus the molecular weight of the hit compounds. (c) Venn diagram illustrating confirmed LYP inhibitors, as well as hits from similar screening campaigns, using the same chemical library, for the related phosphatases HePTP, VHR, and MKP-3.
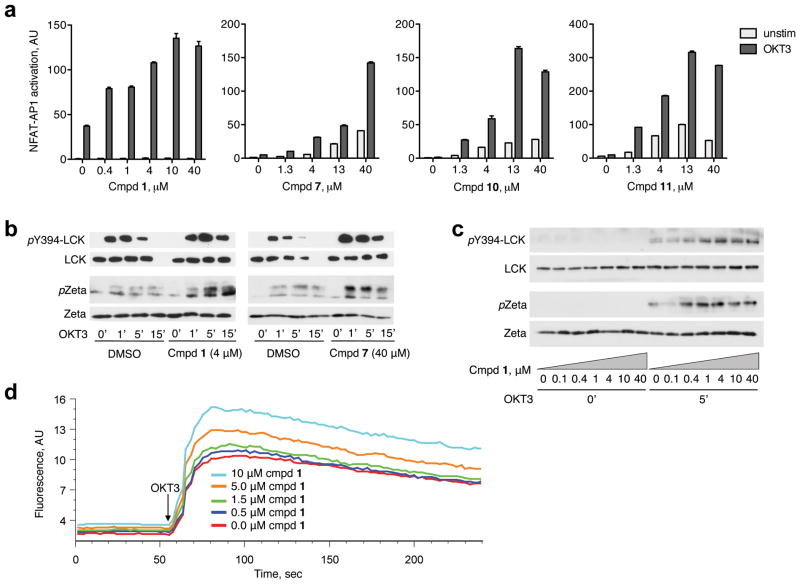
To test if the compounds inhibit endogenous LYP, we utilized a dual luciferase reporter assay encompassing firefly luciferase under control of the proximal IL-2 promoter (containing NFAT and AP1 sites and hence being induced by TCR signaling) and Renilla luciferase under the control of a null-promoter (representing baseline transcriptional activity)18. For these experiments, we employed the Jurkat TAg T cell line, which is homozygous for LYP*R620 and expresses LYP at levels comparable to those seen in primary human T cells (data not shown). Briefly, cells were treated with candidate compounds (40 μM) or DMSO (vehicle control) for 45 min, then TCR-stimulated or not, followed by dual luciferase assays (Supplementary Fig. 3). All compounds giving Renilla luciferase readings deviating >20% from the DMSO control samples were excluded because this suggests the compounds have unspecific effects. In contrast, compounds giving firefly: Renilla luciferase ratios >2-fold higher than the corresponding ratios for the DMSO control samples were selected for retesting in a dose-response format. Among the 13 compounds subjected to retesting, inhibitors 1, 7, 10, and 11 augmented TCR-induced activation of the proximal IL-2 promoter in a dose-dependent manner (Fig. 3a) and were chosen for further analyses.
Figure 3. Compound 1 (LTV-1) is a potent LYP inhibitor in T cells.
(a) Jurkat TAg T cells cotransfected with plasmids encoding firefly luciferase (under control of a promoter containing NFAT and AP1 sites) and Renilla luciferase (containing a null promoter) were pretreated with various concentrations of selected compounds or DMSO (vehicle control) and then stimulated through the TCR (OKT3). Dual luciferase assays were conducted, and the level of NFAT-AP1 activation for each sample was calculated as the ratio between firefly and Renilla luciferase. Each sample was run in duplicate and is presented as average ± half range. (b) Jurkat TAg T cells were pretreated with the indicated compounds (40 μM) or DMSO and then TCR stimulated (OKT3) for 0-1-5-15 min. Reactions were stopped by adding lysis buffer, and cell extracts were subjected to immunoblotting with the indicated antibodies. (c) Experiment as in (b), but cells were pretreated with different concentrations of compound 1 and TCR stimulations were either 0 or 3 min. (d) Analysis of TCR-induced calcium fluxes in Jurkat TAg T cells loaded with Fluo-4 and pre-treated with various concentrations of compound 1.
To probe the phosphorylation states of LYP’s direct substrates LCK and ζ-chain, we pre-treated Jurkat TAg T cells with compounds or DMSO, followed by TCR stimulation for various times. Compounds 1 and 7 (at 4 and 40 μM, respectively) clearly augmented phosphorylation of LCK-Y394 and the ζ-chain in response to TCR stimulation (Fig. 3b), while no substantial effects were observed for compounds 10 and 11 (Supplementary Fig. 4). More importantly, 1 enhanced TCR-signaling in a dose-dependent manner, showing clear increases on phosphorylation levels for both LCK-Y394 and ζ-chain at concentrations as low as 0.4 μM, with the strongest effects observed at about 4 μM (Fig. 3c). These results corresponded well with the data obtained in the reporter assays, confirming that the downstream effects of the inhibitor could be attributed to augmented tyrosine phosphorylation of LYP targets. We also tested if compound 1 affected TCR-induced calcium mobilization, an intermediate signaling readout (Fig. 3d). Jurkat TAg T cells pretreated with various concentrations of 1 exhibited a dose-dependent increase in calcium flux in response to TCR stimulation. Since LYP activity inversely correlates with TCR-induced calcium mobilization8, this further supports that 1 is specific for LYP. Compound 1 also lowered the threshold for TCR signaling (Supplementary Fig. 5); to obtain a given level of T cell activation, the stimulatory anti-CD3 concentration had to be approximately 0.5 log steps higher for the control cells compared to cells pretreated with inhibitor (5 μM). Inhibitor-treated cells also reached substantially higher maximum response levels. Based on all these results, compound 1, which we from now on refer to as LTV-1, was chosen for further analysis.
Structure-activity-relationship (SAR) studies with LTV-1
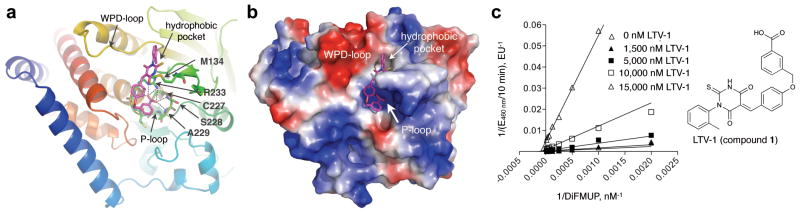
To explore the inhibition of LYP at the molecular level, we performed SAR studies around the thiobarbituric acid scaffold of LTV-1 (analogs 34–51, Supplementary Table 3). We found that the majority of LTV-1 analogs inhibited LYP with IC50 values below 20 μM. However, we noticed that subtle changes in the substitution pattern of the toluene ring caused a substantial decrease in inhibition compared to LTV-1 (40, IC50 = 12.1 μM; 44, IC50 = 17.7 μM). This indicated that LTV-1 must interact specifically with the protein surface so that a small structural change could prevent the compound from tightly binding to the protein. We used the ICM in silico docking algorithm19 and crystal structures of LYP in both the open and closed conformations20 to dock LTV-1 and analogs into the LYP active site. While docking to the closed state failed, LTV-1 could be precisely docked into LYP’s open conformation with an excellent ICM score of −43.9, the best docking score among all analogs (Fig. 4a/b). The docking studies revealed a number of characteristics for the LYP: inhibitor interaction. First, the benzoic acid moiety of LTV-1 binds the phosphate-binding loop (P-loop) via a dense network of hydrogen bonding interactions, mimicking the phosphotyrosine moiety of the natural substrate (Fig. 4a). Second, the ether group interacts with the side chain of Thr275 via van der Waals forces, whereas the benzylidene ring makes cation-π interactions with the guanidinium group of Arg233. Third, the toluene ring specifically occupies a surface depression next to the P-loop, with the methyl group in an ortho-position fitting perfectly into a small hydrophobic pocket (Fig. 4b). Analogs with this methyl group in meta- or para-position were not only less active but also exhibited docking poses with much less favorable interactions, and, consequently, had three-fold lower docking scores. Thus, the results of the in silico docking studies were in excellent agreement with the SAR studies. Importantly, when we tested the best 11 analogs (IC50 < 20 μM) in T cells, all compounds increased TCR-induced activation of the proximal IL-2 promoter in a dose-dependent manner, further supporting the specific effect of LTV-1 and related chemical compounds (Supplementary Fig. 6). Michaelis-Menten kinetic studies with recombinant LYP and varying substrate and inhibitor concentrations revealed a competitive to mixed inhibition pattern for LTV-1 with a calculated Ki of 0.384 ± 0.061 μM (Fig. 4c)
Figure 4. LTV-1 binds to the LYP active site in open conformation.
(a) Ribbon diagram of LYP with LTV-1 (magenta) docked into the active site of the crystal structure of LYP in its atypical open conformation (PDB code 2P6X). Dashed lines indicate hydrogen-bonding interactions of the benzoic acid moiety of 1 with residues of the P-loop. Docking was done with ICM Pro (Molsoft, LLC); the image was prepared with PyMOL (http://pymol.sourceforge.net). (b) Surface representation of the same docking solution as in (a) colored by electrostatic potential (blue, most positive; red, most negative). The image was prepared with PyMOL (http://pymol.sourceforge.net). (c) Michaelis-Menten enzyme kinetic analysis of LTV-1 with recombinant LYP and 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP) as fluorogenic phosphatase substrate. The Lineweaver-Burk plot was generated with GraphPad Prism (v5, GraphPad Software, Inc.).
Specificity of LTV-1 in vitro and in human T cells
We tested LTV-1 against a panel of very closely related PTPs, including PTP-PEST, SHP1, CD45, TCPTP, and PTP1B. Along with LYP, PTP-PEST and SHP1 are negative regulators of proximal TCR signaling and also share physiological substrates1,12. CD45 is the most proximal positive regulatory PTP of the TCR signaling cascade1, while the structurally related TCPTP and PTP1B control cytokine receptor signaling21, and the ubiquitously expressed PTP1B is also a crucial regulator of insulin and leptin signaling22. Using an in vitro phosphatase assay, selectivity of LTV-1 for LYP ranged from 3-fold over TCPTP (IC50 = 1.52 μM) and PTP1B (IC50 = 1.59 μM), 46-fold over SHP1 (IC50 = 23.2 μM), and 59-fold over CD45 (IC50 = 30.1μM), up to greater than 200-fold over PTP-PEST (IC50 > 100 μM). Notably, among all PTPs, the catalytic domain of PTP-PEST has the highest sequence identity (62%) to that of LYP, with nearly identical active sites (Supplementary Fig. 7). We also tested LTV-1 against recombinant PEP, the mouse ortholog of LYP; LTV-1 was 15 times less active against PEP (IC50 = 7.56 μM).
To evaluate the specificity of LTV-1 in a physiological environment, we used Jurkat TAg T cells overexpressing LYP*R620, PTP-PEST, SHP1, TCPTP, PTP1B, or LYP*W620 and conducted reporter assays for T cell activation (Fig. 5a and Supplementary Fig. 8). As observed previously8, LYP*R620 inhibited TCR-induced NFAT-AP1 activation in a dose-dependent manner. This effect was mitigated when cells were treated with LTV-1 (5 μM) prior to TCR stimulation (Fig. 5a). When similar experiments were conducted on cells overexpressing either PTP-PEST or SHP1 (Supplementary Fig. 8a/b), TCR signaling was severely reduced, even in cells pretreated with LTV-1 (5 μM), suggesting that neither PTP-PEST nor SHP1 were inhibited by LTV-1 in these cellular assays. TCPTP overexpression only mildly inhibited TCR signaling, and this weak effect could not be overcome with LTV-1 pretreatment (Supplementary Fig. 8c). Overexpression of PTP1B, which is not believed to act downstream of the TCR, moderately reduced NFAT-AP1 activation, perhaps because of unspecific effects of elevated PTP activity (Supplementary Fig. 8d). At low PTP1B expression levels, LTV-1 was able to reverse this effect, reflecting the observed activity of LTV-1 against recombinant PTP1B. Importantly, we also tested if LTV-1 could inhibit the activity of the disease-associated LYP*W620 (Supplementary Fig. 8e). This mutant strongly lowered TCR-induced NFAT-AP1 activation in the control samples, but signaling was restored in cells pretreated with LTV-1 (5 μM).
Figure 5. LTV-1 specifically inhibits LYP in T cells.
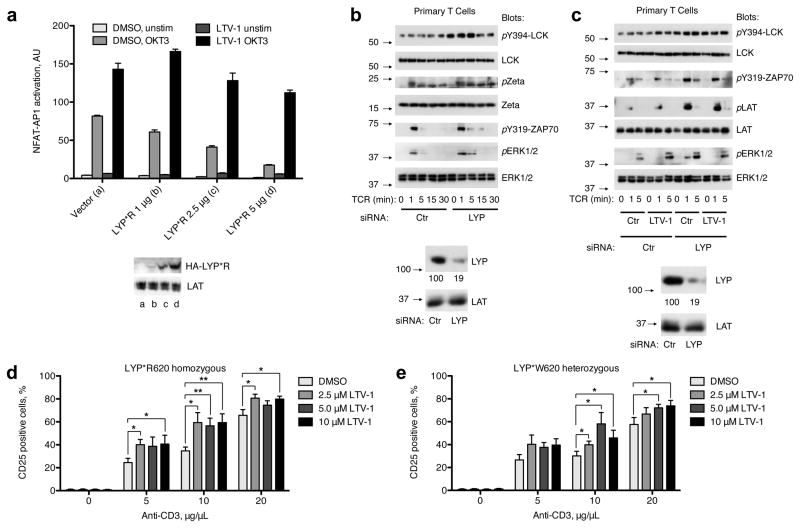
(a) NFAT-AP1 reporter assay as described in Fig. 3a but with cells expressing increasing amounts of LYP*R620. Cells were treated with LTV-1 (5 μM) or DMSO (vehicle control) prior to TCR stimulation (OKT3). For the dual luciferase assays, each sample was run in triplicate and is presented as average ± SD. Blots showing expression levels of transfected LYP*R620 and amounts of LAT (loading control) are also presented. Data are representative of 3 or more experiments. (b) Purified human T cells (LYP*R620 homozygous) were transfected with either control or LYP-specific siRNA. Two days later, cells were TCR-triggered (CD3 cross-linking) for the indicated periods of times, followed by immunoblotting with the indicated antibodies. Blots verifying LYP knockdown are also shown, numbers (arbitrary units) below the LYP blot represent LYP expression relative to LAT expression for the same sample. Data are representative of four independent experiments. (c) Cells and experiment as in (a), but the last 45 min prior to TCR stimulation, cells were incubated with either DMSO or LTV-1 (5 μM). Data are representative of three independent experiments. (d/e) Purified human CD4+ T cells with indicated PTPN22 genotype were pre-treated for 45 min at 37°C with increasing doses of LTV-1. Cells were then activated for 40 h by plate-bound anti-CD3 (various doses) and soluble anti-CD28 (1 μg/mL), and CD25 expression was assessed by FACS. The percentage of activated CD25+ cells is presented as average ± SEM for five different healthy donors, *p < 0.05, **p < 0.01, ***p < 0.001.
To determine if LTV-1 specifically inhibited LYP in primary human T cells, we used purified CD4+ T cells (LYP*R620 homozygous) where LYP had been acutely eliminated by siRNA-mediated knockdown. As expected, human T cells with severely reduced LYP expression (residual expression < 20% of normal levels) displayed augmented and/or sustained TCR-induced responses such as LCK-Y394 phosphorylation, ζ-chain phosphorylation, ZAP70-Y319 phosphorylation, and ERK1/2 activation, demonstrating again that LYP is an inhibitor of TCR signaling (Fig. 5b). Importantly, while LTV-1 pretreatment or LYP knockdown resulted in enhanced TCR-mediated signaling compared to controls, the combined action of LTV-1 and LYP knockdown did not increase signaling beyond what was observed for each alone (Fig. 5c). Together, these data suggest that LTV-1 augments TCR signaling by specifically inhibiting LYP activity in intact cells.
To verify that LTV-1 is not cytotoxic, we tested the compound in HeLa cells, which do not express LYP, as well as in Jurkat TAg T cells and peripheral blood mononuclear cells (PBMC) (Supplementary Methods, Supplementary Fig. 9). For all cells tested, LTV-1 had no notable effect on cell viability (metabolic rate assay), even at the highest concentration tested (40 μM). Thus, LTV-1 has no cytotoxic effects at the concentrations required for efficacy.
Effects of LTV-1 on TCR signaling in primary human T cells
To address whether pharmacological inhibition of LYP results in increased downstream responses in primary human T cells, we examined the expression of the early activation marker CD25. Purified CD4+ T cells from healthy donors (homozygous for the normal C1858 allele) were pre-treated with various concentrations of LTV-1, cultured in the presence of anti-CD3 and anti-CD28 antibodies, and CD25 expression was analyzed by flow cytometry. CD25 up-regulation positively correlated with increased anti-CD3 doses, and importantly, pre-treatment of cells with LTV-1 consistently gave higher readings for all anti-CD3 doses tested (Fig. 5d). Thus, pharmacological inhibition of LYP catalytic activity in primary human cells increases TCR-induced T cell activation. Next, we used purified CD4+ T cells from healthy donors carrying the disease-associated LYP allele (heterozygous for the C1858T SNP). Pretreatment of these cells with LTV-1 significantly up-regulated the CD25 levels when cells were stimulated with an intermediate or high dose of anti-CD3, but not for the lowest dose (Fig. 5e). Importantly, and in contrast to a recent report suggesting that LYP*W620 is an unstable protein compared to LYP*R62023, we found no differences in LYP expression levels in primary CD4+ T cells from donors homozygous for either LYP*R620 or LYP*W620 (Supplementary Fig. 10). In conclusion, these data show that primary T cells expressing the disease-associated allele are also sensitive to LTV-1 treatment.
LYP down-modulates TCR signaling when dissociated from CSK
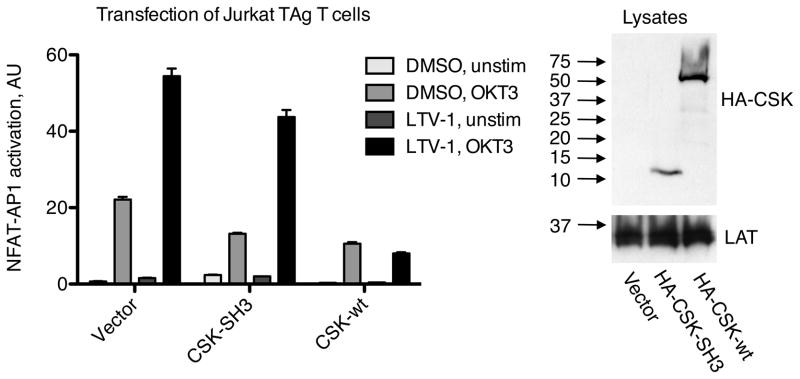
Since dissociation of the LYP/CSK complex paralleled the decline in TCR-induced tyrosine phosphorylation, and because forced dissociation of the LYP/CSK complex by overexpressing the CSK-SH3 domain resulted in lower TCR signaling, we hypothesized that LYP mediates the inhibitory effect of the overexpressed CSK-SH3 domain. To test this, we measured NFAT-AP1 activation in Jurkat TAg T cells overexpressing the CSK-SH3 domain or wild-type CSK in the presence or absence of LTV-1. Critically, pretreatment with LTV-1 restored TCR-induced NFAT-AP1 activation in cells overexpressing the CSK-SH3 domain but not in cells overexpressing wild-type CSK. (Fig. 6). This clearly indicates that overexpression of the CSK-SH3 domain and full-length CSK inhibited TCR signaling through two separate mechanisms, with the CSK-SH3 domain inhibiting in a LYP-dependent manner and full-length CSK inhibiting via a LYP-independent mechanism.
Figure 6. LYP down-modulates TCR signaling when dissociated from CSK.
Jurkat TAg T cells were cotransfected with plasmids encoding firefly luciferase (under control of a promoter containing NFAT and AP1 sites) and Renilla luciferase (containing a null promoter) in addition to empty vector or plasmids encoding either HA-tagged wild-type CSK or HA-tagged CSK-SH3. Later, cells were pretreated with LTV-1 (5 μM) or DMSO, followed by TCR stimulation (OKT3). Subsequently, dual luciferase assays were performed, and the level of NFAT-AP1 activation for each sample was calculated as the ratio between firefly and Renilla luciferase. Data are presented as average ± SD (triplicate measurements). Blots verifying expression of transfected wild-type CSK and CSK-SH3 are also shown. Data are representative of four independent experiments.
DISCUSSION
A genetic variant of LYP has recently been implicated in multiple autoimmune diseases3. In fact, in Caucasian populations, LYP currently ranks third and second in terms of single-gene contribution to the etiology of type 1 diabetes and rheumatoid arthritis, respectively24. LYP and CSK are thought to block TCR signaling in a co-operative manner. Here, we show that LYP dissociates from CSK after TCR stimulation, leading to increased recruitment of LYP to lipid rafts, where key substrates of LYP are localized. To test whether down-modulation of TCR signaling is directly related to the phosphatase activity of LYP molecules that are dissociated from CSK, we developed a potent and specific chemical probe for LYP. After confirming that the probe specifically inhibits LYP activity in human T cells, we used this compound to demonstrate that LYP mainly down-regulates TCR signaling when removed from CSK. This shows that the pool of CSK molecules bound to LYP exerts a positive regulatory role regarding TCR signaling. Our findings also provide a novel explanation for why the disease-associated variant of LYP, LYP*W620, is a stronger inhibitor of TCR signaling.
The development of specific PTP modulators continues to be a challenging task16,20,25,26. Here, we used HTS to identify new lead structures for LYP. As expected, most hit compounds were not selective for LYP but instead inhibited multiple PTPs. However, 33 compounds selective for LYP were identified and further characterized using a reporter assay for T cell activation as a primary screen in cells. Results from this assay allowed us to take only a small number of compounds into more elaborate testing of activity and specificity in Jurkat TAg T cells and primary human T cells. These studies led to the identification of LTV-1, which inhibited LYP in a dose-dependent manner at low- and sub-micromolar concentrations in T cells. LTV-1 increased the phosphorylation levels of the direct physiological targets of LYP, augmented calcium mobilization, activated the proximal IL-2 promoter, and enhanced the expression of the early activation marker CD25. Importantly, the downstream effects of LTV-1 on T cell activation were also shown to be due to direct inhibition of LYP. For example, T cells ectopically expressing different PTPs (e.g. LYP, PTP-PEST, SHP1) with known inhibitory effects on TCR signaling clearly demonstrated that LTV-1 was specific in inhibiting LYP. This occurred despite LYP’s high sequence similarity with PTP-PEST and SHP1. Furthermore, a sound SAR around the chemical scaffold of LTV-1 was established for LYP, and several analogs of LTV-1 showed efficacy in T cells at low micromolar concentrations. In addition to inhibiting the major allele LYP*R620, LTV-1 also inhibited the activity of the autoimmunity-associated allele LYP*W620, suggesting similar active site conformations in the two variants.
Under physiological conditions, LTV-1 is the most potent LYP inhibitor reported to date, and, more importantly, was found to be selective for inhibiting LYP over the closely related phosphatases PTP-PEST, SHP1, CD45, TCPTP, and PTP1B. Selective inhibition of LYP over PTP1B has been particularly challenging, and only two other small molecules have been reported to selectively inhibit LYP over PTP1B - 2.6-fold27 and 2.3-fold18, respectively. Compared to these inhibitors, LTV-1 is an order of magnitude more potent, while exhibiting a slightly better degree of selectivity. SAR analysis and in silico docking studies suggest that LTV-1 binds LYP specifically to the open state, interacting with both the P-loop and a small hydrophobic pocket that is only exposed in LYP’s open conformation. Open state binding, in which a ligand locks the flexible WPD-loop in its inactive open conformation, has only recently been described28–30. Notably, LYP is among the few members of the PTP family in which the WPD-loop adopts an exceptionally open conformation, resulting in very unique surface properties within the active site20. The three-dimensional structure of LYP’s closest relative, PTP-PEST, is not yet known. However, considering that their amino acid sequences within the active site are almost identical, the excellent selectivity of LTV-1 for LYP over PTP-PEST (200-fold) may be the result of substantial differences between their open state conformations. The notable lower selectivity of LTV-1 over both TCPTP and PTP1B, enzymes with lower sequence similarity with LYP, suggests that conformational resemblance between the PTP active sites may play a greater role for inhibitor specificity than amino acid sequence similarity. This notion is further supported by the fact that LTV-1 is less active against PEP, which shares 100% sequence identity with LYP within the active site.
Collectively, our data obtained with primary human T cells suggest a model where dissociation of the LYP/CSK complex is necessary for LYP-dependent down-modulation of TCR-induced responses (Fig. 7). This model is supported by three important observations. First, in resting T cells most LCK molecules were found in lipid rafts together with 5–10% of the CSK molecules. In contrast, LYP was not found in rafts of resting cells. Second, in response to TCR stimulation the LYP/CSK complex dissociated with kinetics paralleling the decline in the TCR signal. At the same time, CSK dissociated from lipid rafts while LYP was recruited, indicating that LYP partitioning into rafts occurs independently of CSK. The latter is further supported by our finding that the disease-associated LYP*W620 partitioned into lipid rafts more efficiently than LYP*R620. Finally, disruption of the LYP/CSK complex by overexpressing the CSK-SH3 domain resulted in weakened TCR signaling that could be restored by pretreatment of cells with our LYP inhibitor. The dynamic interaction between LYP*R620 and CSK, and its physiological consequences, also provide an explanation for the diminished TCR signaling caused by the risk allele LYP*W620, which cannot bind CSK. However, we cannot exclude the possibility that the R620W mutation creates a docking site for a novel set of proteins, thereby causing altered sub-cellular localization of LYP. In addition, LYP*R620 and to a lesser extent LYP*W620 can be phosphorylated on Y536, resulting in a reduction in the PTP activity31.
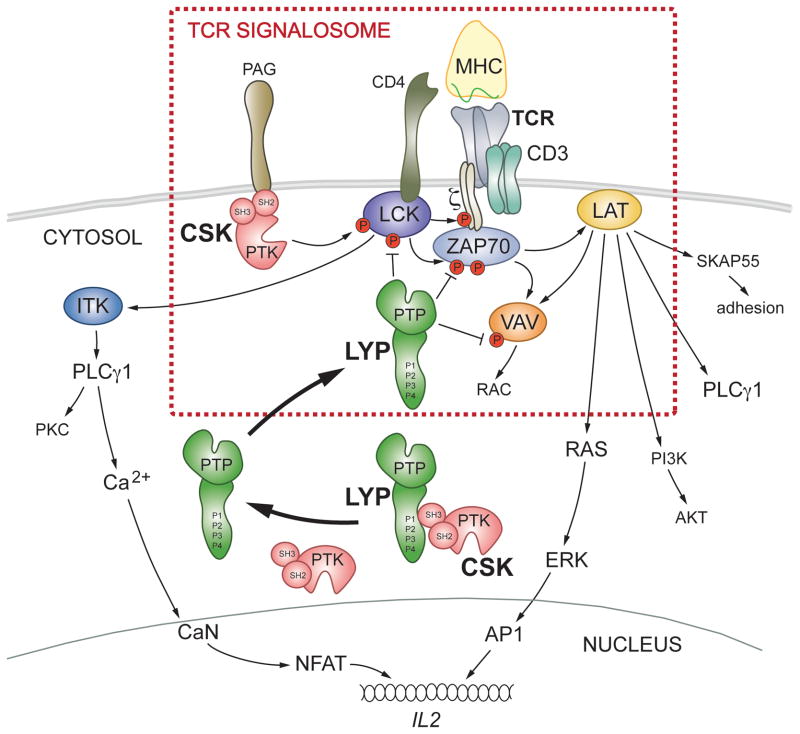
Figure 7. Proposed model of how LYP and CSK control TCR-induced signaling.
The combined binding of antigenic peptide/MHC class II molecule to the TCR and the co-receptor CD4 initiates a signaling cascade eventually resulting in formation of a multimeric signaling complex referred to as the ‘TCR signalosome’. The ‘TCR signalosome’ is located in the plasma membrane and is constituted of lipid raft resident proteins (e.g. LCK and LAT) as well as proteins that are more loosely associated with rafts (e.g. the TCR itself, CD3/ζ-chains, and ZAP70). Under resting conditions, LYP and CSK form a complex, thereby preventing LYP from gaining access to its substrates. TCR stimulation induces formation of the ‘TCR signalosome’ but also leads to dissociation of the LYP/CSK complex and subsequent recruitment of LYP to lipid rafts, where LYP can dephosphorylate and hence inactivate important players in the ‘TCR signalosome’. The risk allele LYP*W620 cannot bind CSK, and thus is not restricted from entering rafts, even under resting conditions.
Previous studies using primary T and B cells from genotyped healthy donors and patients have indicated that LYP*W620 is a more potent inhibitor of both TCR and BCR signaling compared to the normal allele LYP*R6208–10. In contrast, LYP*W620 was recently reported as a hypomorph, i.e. being a weaker inhibitor of TCR signaling compared to LYP*R62032. This conclusion was based on studies with Jurkat T cells co-transfected with GFP, CSK, and LYP. Since LYP and CSK form a complex, the authors argued that overexpression studies with LYP should be conducted together with overexpressed CSK. However, the data presented with LYP/CSK in this paper, combined with existing data on PEP/CSK4,13,33, demonstrate that there is a significant molar excess of CSK over LYP/PEP in both human and murine T cells. While this suggests that there is no need for ectopical expression of CSK when transfected LYP or PEP are studied, the conclusion that LYP is a hypomorph was based on their analysis of the GFPhigh cells32, where LYP expression levels most likely were supra-physiological. In fact, in their GFPlow cell population, which constitute a modest level of LYP overexpression, phospho-ERK signals were slightly lower in cells expressing LYP*W620 compared to LYP*R620, suggesting that LYP*W620 is indeed a stronger inhibitor of TCR signaling in these cells. The same report also showed that transfected CSK inhibited TCR signaling more potently when co-transfected with LYP*R620 relative to LYP*W620, presumably because CSK and LYP*R620 can form a complex while CSK and LYP*W620 cannot32. However, based on our data an alternative explanation can be provided, namely that CSK and LYP control TCR signaling with different timing. While raft-associated CSK is important for setting the threshold for TCR signaling in resting T cells, LYP*R620 is recruited to rafts in order to down-modulate signaling in activated T cells. LYP*W620 partitions into rafts more efficiently than LYP*R620 and will negatively regulate the raft-resident Src family kinases LCK and FYN, perhaps even in resting cells. As a consequence, and because recruitment of CSK to rafts depends on LCK and FYN-driven phosphorylation of CSK-binding proteins in rafts, the dynamic regulation of CSK in rafts may be different in cells expressing LYP*W620.
Our model for LYP/CSK in human T cells is not entirely compatible with observations made with mouse T cells, where PEP and CSK block TCR-induced responses in a co-operative manner through formation of a PEP/CSK complex33,34. This discrepancy may be explained by differences between the LYP and PEP protein structures, which share 89% amino acid identity in their PTP domains but only 61% in their non-catalytic portions. Furthermore, a recent characterization of PEP-R619W knock-in mice (mutation corresponding to LYP-R620W) revealed that these mice, even when homozygous for the mutation, remain healthy and do not show any signs of autoimmunity or organ pathology apart from slight enlargement of thymi and spleens23. In comparison, the LYP-R620W mutation is causative for various autoimmune diseases in humans, with typically 2 or 4-fold increased risk for autoimmunity among carriers of one or two LYP*W620 alleles, respectively35. Interestingly though, PEP-R619W protein in the PEP-R619W knock-in mice was unstable compared to wild-type PEP, presumably through calpain- and proteasome-mediated cleavage and degradation23. The authors extended these findings to LYP and proposed that LYP*W620 is also an unstable protein. In contrast, we found no differences in LYP expression levels in primary CD4+ T cells from healthy donors homozygous for either LYP*R620 or LYP*W620. Currently, our only explanation for these discrepancies is that there may be differences between the antibodies used in these two studies.
In conclusion, we developed a specific chemical probe for LYP and used this probe to show that LYP mainly down-modulates TCR-induced signaling when dissociated from CSK. Our LYP inhibitor also represents a starting point for developing a LYP-based treatment for autoimmune diseases and provides a new tool for further studies aimed at elucidating how LYP contributes to the development of autoimmunity.
METHODS
Reagents, antibodies, plasmids, recombinant PTPs
OMFP and PEG8000 were from Sigma, DiFMUP from Invitrogen, LTV-1 from ChemBridge. The identity and purity (>95%) of LTV-1 was verified by LC/MS and 1H-NMR. All other compounds and analogs were from ChemBridge (>95% purity). All siRNAs were purchased from Qiagen. Antibodies were as before8,15, anti-SRC phosphotyrosine-416 (cross-reacting with LCK phosphotyrosine-394) was from from Cell Signaling Technology, anti-LYP (goat) was from R&D (a characterization of this antibody is presented in Supplementary Fig. 11). Phosphotyrosine content in ζ-chain and LAT was assessed with 4G10. Plasmids were as before8,12,18,21 apart from CSK-SH3, which was generated from wild-type CSK by site-directed mutagenesis (Stratagene, introduction of stop codon corresponding to amino acid residue 82 in wild-type CSK). Recombinant LYP catalytic domain (residues 2–309) was expressed and purified as described29. Recombinant PTP-PEST, SHP1, CD45, and PTP1B were from Biomol.
Chemical library screening and counter screening
50,000 drug-like molecules from the DIVERSet™ library (ChemBridge) were screened in a 384-well format in vitro assay. Compound working concentration was 0.025 mg/mL in 5% DMSO. Each reaction (25 μL) contained 6.5 nM LYP, 60 μM OMFP (Km value for LYP), 0.005 mg/mL compound, 20 mM Bis-Tris pH 6.0, 1 mM dithiothreitol, 1% PEG8000, and 1.6% DMSO. Reactions were initiated by addition of OMFP after preincubating the enzyme with compounds for 10 min at room temperature. After 60 min reactions were quenched by addition of 25 μL 0.2 M NaOH, and the OMFP hydrolysis was determined by measuring the fluorescence intensity of the dephosphorylated product 3-O-methylfluorescein (excitation 485 nm, emission 528 nm) using an EnVision 2103 reader (Perkin Elmer). Nonenzymatic hydrolysis of substrate was corrected by measuring control samples not containing enzyme. Other controls included a positive control (1 mM vanadate added) and a negative control (no compound added). The ratio of inhibition compared to the negative control was determined for each compound. Compounds with >50% inhibition were cherry-picked and re-screened in a 10-point dose-response (5×10−3 mg/mL to 9.8×10−6 mg/mL). Hits with dose-dependent inhibition of LYP with IC50 values <0.005 mg/mL were compared to hits from screenings with the PTPs HePTP, VHR, and MKP-3, all of which were carried out under similar conditions. Common hits between LYP and either HePTP, VHR, or MKP-3 were filtered out for non-selective inhibition. The datasets for HePTP, VHR, and MKP-3 were retrieved from the PubChem website (http://pubchem.ncbi.nlm.nih.gov/), BioAssay IDs 521 (HePTP), 1992 (VHR), and 425 (MKP-3).
IC50 and Ki measurements
LYP-catalyzed hydrolysis of DiFMUP was assessed in 96-well format under similar conditions as described above. At various compound concentrations, the initial rate at various (0.5-1-2-4-8-16-32-64 μM) or fixed (equal to the corresponding Km value for each PTP) substrate concentrations was determined using an FLx800 reader (Biotek, excitation 360 nm, emission 460 nm). Nonenzymatic hydrolysis of substrate was corrected by measuring control samples not containing enzyme. The IC50 and Ki values were calculated as described previously36.
Cells, transfections, cytokine measurements, T cell activation reporter assays
CD4+ T cells were purified from buffy coats from healthy donors using EasySep human CD4+ T cell enrichment kit (Stemcell Technologies). Genotyping of PTPN22 SNPs were performed both by TaqMan allelic discrimination and by sequencing as described previously37. Transfection of cells (Jurkat TAg T cells or primary T cells) and cytokine secretion assays (ELISA, R&D) were performed as before8. T cell activation reporter assays were conducted as described previously18. Whenever cells were pre-treated with small-molecule compound or DMSO (vehicle control), this was performed at 37°C for 45 min prior to TCR stimulation.
TCR time courses, cell lysis, immunoprecipitations
Cells pre-treated with either small-molecule compound or DMSO were TCR stimulated (500 ng/ml OKT3 for Jurkat TAg T cells, for primary T cells biotinylated OKT3 was used followed 2 min later by cross-linking with 25 μg/ml avidin) for the indicated time periods. Cells were lysed in standard lysis buffer (50 mM HEPES pH 7.4, 100 mM NaCl, 1% Triton X-100, 50 mM NaF, 5 mM Na3VO4, 1 mM PMSF, 5 mM EDTA). For immunoprecipitations, cells were lysed in standard lysis buffer containing n-octyl-β-D-glucopyranoside (50 mM) and otherwise performed as before18.
Lipid raft purification
Cells (stimulated or not) were rested on ice for 1 min, lysed in 1 ml standard lysis buffer (with 1% Brij-58 instead of 1% Triton X-100), and subjected to sucrose gradient centrifugation in a total volume of 5 ml as described previously15. After centrifugation, 9 fractions of 500 μl were collected from the top; lipid rafts were found in the upper three fractions.
Calcium flux assay
Jurkat TAg T cells were loaded with Fluo-4 AM (1 μM) as recommended by the manufacturer (Invitrogen). After incubation with LYP inhibitor or DMSO, cells were rested on ice for 30 min, followed by calcium measurements on a BD Biosciences FACSCanto II using the following protocol: cells were pre-equilibrated at 37 °C for 5 min, then baseline levels were recorded (1000–5000 events/sec) for 1 min, followed by addition of OKT3 (5 μg/mL) and recording for another 3 min.
Assays for CD25 up-regulation and assessment of LYP expression in primary CD4+ T cells
T cells were pre-treated with LYP inhibitor in serum-free medium. Cells were then seeded in wells and stimulated or not in complete growth medium with coated anti-CD3 (5 μg/mL unless otherwise stated, clone UCHT1 from BD Pharmingen) and soluble anti-CD28 (1 μg/mL of CD28.2 clone from BD Pharmingen) for 40 h. Later, CD25 expression was analyzed on a BD Biosciences FACSCalibur. For assessment of LYP expression in primary human T cells, purified CD4+ T cells (from donors homozygous for either LYP*R620 or LYP*W620) were stained with a LYP-specific antibody, followed by assessment of LYP expression levels on a BD Biosciences FACSCanto II.
Software, statistical analysis and densitometric scanning analysis
Flow cytometry data were analyzed with Cell Quest (BD Bioscience) and FlowJo (TreeStar) software. Statistical analysis was performed using t-test (unpaired or paired) as implemented in GraphPad Prism (v5, GraphPad Software, Inc.) or Excel (Microsoft). The program Scion Image (Scion Corporation) was used for densitometric scanning analysis.
Supplementary Material
Acknowledgments
We thank Dr. Layton Smith and the Exploratory Pharmacology Core at SBMRI for support and thoughtful discussions, Dr. Sandra Ormenese and the GIGA-imaging core facility for their help with flow cytometry, and Gladys M. Tjørhom, Siri T. Flåm, and Jorun Solheim for technical assistance. This work was supported by the US National Institutes of Health (Grant 1R21CA132121 to L.T.), the Norwegian Cancer Society (T.V.), the American Cancer Society (Research Scholar Grant RSG-08-067-01-LIB to R.P.), the Oxnard Foundation (Research Grant to T.M./R.C.R.), the Belgian Research National Scientific Fund (MIS Young Investigator Research Grant to S.R.), and the Liège University (Startup Grant to S.R.).
Footnotes
Author contributions
T.V., W.H.L., L.D., S.W., R.D., B.A.L., S.R., and L.T., designed research, performed research, analyzed data, wrote the paper; S.V., L.Y., L.M., D.F., and J.L., designed research, performed research, analyzed data; K.T. and M.L.T., contributed new reagents or analytical tools; R.P., designed research, analyzed data, wrote the paper; T.M. and R.C.R., designed research.
Competing interests statement
The authors declare no competing interests.
Supplementary Methods are available.
References
- 1.Mustelin T, Vang T, Bottini N. Protein tyrosine phosphatases and the immune response. Nat Rev Immunol. 2005;5:43–57. doi: 10.1038/nri1530. [DOI] [PubMed] [Google Scholar]
- 2.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vang T, et al. Protein tyrosine phosphatases in autoimmunity. Annu Rev Immunol. 2008;26:29–55. doi: 10.1146/annurev.immunol.26.021607.090418. [DOI] [PubMed] [Google Scholar]
- 4.Cloutier JF, Veillette A. Association of inhibitory tyrosine protein kinase p50csk with protein tyrosine phosphatase PEP in T cells and other hemopoietic cells. EMBO J. 1996;15:4909–4918. [PMC free article] [PubMed] [Google Scholar]
- 5.Gregorieff A, Cloutier JF, Veillette A. Sequence requirements for association of protein-tyrosine phosphatase PEP with the Src homology 3 domain of inhibitory tyrosine protein kinase p50(csk) J Biol Chem. 1998;273:13217–13222. doi: 10.1074/jbc.273.21.13217. [DOI] [PubMed] [Google Scholar]
- 6.Ghose R, Shekhtman A, Goger MJ, Ji H, Cowburn D. A novel, specific interaction involving the Csk SH3 domain and its natural ligand. Nat Struct Biol. 2001;8:998–1004. doi: 10.1038/nsb1101-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bottini N, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 8.Vang T, et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet. 2005;37:1317–1319. doi: 10.1038/ng1673. [DOI] [PubMed] [Google Scholar]
- 9.Rieck M, et al. Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. J Immunol. 2007;179:4704–4710. doi: 10.4049/jimmunol.179.7.4704. [DOI] [PubMed] [Google Scholar]
- 10.Arechiga AF, et al. Cutting edge: the PTPN22 allelic variant associated with autoimmunity impairs B cell signaling. J Immunol. 2009;182:3343–3347. doi: 10.4049/jimmunol.0713370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cote JF, et al. PSTPIP is a substrate of PTP-PEST and serves as a scaffold guiding PTP-PEST toward a specific dephosphorylation of WASP. J Biol Chem. 2002;277:2973–2986. doi: 10.1074/jbc.M106428200. [DOI] [PubMed] [Google Scholar]
- 12.Arimura Y, Vang T, Tautz L, Williams S, Mustelin T. TCR-induced downregulation of protein tyrosine phosphatase PEST augments secondary T cell responses. Mol Immunol. 2008;45:3074–3084. doi: 10.1016/j.molimm.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davidson D, Cloutier JF, Gregorieff A, Veillette A. Inhibitory tyrosine protein kinase p50csk is associated with protein-tyrosine phosphatase PTP-PEST in hemopoietic and non-hemopoietic cells. J Biol Chem. 1997;272:23455–23462. doi: 10.1074/jbc.272.37.23455. [DOI] [PubMed] [Google Scholar]
- 14.Chow LM, Fournel M, Davidson D, Veillette A. Negative regulation of T-cell receptor signalling by tyrosine protein kinase p50csk. Nature. 1993;365:156–160. doi: 10.1038/365156a0. [DOI] [PubMed] [Google Scholar]
- 15.Vang T, et al. Activation of the COOH-terminal Src kinase (Csk) by cAMP-dependent protein kinase inhibits signaling through the T cell receptor. J Exp Med. 2001;193:497–507. doi: 10.1084/jem.193.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tautz L, Mustelin T. Strategies for developing protein tyrosine phosphatase inhibitors. Methods. 2007;42:250–260. doi: 10.1016/j.ymeth.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 17.Willett P. Similarity-based virtual screening using 2D fingerprints. Drug Discov Today. 2006;11:1046–1053. doi: 10.1016/j.drudis.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Vang T, et al. Inhibition of lymphoid tyrosine phosphatase by benzofuran salicylic acids. J Med Chem. 2011;54:562–571. doi: 10.1021/jm101004d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abagyan R, Totrov M, Kuznetsov D. ICM - A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J Comp Chem. 1994;15:488–506. [Google Scholar]
- 20.Barr AJ, et al. Large-scale structural analysis of the classical human protein tyrosine phosphatome. Cell. 2009;136:352–363. doi: 10.1016/j.cell.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinonen KM, Bourdeau A, Doody KM, Tremblay ML. Protein tyrosine phosphatases PTP-1B and TC-PTP play nonredundant roles in macrophage development and IFN-gamma signaling. Proc Natl Acad Sci U S A. 2009;106:9368–9372. doi: 10.1073/pnas.0812109106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bourdeau A, Dube N, Tremblay ML. Cytoplasmic protein tyrosine phosphatases, regulation and function: the roles of PTP1B and TC-PTP. Curr Opin Cell Biol. 2005;17:203–209. doi: 10.1016/j.ceb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, et al. The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness. Nat Genet. 2011;43:902–907. doi: 10.1038/ng.904. [DOI] [PubMed] [Google Scholar]
- 24.Todd JA, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tautz L, Pellecchia M, Mustelin T. Targeting the PTPome in human disease. Expert Opin Ther Targets. 2006;10:157–177. doi: 10.1517/14728222.10.1.157. [DOI] [PubMed] [Google Scholar]
- 26.Vintonyak VV, Antonchick AP, Rauh D, Waldmann H. The therapeutic potential of phosphatase inhibitors. Curr Opin Chem Biol. 2009;13:272–283. doi: 10.1016/j.cbpa.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 27.Yu X, et al. Structure, inhibitor, and regulatory mechanism of Lyp, a lymphoid-specific tyrosine phosphatase implicated in autoimmune diseases. Proc Natl Acad Sci U S A. 2007;104:19767–19772. doi: 10.1073/pnas.0706233104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S, et al. Targeting inactive enzyme conformation: aryl diketoacid derivatives as a new class of PTP1B inhibitors. J Am Chem Soc. 2008;130:17075–17084. doi: 10.1021/ja8068177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu S, Bottini M, Rickert RC, Mustelin T, Tautz L. In Silico Screening for PTPN22 Inhibitors: Active Hits from an Inactive Phosphatase Conformation. ChemMedChem. 2009;4:440–444. doi: 10.1002/cmdc.200800375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, et al. Salicylic acid based small molecule inhibitor for the oncogenic Src homology-2 domain containing protein tyrosine phosphatase-2 (SHP2) J Med Chem. 2010;53:2482–2493. doi: 10.1021/jm901645u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiorillo E, et al. Autoimmune-associated PTPN22 R620W variation reduces phosphorylation of lymphoid phosphatase on an inhibitory tyrosine residue. J Biol Chem. 2010;285:26506–26518. doi: 10.1074/jbc.M110.111104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zikherman J, et al. PTPN22 deficiency cooperates with the CD45 E613R allele to break tolerance on a non-autoimmune background. J Immunol. 2009;182:4093–4106. doi: 10.4049/jimmunol.0803317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cloutier JF, Veillette A. Cooperative inhibition of T-cell antigen receptor signaling by a complex between a kinase and a phosphatase. J Exp Med. 1999;189:111–121. doi: 10.1084/jem.189.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gjorloff-Wingren A, Saxena M, Williams S, Hammi D, Mustelin T. Characterization of TCR-induced receptor-proximal signaling events negatively regulated by the protein tyrosine phosphatase PEP. Eur J Immunol. 1999;29:3845–3854. doi: 10.1002/(SICI)1521-4141(199912)29:12<3845::AID-IMMU3845>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 35.Gregersen PK, Olsson LM. Recent advances in the genetics of autoimmune disease. Annu Rev Immunol. 2009;27:363–391. doi: 10.1146/annurev.immunol.021908.132653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tautz L, et al. Inhibition of Yersinia tyrosine phosphatase by furanyl salicylate compounds. J Biol Chem. 2005;280:9400–9408. doi: 10.1074/jbc.M413122200. [DOI] [PubMed] [Google Scholar]
- 37.Viken MK, et al. The PTPN22 promoter polymorphism -1123G>C association cannot be distinguished from the 1858C>T association in a Norwegian rheumatoid arthritis material. Tissue Antigens. 2007;70:190–197. doi: 10.1111/j.1399-0039.2007.00871.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.