Abstract
Polycomb Group (PcG) proteins are concentrated in nuclear foci called PcG bodies. Although the some of these foci are due to the tendency of PcG binding sites in the genome to occur in linear clusters, distant PcG sites can contact one another and in some cases congregate in the same PcG body when they are repressed. Experiments using transgenes containing PcG binding sites reveal that co-localization depends on the presence of insulator elements rather than of Polycomb Response Elements (PREs) and that it can occur also when the transgenes are in the active state. A model is proposed according to which insulator proteins mediate shuttling of PcG target genes between PcG bodies when repressed to transcription factories when transcriptionally active.
Introduction
Nuclear bodies
In the past ten years, many types of “bodies” have been identified in the nucleus, using different techniques but most often imaging nuclei stained with antibodies specific for a nuclear protein. For our purposes, the most interesting are those involving chromatin-binding proteins. Examples are PML bodies, nuclear stress bodies, insulator bodies, Polycomb or PcG bodies, transcription factories (for a recent review, see ref. 1). These suggest the possibility that parts of the genome that bind a common protein factor might gather together in the nucleus to interact, to share common machinery, to create local concentrations of specific factors, etc. The physical nature and significance of these bodies has often been debated and it has been questioned whether they are true structures actively assembled rather than passive or accidental local protein accumulations. The present discussion concerns primarily Polycomb bodies although some of the other bodies may be also implicated.
Habeas Corpus
Polycomb complexes have been traditionally thought of as somehow sticky. This is in part a legacy from the time when they were thought to act like heterochromatin complexes, which in turn were thought to polymerize along the chromatin and form condensed structures impervious to transcription factors. This view was influential in the 1990s and continues to be cited although there has not been much evidence to support it. There is, however, considerable evidence showing that Polycomb Group (PcG) proteins are found aggregated in nuclear foci rather than dispersed in the hundreds or thousands of genomic binding sites known by genome-wide ChIP analyses. Microscopy with antibody staining or live imaging of fluorescently labeled proteins have shown that PcG proteins are in fact localized in a relatively small number of nuclear concentrations that have been called Polycomb Bodies or PcG Bodies [2,3,4,5 and many others] (Figure 1). The numbers and sizes of such bodies vary in different cell types. They are said to be fewer and larger in embryonic stem cells and become smaller and more numerous when these cells are induced to differentiate [6]. In certain cell lines, aberrant concentrations are seen: V2OS and other transformed tumor cells have just a few but very large PcG bodies associated with the periphery of heterochromatin or centromeric satellites [2,5]. Although these cell lines clearly represent abnormal states, they raise questions about the nature and diversity of morphologies that may require different explanations.
Figure 1. Polycomb bodies.
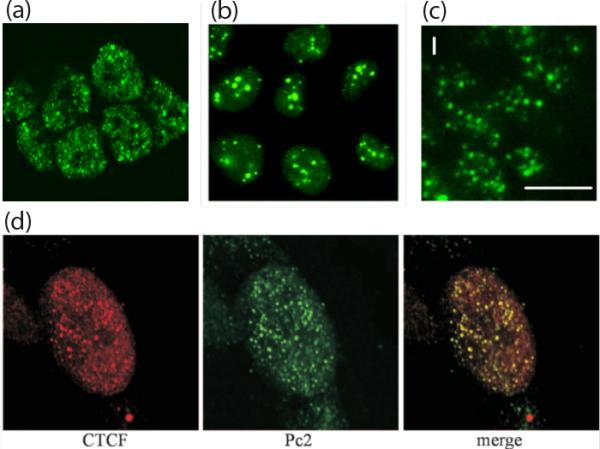
(a) Human embryonic stem cell line stained with antibody against the PcG protein Ring1B (reproduced from ref. 50, with permission). (b) Polycomb bodies visualized in the human cancer cell line U2OS expressing GFP-BMI1 (reproduced from ref. 5, with permission). (c) Drosophila live imaginal disc cells expressing GFP-PH (reproduced from Development, ref. 3, with permission). (d) HeLa cell nucleus stained with anti-CTCF (red) and anti-PC2/CBX4 (green). The two images are merged to show remarkable co-localization (reproduced from ref. 27**, with permission).
Are there different kinds of PcG bodies? Are they sites where PcG proteins are stored or even sequestered from the rest of the nuclear volume or do they represent PcG complexes bound to chromatin sites? If the latter, are these functional chromatin targets, i.e. genes that bind PcG proteins and are thereby repressed? In that case, what are the mechanisms that bring them together in the nucleus? And, finally, has this clustering a functional importance? These are questions that we will briefly review in this article.
Genomic distribution of PcG target sites
One possible explanation for these PcG bodies has nothing to do with the properties of PcG complexes but with the genomic distribution of their target sites. It is clear that some of the major targets of PcG regulation are gene clusters: multiple genes that bind PcG complexes and are physically adjacent in the linear genome map. A general tendency of PcG target genes to be enriched in genomic vicinities has been pointed out [7] and can be visualized in a Hilbert folding map of the PcG binding sites along a Drosophila chromosome arm (Figure 2A). Examples of these PcG target concentrations in Drosophila are the two Hox clusters ANT-C and BX-C, the two largest concentrations of PcG targets in the Drosophila genome, separated by about 10 Mb. Other examples are the cluster of NK-homeodomain protein genes (at least six PRE-containing genes including tinman, bagpipe, etc.); the GATA cluster (at least four PRE-containing genes including pannier, serpent, GATAe); the cnc cluster (at least four PRE-containing genes including hh, cnc, pnt). In each cluster, multiple genes, each a PcG target, are located in a chromosome domain of the order of 200 kb. Similar examples, often involving genes homologous to the Drosophila targets, are found in mammalian genomes. It is evident in these cases that PcG complexes are concentrated in certain nuclear neighborhoods simply because their targets are clustered in genomic neighborhoods. Furthermore, PcG binding sites that are isolated either because they are not closely linked with others on the genome or because they have transiently looped away from the other PcG sites would stain more weakly and therefore tend to be neglected by comparison with the strong signals caused by the clustered PcG sites. Thus, at least a fraction of PcG bodies are simply the result of genomic clustering of target genes. This, however, is not the whole story.
Figure 2. Constitutive bodies and dynamic bodies.
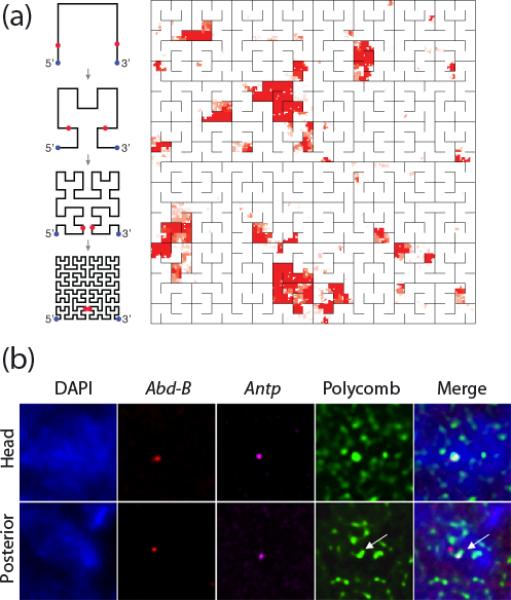
(a) Hilbert folding representation of Polycomb binding sites on chromosome 3R of Drosophila. Some 75-100 (depending on how they are counted) PC binding sites correspond to this map that shows a high degree of clustering along the chromosome. The diagrams on the left explain how the iterative Hilbert folding (for two dimensions, called Peano folding; for applications to genomic data see ref. 51) of the linear chromosome arm is achieved. Figure kindly provided by P. Kharchenko. (b) The Drosophila Antp gene (magenta) and the Abd-B gene (red) visualized by FISH co-localize within a PcG body (Polycomb) in nuclei from the embryonic head region, where both are repressed by PcG complexes. In nuclei from the posterior region, the active Antp gene moves out of the PcG body but the repressed Abd-B gene remains associated with the PcG body (reproduced from ref. 10**, with permission)
Gathering the Polycombs
There is a large residue of PcG bodies that clearly involves genes that map at large distances from one another or even on different chromosomes but nevertheless give evidence of interacting or physically co-localizing in the nucleus.
To begin with the Hox genes, the Drosophila Antp gene (part of the ANT-C) has been shown to co-localize with the Abd-B gene (part of the BX-C) although the two are separated by some 10 Mb along chromosome 3R [8,9**,10**] (Figure 2B). The co-localization is visible by FISH in a fraction of nuclei ranging from 15% to 28% when both genes are repressed by PcG complexes, compared to 5-15% when one is active and the other repressed or in Pc- background. Since Abd-B is associated with a PcG body in 87% of nuclei [9**], this means that the two Hox clusters co-localize about one fifth of the time. This relatively low frequency suggests that the association is not a stable one or that it varies with time or during the cell cycle or from one cell cycle to the next. Similar experiments by Fedorova et al. [11] concluded, however, that the frequencies of co-localization were less compelling and were in any case dependent on the tissue and fly stock used. Furthermore, a transgene containing the Fab-7 PcG binding site was found within a PcG body in only 54% of nuclei. This suggests that a certain percentage of the time individual Polycomb binding sites are not detected or may even dissociate from PcG complexes. Imaging with live cells suggests, however, that the associations of distant PcG binding sites have half-lives of at least a few minutes [12,13**], thus comparable or longer than the rates of exchange of PcG component proteins [3,5]. Most importantly, co-localization between Antp and Abd-B is lost when one is repressed and the other is active [9**,10**]. The authors concluded that the PcG complexes might be responsible for keeping them together. A more cautious interpretation might be simply that active and repressed PcG target genes occupy different nuclear environments.
That some type of dynamic change in the spatial organization of Hox clusters can take place has been shown for the mammalian Hoxb and Hoxd clusters during stem cells differentiation (14,15]. In these cells, locus-wide changes in histone modifications (H3K9 acetylation and H3K4 methylation) and physical decondensation are detected first, followed by a progressive looping out of individual genes out of the chromosome territory and by programmed transcriptional activation. It would be important to know at what stage in this process the PcG complexes are lost from which Hox genes.
Do Polycomb target genes interact with one another?
The idea that certain kinds of genes might associate with one another although remotely located in the genome was first aired to account for the behavior of certain transgenes in Drosophila, which will be discussed further below. More recently, genome-wide 4C and related approaches have been used to ask if PcG target genes tend to contact one another [10**,16**]. Tolhuis et al. took a number of individual PcG targets as baits to determine the frequency of crosslinking to other sites in the genome [16**]. The results show that the most frequent partners are most of the other PcG sites on the same chromosome arm but long-range contacts are rare with non-PcG sites. When non-PcG target sites are used as bait, interaction partners are also found preferentially on the same chromosome arm but they are generally not PcG binding sites. The conclusion that PcG binding sites come frequently and specifically in contact with one another has been taken as evidence that the PcG complexes are likely to be involved in these interactions.
Many of the experiments that initially revealed interactions between remote PcG targets in the nucleus were done using transgenes containing Polycomb Response Elements (PREs). This approach is common in Drosophila studies since Drosophila PREs have been well characterized as DNA fragments of up to a few hundred bp that suffice to recruit PcG complexes, produce a domain of H3K27 trimethylation, and repress nearby genes. Although the degree of silencing of the reporter gene (often the white gene, responsible for eye pigmentation) is variable from one transgene insertion site to another, it is usually possible to observe pairing-dependent silencing [17,18,19]. That is, when the fly is homozygous for a transgene insertion, the homologous pairing of the two copies results in stronger silencing and less expression than with a single copy of the transgene. This is not necessarily evidence for clustering of PcG sites, since homologous chromosomes are paired during interphase in insects like Drosophila, but it does demonstrate that a functional effect results from the proximity of two or more PcG binding sites. However, in some cases, two transgenes inserted at different chromosomal sites, sometimes on different chromosomes, show the same type of interaction. That is, they are both more repressed when present in the same nucleus than each copy is in the absence of the other.
Transgenes containing the Fab-7 or the Mcp PRE regions from the BX-C have been used to show that two copies of the transgenes inserted at sites remote from one another can often functionally interact in this fashion (17,18,19]. That this functional interaction involves physical interaction between the two transgene copies and between the transgenes and the endogenous element was demonstrated by imaging, as well as by the 3C technique [8,12,13**]. Here also, the long-distance interactions are more frequently observed between insertions on the same chromosome arm but they could be detected also between sites on different chromosomes. Surprisingly, however, these interactions could not be demonstrated for some other PRE elements, including the very strong silencer bxd PRE. Earlier, Sigrist and Pirrotta had found that such interactions could be readily obtained between transgenes containing the bxd PRE if they also contained the gypsy Su(Hw) insulator [20]. This is highly significant because both Mcp and Fab-7 elements have been shown to contain an insulator activity closely associated but separable from the PRE [21,22]. To clarify this relationship, Li et al. tested a variety of transgenes comparing the bxd PRE and the Mcp element, and the Mcp or Fab-7 elements with either PRE or insulator moieties deleted [13**]. The clear conclusion was that none of the three PREs shows long-distance interactions by itself but both Mcp and Fab-7 insulators mediate such interactions independently of the presence of a PRE. The conclusion, therefore, was that the long-distance interactions between Mcp or Fab-7 transgenes or between these and the endogenous Mcp or Fab-7 are not mediated by PcG complexes but by insulator elements. The frequencies of nuclei exhibiting co-localization was relatively low (6-8%) but well above the frequencies seen in the absence of insulator (0.1-0.5%).
Insulators are now well known to mediate chromatin looping interactions both in flies and in mammals (see article by V. Corces in this volume) but these results appear to exclude a role for PcG proteins in the long-distance interactions. Both the ANT-C and the BX-C Hox clusters in Drosophila contain multiple binding sites for the insulator proteins dCTCF and CP190. These have been found to be involved in the extensive looping and infra-locus contacts among the numerous regulatory elements, including the Mcp and Fab-7 elements [23,24,25,26]. These BX-C insulator elements and similar elements found in ANT-C could account for many of the co-localization effects reported for these two loci. Suggestively, the mammalian CTCF protein forms bodies that co-localize with PcG bodies in HeLa cell nuclei [27*] (Figure 1D). Does this mean that PcG proteins play no role in the formation of PcG bodies? It is important here to distinguish between the two kinds of observation: the co-localization between genes or transgenes and the concentrations of PcG proteins into PcG bodies. The nuclear environment in which two Mcp transgenes meet need not always be a PcG body. Given the low frequency of co-localization, it is clear that the association is relatively short-lived and is not incompatible with other interactions and co-localizations. For example, when a transgene is transcriptionally active, it is unlikely to be found within a PcG body. This question is raised in particular by the experiments of Vazquez et al. [12] who used a transgene containing the Mcp element (PRE + insulator) as well as the eye enhancer of the white gene and observed a very high frequency of co-localization (up to 90%) but only in the eye imaginal disc, in which the transgene is transcriptionally active as witnessed by the strong eye pigmentation. Convergence of the transgene copies to a single focus was seen even in nuclei containing four different Mcp transgenes, demonstrating the remarkable stability of this co-localization in the post-mitotic eye cells that express the white gene.
Recent experiments have shown that this high frequency co-localization is different from the low frequency co-localization seen with the insulator alone: in addition to the insulator, it requires transcriptional activity as well as the PRE (H-B Li and V Pirrotta, unpublished results). The PRE is involved not because it binds PcG proteins but because, like all PREs, it binds Trithorax and is also a Trithorax Response Element (TRE). Mutations in Trithorax or dCTCF but not Polycomb mutations abolish high level co-localization. These results indicate that co-localization of the transgenes is taking place at a site containing high transcriptional activity. A most likely candidate is a transcription factory. It is evident in any case why Antp and Abd-B no longer co-localize when one of the two is transcriptionally active [9**,10**]. They are targeted to two different places in the nucleus.
Transcription factories
Transcription factories are nuclear sites that have been posited to account for the localization of RNA pol II and most transcriptional activity to foci that are relatively few in number compared to the number of active genes in the nucleus (for review see ref. 28 and the article by P. Fraser in this volume). Multiple genes are transcribed in a single transcription factory and, in some cases, genes that are remotely located have been found to share preferentially the same transcription factory [29]. In some cases, trans-activation between remote genes has been shown to depend on CTCF [30*] and a comprehensive mapping of the CTCF interactome in mouse embryonic stem cells has revealed pervasive interactions among proximal and remote regulatory elements and promoters [31*]. It is not clear how these interactions are targeted but one possibility is that activators may play a role [32**], in which case genes that share transcriptional activators may tend to co-localize in the same transcription factory. Alternatively, insulators might continue to play a key role but would be modified by transcriptional activators to switch their preference from PcG bodies to transcription factories (see model in Figure 3). It is interesting therefore that CTCF has been found as a component of mammalian transcription factories [52]. A particularly attractive possibility is that Trithorax and/or its functional partner ASH1, which bind to derepressed PcG target genes [33] might direct the insulators to a subset of transcription factories. Together, ASH1 and TRX are responsible for conveying the epigenetic memory of the transcriptionally active state of a gene from one cell cycle to the next [34,35]. We speculate that this memory function might be achieved by a longer-term association between the derepressed gene and a transcription factory. TRX/MLL is in fact known to remain associated with chromatin in condensed mitotic chromosomes, particularly favoring genes that had been transcriptionally active during interphase [36]. Just as the spatial vicinity of two PREs produces a pairing-dependent enhancement of silencing, the vicinity of two derepressed PcG target genes might result in enhanced transcriptional activity or stability of the active state. A remarkable report concerning the transcriptional activity of the Drosophila Hox gene Ubx demonstrated in fact that, when two copies of the gene are homologously paired, transcription from each allele is enhanced, while chromosome rearrangements that prevent pairing reduce transcription [37].
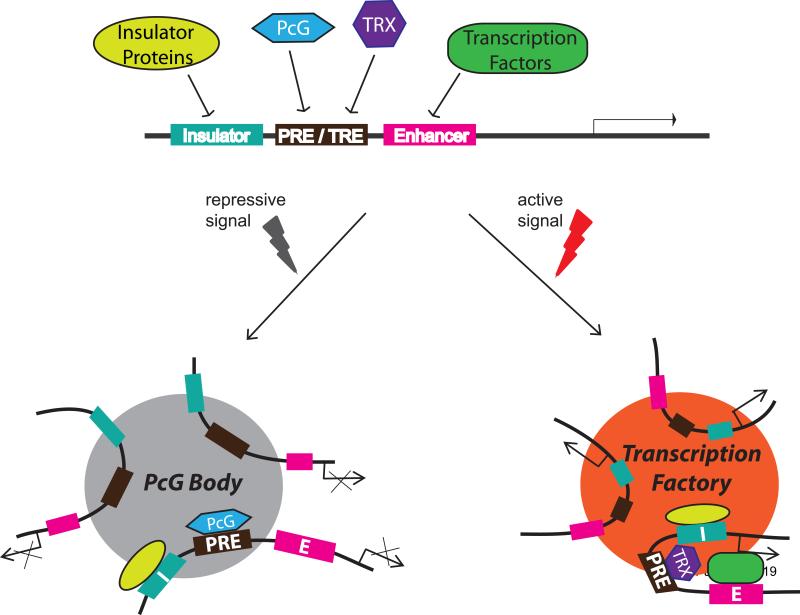
Figure 3. A shuttling model for the assembly of PcG target genes into nuclear bodies.
A generalized PcG target gene is presumed to be associated with one or more of each Enhancer, PRE, and Insulator elements. In this speculative model the insulator drives the association of such PcG target genes into PcG bodies when they are repressed. When the gene switches to the transcriptionally active state, it generally forms a domain that binds N-ter Trithorax and ASH1 proteins [33]. The insulator now targets the active gene to a Trithorax transcription factory. The switch in insulator preference for transcription factory vs PcG body localization might depend on modifications such as, for example, acetylation vs sumoylation.
RNAi involvement?
Grimaud et al. found that the physical interactions between remote copies of the Fab-7 element were dependent on components of the RNAi machinery [9**]. Dicer-2, AGO1, Aub, Piwi-1 and Piwi-2 are all needed for the long-distance interactions. Dicer-2 is needed for full repressive activity and for long-distance interactions but not for the recruitment of PcG proteins to the Fab-7 PRE, supporting the idea that PcG complexes do not directly cause the interactions. Differences in the effects of different RNAi components suggests that they may play somewhat different roles in these processes. A recent paper adds perplexing complexities to this picture, if confirmed. Another RNAi component, AGO2, is found associated with many PREs in Drosophila [38*]; it is also associated with dCTCF binding sites and is needed for at least some CTCF-mediated insulator functions. In contrast with the other RNAi proteins, AGO2 appears to have a weak anti-repressive role at PREs. These properties are not related to the RNAi role of AGO2 since they are not affected by mutations in the catalytic site that abolish the RNAi-related function.
It appears therefore that different components of the RNAi machinery are involved in PcG repression and in long-distance interactions but in different, possibly antagonistic senses. Some of these observations may be explained by the involvement of insulator-binding proteins in PcG long-distance interactions. RNAi components are required for the action of another insulator protein, Su(Hw), and an RNA helicase Rm62 is found associated with Su(Hw) insulator complexes [39]. Similarly, the DEAD-box RNA helicase p68 has been found to be associated with mammalian CTCF, together with a long ncRNA [40]. The combination of these results could be used to argue that looping interactions that mediate insulator function and long-distance interactions between Fab-7 or Mcp transgenes depend in some way on RNAi proteins and probably on some RNA component. The insulator-mediated looping together of PREs enhances their repressive activity. Nothing is known about the effects of co-localization on the expression of the associated genes when they are derepressed.
The role of RNA in insulator function and long-distance interactions, if there is one, is unclear. One possibility is that it could serve as a scaffold for the assembly of large complexes, whether insulator or PcG super-complexes, that underlie insulator bodies and PcG bodies. The existence of a possible connection between PcG bodies and RNAs is also hinted at by several recent reports of noncoding RNA molecules involved in the recruitment of mammalian PcG complexes to their target loci (see for example ref. 41,42,43,44]. One of these is the HOTAIR RNA that binds the Polycomb complex PRC2 and recruits it to the HoxD locus [42]. Another is the ANRIL ncRNA from the INK4/ARF locus that binds PRC1 and is needed for repression of that locus [44]. It appears therefore that both PRC1 and PRC2 have the potential to bind to a ncRNA although the mechanics of targeting to a specific chromatin locus remain unclear. No such evidence is yet available in Drosophila, where PcG complexes are known to be recruited in some cases by DNA sequences less than 100 bp in length. However, Drosophila PREs do not all have the same properties and it remains very possible that some involve ncRNAs in their function. Whether or not some Drosophila genes recruit PcG complexes through a ncRNA, the existence of PcG protein/RNA interactions justifies envisioning that RNA molecules might serve as scaffolds for higher order assemblies of PcG-binding loci. Such a role of both coding and non-coding RNAs has been described for the assembly of stress bodies, paraspeckles, histone locus bodies and Cajal bodies [45] (Shevtsov and Dundr, 2011) although, admittedly, the function of these bodies is specific for RNA-related processes.
Conclusion
Overall, then, the clustering of PcG target sites in the nucleus can be the result of different mechanisms. The most obvious is that due to existence of numerous multi-gene clusters, which probably account for the largest PcG bodies. Another mechanism is that mediated by insulator-like elements. PcG complexes bound to a target gene might modify the nearest insulator proteins to target a PcG body. Many PcG target genes are closely associated with insulator elements. This could easily explain the co-localization of the two Hox gene clusters in Drosophila. Less easy to interpret are the 4C interactions between PcG targets on a chromosome arm [16**]. These are far too specific to be accounted for by interactions among all insulators since there are hundreds of insulator protein binding sites per chromosome arm. It is possible therefore that PcG proteins might contribute to association. One way to effect that would be through post-translational modifications of the insulator proteins. CTCF has been found to be sumoylated and closely associated with PcG bodies [27*] (Figure 1D) and mammalian Polycomb2/CBX4 protein has been shown to be a SUMO E3 ligase [46,47]. When the target gene switches to the active state, TRX/ASH1 and the CBP acetylase associated with them [48,49] might produce an alternative modification of CTCF, for example, acetylation, that would now favor association with transcription factories. This model is illustrated schematically in Figure 3. Clearly this is highly speculative and many questions remain to be investigated before the nuclear distribution of PcG proteins and PcG targets can be understood.
Acknowledgments
We are grateful to P. Kharchenko for kindly providing the map in Figure 2A. Our research relevant to this review was funded by grant R01 GM082918 to VP from the U.S. National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saurin AJ, Shiels C, Williamson J, Satijn DPE, Otte AP, Sheer D, Freemont PS. The human Polycomb Group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J Cell Biol. 1998;142:887–898. doi: 10.1083/jcb.142.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ficz G, Heintzmann R, Arndt-Jovin DJ. Polycomb group protein complexes exchange rapidly in living Drosophila. Development. 2005;132:3963–3976. doi: 10.1242/dev.01950. [DOI] [PubMed] [Google Scholar]
- 4.Dietzel S, Niemann H, Brückner B, Maurange D, Paro R. The nuclear distribution of Polycomb during Drosophila melanogaster development shown with a GFP fusion protein. Chromosoma. 1999;108:83–94. doi: 10.1007/s004120050355. [DOI] [PubMed] [Google Scholar]
- 5.Hernandez-Munoz I, Taghavi P, Kuijl C, Neefjes J, van Lohuizen M. Association of BMI1 with Polycomb Bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1. Mol Cell Biol. 2005;25:11047–11058. doi: 10.1128/MCB.25.24.11047-11058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren X, Vincenz C, Kerppola TK. Changes in the distributions and dynamics of Polycomb Repressive Complexes during embryonic stem cell differentiation. Mol Cell Biol. 2008;28:2884–2895. doi: 10.1128/MCB.00949-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2010;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bantignies F, Grimaud C, Lavrov S, Gabut M, Cavalli G. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 2003;17:2406–2420. doi: 10.1101/gad.269503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9**.Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi components are required for nuclear clustering of Polycomb Group Response Elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [The authors demonstrate that components of the RNAi machinery are involved in the Fab-7 transgene-mediated long-range interactions. This was the first demonstration that Antp and Abd-B could interact with one another when both are repressed.] [DOI] [PubMed] [Google Scholar]
- 10**.Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–226. doi: 10.1016/j.cell.2010.12.026. [Using 3D-FISH and 4C techniques, the authors show that Antp and Abd-B, respectively in the ANT-C and BX-C homeotic gene clusters, 10 Mb distant from one another in the Drosophila genome, co-localize inside a PcG body when both are repressed but do not co-localize when one is silenced and the other is active.] [DOI] [PubMed] [Google Scholar]
- 11.Fedorova E, Sadoni N, Dahlsveen I, Koch J, Kremmer E, Eick D, Paro R, Zink D. The nuclear organization of Polycomb/Trithorax group response elements in larval tissues of Drosophila melanogaster. Chromosome Res. 2008;16:649–673. doi: 10.1007/s10577-008-1218-6. [DOI] [PubMed] [Google Scholar]
- 12.Vazquez J, Müller M, Pirrotta V, Sedat JW. The Mcp element mediates stable long-range chromosome-chromosome interactions in Drosophila. Mol Biol Cell. 2006;17:2158–2165. doi: 10.1091/mbc.E06-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13**.Li H-B, Muller M, Bahechar IA, Kyrchanova O, Ohno K, Georgiev P, Pirrotta V. Insulators, not Polycomb Response Elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol Cell Biol. 2011;31:616–625. doi: 10.1128/MCB.00849-10. [3C, in-vivo imaging and functional interactions were used to show that Fab-7 and Mcp transgenes co-localize with each other and with endogenous partners at low frequency, and this interaction depend on the insulator part, not the PRE part of the transgenes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134:909–919. doi: 10.1242/dev.02779. [DOI] [PubMed] [Google Scholar]
- 16**.Tolhuis B, Blom M, Kerkhoven RM, Pagie L, Teunissen H, Nieuwland M, Simonis M, de Laat W, van Lohuizen M, van Steensel B. Interactions among Polycomb domains are guided by chromosome architecture. PLoS Genet. 2011;7:e1001343. doi: 10.1371/journal.pgen.1001343. [Using a 4C approach, the authors show that PcG target genes preferentially interact with each other, and those interactions are limited to genes on the same chromosome arms.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kassis JA, VanSickle EP, Sensabaugh SM. A fragment of engrailed regulatory DNA can mediate transvection of the white gene in Drosophila. Genetics. 1991;128:751–761. doi: 10.1093/genetics/128.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagstrom K, Müller M, Schedl P. A Polycomb and GAGA dependent silencer adjoins the Fab7 boundary in the Drosophila bithorax complex. Genetics. 1997;146:1365–1380. doi: 10.1093/genetics/146.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller M, Hagstrom K, Gyurkovics H, Pirrotta V, Schedl P. The mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics. 1999;153:1333–1356. doi: 10.1093/genetics/153.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sigrist CJA, Pirrotta V. Chromatin insulator elements block the silencing of a target gene by the Drosophila Polycomb Response Element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics. 1997;147:209–221. doi: 10.1093/genetics/147.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagstrom K, Müller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 22.Gruzdeva N, Kyrchanova O, Parshikov A, Kullyev A, Georgiev P. The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer-promoter communication. Mol Cell Biol. 2005;25:3682–3689. doi: 10.1128/MCB.25.9.3682-3689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- 24.Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. CTCF genomic binding sites in Drosophila and the organisation of the Bithorax Complex. PLoS Genet. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cleard F, Moshkin YM, Karch F, Maeda RK. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat Genet. 2006;38:931–935. doi: 10.1038/ng1833. [DOI] [PubMed] [Google Scholar]
- 26.Smith CT, Wickramasinghe P, Olson A, Loukinov D, Lin L, Deng J, Xiong Y, Rux J, Sachinandam R, Sun H, et al. Genome wide ChIP-chip analyses reveal important roles for CTCF in Drosophila genome organization. Dev Biol. 2009;328:518–528. doi: 10.1016/j.ydbio.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27*.MacPherson MJ, Beatty LG, Zhou W, Du M, Sadowski PD. The CTCF insulator protein is posttranslationally modified by SUMO. Mol Cell Biol. 2009;29:714–725. doi: 10.1128/MCB.00825-08. [The authors show that CTCF is a target of SUMOylation, and this modification contributes to its repressive function. They also show that CTCF bodies co-localize with PcG bodies.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 29.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 30*.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [Active globin genes preferentially associate with other Klf1-regulated genes, on the same or different chromosomes, in a few specialized transcription factories.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31*.Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CWH, Ye C, Ping JLH, Mulawadi F, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [The authors generated the CTCF-chromatin interactome in mouse ESC cells, revealing five distinct CTCF-associated chromatin domains.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32**.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [This report shows that CTCF can mediate interchrosomosome interactions by bringing an enhancer on chromosome 7 to regulate gene expression on chromosome 11, a perfect example of what used to be called “transvection”.] [DOI] [PubMed] [Google Scholar]
- 33.Schwartz YB, Kahn TG, Stenberg P, Ohno K, Bourgon R, Pirrotta V. Alternative epigenetic chromatin states of Polycomb target genes. PLoS Genet. 2010;6:e1000805. doi: 10.1371/journal.pgen.1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poux S, Horard B, Sigrist CJA, Pirrotta V. The Drosophila Trithorax protein is a coactivator required to prevent re-establishment of Polycomb silencing. Development. 2002;129:2483–2493. doi: 10.1242/dev.129.10.2483. [DOI] [PubMed] [Google Scholar]
- 35.Klymenko T, Müller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Reports. 2004;5:373–377. doi: 10.1038/sj.embor.7400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blobel GA, Kadauke S, Wang E, Lau AW, Zuber J, Chou MM, Vakoc CR. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Molecular Cell. 2009;36:970–983. doi: 10.1016/j.molcel.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldsborough AS, Kornberg TB. Reduction of transcription by homologue asynapsis in Drosophila imaginal discs. Nature. 1996;381:807–810. doi: 10.1038/381807a0. [DOI] [PubMed] [Google Scholar]
- 38*.Moshkovich N, Nisha P, Boyle PJ, Thompson BA, Dale RK, Lei EP. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 2011;201125:1686–1701. doi: 10.1101/gad.16651211. [The authors find that AGO2 binds to many genomic sites of CTCF/CP190, physically interacts with CTCF/CP190, and is needed for chromosome looping and insulator function.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lei EP, Corces VG. A long-distance relationship between RNAi and Polycomb. Cell. 2006;124:886–888. doi: 10.1016/j.cell.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 40.Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543–2555. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide Identification of Polycomb-Associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad Sci. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou M-M. Molecular Interplay of the Noncoding RNA ANRIL and Methylated Histone H3 Lysine 27 by Polycomb CBX7 in Transcriptional Silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- 46.Kagey MH, Melhuish TA, Wotton D. The Polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 47.Kagey MH, Melhuish TA, Powers SE, Wotton D. Multiple activities contribute to Pc2 E3 function. EMBO J. 2005;24:108–119. doi: 10.1038/sj.emboj.7600506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, Nakamura T, Canaani E, Croce CM, Mazo A. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science. 2001;294:1331–1334. doi: 10.1126/science.1065683. [DOI] [PubMed] [Google Scholar]
- 49.Bantignies F, Goodman RH, Smolik SM. Functional interaction between the coactivator drosophila CREB-binding protein and ASH1, a member of the trithorax group of chromatin modifiers. Mol Cell Biol. 2000;20:9317–9330. doi: 10.1128/mcb.20.24.9317-9330.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Stoop P, Boutsma EA, Hulsman D, Noback S, Heimerikx M, et al. Ubiquitin E3 ligase Ring1b/Rnf2 of Polycomb Repressive Complex 1 contributes to stable maintenance of mouse embryonic stem cells. PLoS ONE. 2008;3:e2235. doi: 10.1371/journal.pone.0002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anders S. Visualization of genomic data with the Hilbert curve. Bioinformatics. 2009;25:1231–1235. doi: 10.1093/bioinformatics/btp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Melnik S, Deng B, Papantonis A, Baboo S, Carr IM, Cook PR. The proteomes of transcription factories containing RNA polymerases I, II or III. Nat Meth. 2011;8:963–968. doi: 10.1038/nmeth.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]