Abstract
Background
Diabetic patients have an increased risk of developing atherosclerosis and related complications compared to non-diabetic individuals. The increased cardiovascular risk associated with diabetes is due in part to genetic variations that influence both glucose homeostasis and atherosclerotic lesion growth. Mouse strains C57BL/6J (B6) and BALB/cJ (BALB) exhibit distinct differences in fasting plasma glucose and atherosclerotic lesion size when deficient in apolipoprotein E (Apoe−/− . Quantitative trait locus (QTL) analysis was performed to determine genetic factors influencing the two phenotypes.
Methods and Results
266 female F2 mice were generated from an intercross between B6.Apoe−/− and BALB.Apoe−/− mice and fed a Western diet for 12 weeks. Atherosclerotic lesions in the aortic root, fasting plasma glucose, and body weight were measured. 130 microsatellite markers across the entire genome were genotyped. Four significant QTLs, Ath1 on chromosome (Chr) 1, Ath41 on Chr2, Ath42 on Chr5, and Ath29 on Chr9, and one suggestive QTL on Chr4, were identified for atherosclerotic lesion size. Four significant QTLs, Bglu3 and Bglu12 on Chr1, Bglu13 on Chr5, Bglu15 on Chr12, and two suggestive QTLs on Chr9 and Chr15 were identified for fasting glucose levels on the chow diet. Two significant QTLs, Bglu3 and Bglu13, and one suggestive locus on Chr8 were identified for fasting glucose on the Western diet. One significant locus on Chr1 and two suggestive loci on Chr9 and Chr19 were identified for body weight. Ath1 and Ath42 coincided with Bglu3 and Bglu13, respectively, in the confidence interval.
Conclusions
We have identified novel QTLs that have major influences on atherosclerotic lesion size and glucose homeostasis. The colocalization of QTLs for atherosclerosis and diabetes suggests possible genetic connections between the two diseases.
Keywords: Atherosclerosis, type 2 diabetes, quantitative trait locus, hyperglycemia
Introduction
Type 2 diabetes mellitus (T2DM) is a major risk factor for atherosclerotic cardiovascular disease. Diabetic patients have a two- to four-fold higher risk of developing atherosclerosis and its complications compared with non-diabetic individuals 1. Part of the increased cardiovascular risk associated with diabetes is due to genetic variations that influence both glucose homeostasis and the development of atherosclerosis. A few rare gene mutations result in both early coronary heart disease and T2DM that are observable as Mendelian traits segregatingin families, which include LRP62, ABCA1 3,4, and APOB 5. Recent genome-wide association studies (GWAS) have identified dozens of common genetic variants for both atherosclerosis and T2DM (http://www.genome.gov/GWAStudies/). Several of the variants for coronary heart disease are within genes that are involved in glucose metabolism or are associated with T2DM, such as MTHFD1L and HNF1A. Thus far few studies have been conducted to examine such genetic variants and in those that have the findings are inconsistent 6, 7, 8, 9,10. One major challenge for such studies is the difficulties inherent in establishing causality between genetic variants and complex disease in humans due to small gene effects, complex genetic structure, and environmental influences.
A complementary approach to the identification of genetic components in human disease is to use animal models. One commonly usedrodent model of atherosclerosis is the apolipoprotein E-deficient (Apoe−/−) mouse, which develops all phases of atherosclerotic lesions seen in humans, progressing from the early foam cell stage to the advanced stage with a fibrous cap and necrotic lipid core 11. We have observed that atherosclerosis susceptible C57BL/6 (B6) Apoe−/− mice develop significant hyperglycemia when fed a Western-type diet 12. In contrast, atherosclerosis-resistant BALB/cJ (BALB) Apoe−/− mice are highly resistant to hyperglycemia 13. The concordant differences between the two Apoe−/− strains in susceptibility to atherosclerosis and to hyperglycemia provide an ideal model for investigating genetic connections between the phenotypes. In the present study, we performed quantitative trait locus (QTL) analysis on female mice from an intercross between B6. Apoe−/− and BALB. Apoe−/− mouse strains to investigate the genetic control of atherosclerosis and glucose homeostasis.
Methods
Mice
B6.Apoe−/− mice were purchased from the Jackson Laboratories. BALB. Apoe−/− mice at the N10 generation were generated in our laboratory, as previously described 14. B6. Apoe−/− mice were crossed with BALB. Apoe−/− mice to generate F1s, which were intercrossed by brother-sister mating to generate a large F2 population. Female F2 mice were weaned at 3 weeks of age onto a rodent chow diet, and male F2 mice were euthanized at the time of weaning. At 6 weeks of age, F2 mice were started with a Western diet containing 21% fat, 34.1% sucrose, 0.15% cholesterol, and 19.5% casein (Harlan Laboratories, TD 88137) and maintained on the diet for 12 weeks. All this procedures were in accordance with current National Institutes of Health guidelines and approved by the University Animal Care and Use Committees.
Measurements of plasma glucose
Mice were bled twice: once before initiation of the Western diet and once at the end of the 12 weeks’ high-fat feeding period. Mice were fasted overnight before blood was drawn from the retro-orbital venous plexus with the animals under isoflurane anesthesia. Plasma glucose was measured with a Sigma glucose (HK) assay kit, which was adapted for a microplate assay. Briefly, 10 μl of plasma samples (plasma from high-fat diet fed mice was diluted 1:2 in distill water) were mixed with 90 μl of reagent in a 96-well plate. After a 15-min incubation at room temperature, the absorbance at 340 nm was read on a Molecular Devices (Menlo Park, CA) plate reader.
Aortic lesion analysis
Atherosclerotic lesions in aortic root were measured as previously reported 15. Briefly, the aortic root and adjacent heart were excised en bloc and embedded in optimal cutting temperature compound. 10-μm thick cross sections of the vessel were collected, stained with oil red O and hematoxylin, and counterstained with fast green. Atherosclerotic lesion areas were quantified using an ocular lens with a square-micrometer grid on a light microscope. The lesion areas of five sections with the largest readings were averaged for each mouse and this average was used for statistical analysis.
Genotyping
Genomic DNA was isolated from the tails of mice by using the phenol/chloroform extraction and ethanol precipitation method. A total of 130 microsatellite markers covering all 19 autosomes and the X chromosome at an average interval of 12 cM were typed. Parental and F1 DNA served as controls for each marker.
Statistical analysis
QTL analysis was performed using J/qtl and Map Manager QTX software as previously described 12,15, 16. One thousand permutations of trait values were run to define the genome-wide LOD (logarithm of odds) score threshold required to be significant or suggestive for each specific trait. Loci that exceeded the 95th percentile of the permutation distribution were defined as significant (P<0.05) and those exceeding the 37th percentile were suggestive (P<0.63) according to the criteria recommended by the genetics community in 2003 17.
Results
Trait value distributions
Fasting plasma glucose levels of 266 F2 mice before and after 12 weeks on the Western diet, atherosclerotic lesions in the aortic root, and body weight were measured. As shown in figure 1, values of fasting plasma glucose levels on both chow and Western diets, log-transformed atherosclerotic lesion sizes, and body weight approach normal distributions. These data were analyzed to identify chromosomal regions segregating with the traits. Those loci exhibiting significant linkage and suggestive linkage are presented in Table 1.
Figure 1.
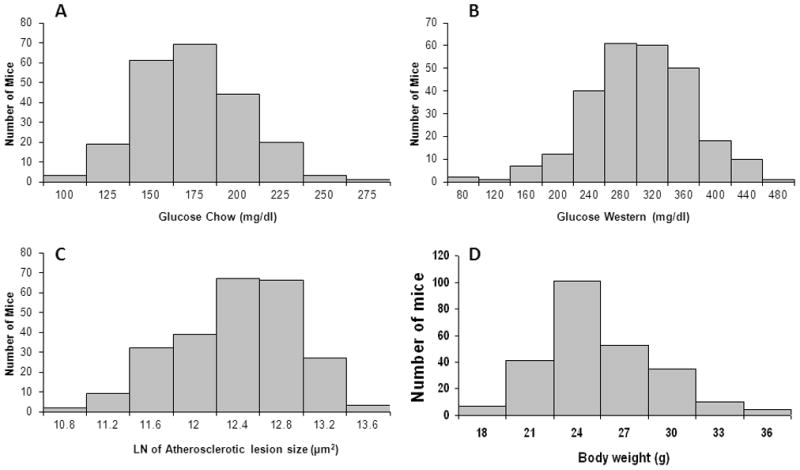
Distributions of fasting plasma glucose before (A) and after (B) 12 weeks on the Western diet, LN (natural log)-transformed atherosclerotic lesion sizes (C), and body weight (D) in 266 female F2 mice derived from B6. Apoe−/− and BALB. Apoe−/− mice.
Table 1.
Significant and suggestive QTLs for atherosclerotic lesion size and fasting plasma glucose levels l in F2 mice derived from B6. Apoe−/− and BALB. Apoe−/− mice.
| Locus namea | Chr | Trait | LOD scoreb | 95% CIc | Peakd | P valuee | High allele | Mode of inheritancef |
|---|---|---|---|---|---|---|---|---|
| Ath1 | 1 | Atherosclerosis | 3.82 | 82.3–92.3 | 89 | 0.019 | B6 | Additive |
| Ath41 | 2 | Atherosclerosis | 3.77 | 26.2–64.2 | 52.2 | 0.026 | B6 | Additive |
| Athsq1 | 4 | Atherosclerosis | 2.8 | 55.6–69.1 | 63.3 | 0.191 | BALB | Additive |
| Ath42 | 5 | Atherosclerosis | 5.69 | 48.5–60.7 | 54.7 | 0.001 | B6 | Dominant |
| Ath29 | 9 | Atherosclerosis | 3.88 | 17.8–31.8 | 19.8 | 0.016 | B6 | Additive |
| Bglu12 | 1 | Glucose, chow | 3.94 | 48.3–78.3 | 60.3 | 0.022 | BALB | Additive |
| Bglu3 | 1 | Glucose, chow | 3.45 | 64.3–84.3 | 74.3 | <0.05 | BALB | Additive |
| Bglu13 | 5 | Glucose, chow | 6.72 | 42.7–56.7 | 47.3 | 0.001 | B6 | Dominant |
| Bglu14 | 9 | Glucose, chow | 3.06 | 17.8–61.8 | 47.8 | 0.11 | BALB | Recessive |
| Bglu15 | 12 | Glucose, chow | 3.43 | 5.5–21.5 | 10.0 | 0.05 | B6 | Additive |
| Fbg-2 | 15 | Glucose, chow | 2.14 | 18.5–33.4 | 26.5 | 0.579 | B6 | Heterosis |
| Bglu3 | 1 | Glucose, West | 4.93 | 76.3–86.3 | 82.3 | 0.001 | BALB | Additive |
| Bgl13 | 5 | Glucose, West | 4.81 | 38.7–58.7 | 48.7 | 0.001 | B6 | Dominant |
| Giq1 | 8 | Glucose, West | 2.90 | 17.7–57.7 | 33.7 | 0.16 | B6 | Additive |
| BW8q1 | 1 | Body weight | 5.42 | 76.3–94.3 | 80.3 | 0.0001 | BALB | Dominant |
| W10q13 | 9 | Body weight | 2.785 | 55.8–69.8 | 67.8 | 0.188 | B6 | Recessive |
| Wtmq9 | 19 | Body weight | 2.898 | 18.3–46.3 | 40.3 | 0.154 | BALB | Dominant |
Plasma glucose levels were measured before (chow) mice were started on the Western diet and at the end of the 12 weeks’ Western diet (West) feeding period. Blood was drawn from overnight-fasted mice.
QTLs were named if they were significant or if they overlapped with previously reported suggestive ones. The nomenclature of Ath was for atherosclerosis QTLs, and Bglu for blood glucose QTLs. The newly identified QTLs were underlined to easily distinguish them from known ones.
LOD scores were obtained from QTL analysis. The significant LOD scores were highlighted in bold. The suggestive and significant LOD score thresholds were determined by 1,000 permutation tests for each trait. Suggestive and significant LOD scores were 2.087 and 3.458, respectively, for atherosclerosis; 2.071 and 3.428 for plasma glucose on the chow diet; 2.084 and 3.423 for glucose on the Western diet, and 2.099 and 3.486 for body weight.
95% Confidence interval in cM obtained from a whole genome QTL scan.
QTL peak position in cM.
The p-values reported represent the level of genome-wide significance as they were generated based on genome-wide permutation tests
Mode of inheritance was defined according to allelic effect at the nearest marker of a QTL.
Atherosclerotic lesion size
Four significant QTLs, located on chromosomes (Chr) 1, 2, 5, and 9, and one suggestive QTL on Chr4, were identified for atherosclerotic lesion sizes (Figure 2). Details of the QTLs detected, including locus name, LOD score, 95% confidence interval (CI), peak location, genome-wide significance P value, high allele, and mode of inheritance are presented in Table 1. The two significant QTLs on Chr1 and Chr9 replicated the previously reported QTLs, Ath1 and Ath29, respectively 15,18. The other two significant QTLs were novel. The Chr2 locus had a significant LOD score of 3.77 and a genome-wide significant P value of 0.026. It peaked at 52.2 cM and did not overlap in the confidence interval with known mouse atherosclerosis QTLs. We named it Ath41 according to the QTL nomenclature for mouse atherosclerotic lesions. The Chr5 locus had a highly significant LOD score of 5.69 and a genome-wide P value of <0.001. Its peak appeared at 54.7 cM. We named it Ath42. The Chr4 locus had a suggestive LOD score of 2.8 and peaked at 63.3 cM. This QTL was partially overlapping with Athsq1, an atherosclerotic lesion locus identified in a (MOLF/Ei x B6.Ldlr−/−) x B6.Ldlr−/− backcross 19. Paradoxically, the BALB allele was associated with increased lesion size while the B6 allele was associated with decreased lesion size (Table 2). In contrast, for the 4 significant QTLs, the B6 allele was the high allele that increased lesion size and the BALB allele was the low one that reduced lesion size. Ath42 affected atherosclerotic lesion size in a dominant mode of inheritance while other QTLs exhibited an additive effect on the trait.
Figure 2.
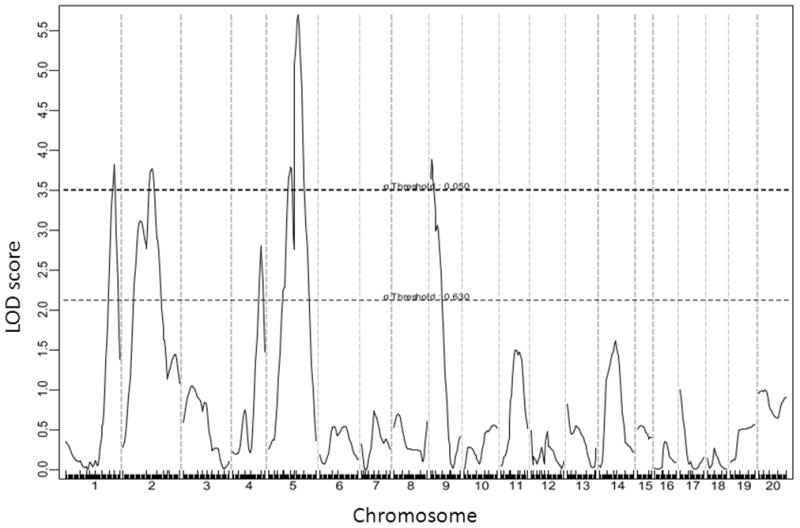
A genome-wide scan to search for loci influencing atherosclerotic lesion sizes of the F2 mice. Chromosomes 1 through 20 are represented numerically on the X-axis. The relative width of the space allotted for each chromosome reflects the relative length of each chromosome. The Y-axis represents the LOD score. Atherosclerotic lesion sizes were determined by averaging the lesion areas of 5 cross-sections with the largest readings for each F2 mouse. Two horizontal dashed lines denote genome-wide empirical thresholds for suggestive (P=0.63) and significant (P=0.05) linkage.
Table 2.
Effects of B6 (B) and BALB (C) alleles in different QTLs on atherosclerosis, plasma lipids, and body weight in the B6. Apoe−/− and BALB. Apoe−/− intercross
| Locus name | Chr | Trait | Peak Marker | BB | CC | BC | P-value |
|---|---|---|---|---|---|---|---|
| Ath1 | 1 | Atherosclerosis | D1mit406 | 249571±123560 (55) | 167588±83413 (64) | 226485±106846 (137) | 5.51E-05 |
| Ath41 | 2 | Atherosclerosis | D2mit328 | 243816±109941 (61) | 186641±116201 (59) | 223779±110268 (135) | 1.76E-02 |
| Athsq1 | 4 | Atherosclerosis | D4mit203 | 189830±108061 (60) | 253744±115878 (72) | 212235±109735 (126) | 3.37E-03 |
| Ath42 | 5 | Atherosclerosis | D5mit41 | 243106±106054 (68) | 165375±99213 (55) | 231499±115933 (127) | 1.68E-04 |
| Ath29 | 9 | Atherosclerosis | D9mit247 | 253591±126554 (68) | 173685±87719 (54) | 217985±109284 (137) | 4.59E-04 |
| Bglu12 | 1 | Glucose, chow | D1mit495 | 149.2 ± 25.8 (60) | 170.7 ± 28.8 (55) | 161.6 ± 29.1 (111) | 2.91E-04 |
| Bglu3 | 1 | Glucose, chow | D1mit354 | 149.7 ± 25.1 (53) | 169.1 ± 29.7 (61) | 160.7 ± 29.7 (112) | 1.78E-3 |
| Bglu13 | 5 | Glucose, chow | D5mit312 | 160.5 ± 28.6 (62) | 141.6 ± 19.5 (49) | 168.4 ± 29.6 (119) | 2.12E-07 |
| Bglu14 | 9 | Glucose, chow | D9mit274 | 159.0 ± 29.9 (60) | 172.8 ± 23.7 (57) | 154.3 ± 28.0 (115) | 2.27E-04 |
| Bglu15 | 12 | Glucose, chow | D12mit84 | 171.7 ± 34.2 (47) | 149.5 ± 26.0 (60) | 160.9 ± 26.8 (125) | 3.41E-04 |
| Fbg-2 | 15 | Glucose, chow | D15mit123 | 166.9 ± 27.4 (45) | 166.7 ± 32.0 (64) | 154.6 ± 27.4 (123) | 6.51E-03 |
| Bglu3 | 1 | Glucose, West | D1mit354 | 252.0 ± 54.2 (57) | 301.5 ± 70.8 (68) | 288.6 ± 61.5 (128) | 4.44E-05 |
|
Bgl13 Giq1 |
5 8 |
Glucose, West Glucose, West |
D5mit155 D8mit68 |
284.3 ± 56.1 (71) 305.0 ± 60.7 (73) |
249.6 ± 71.3 (51) 265.3 ± 54.9 (63) |
299.5 ± 64.0 (135) 283.4 ± 70.3 (126) |
1.74E-05 1.69E-03 |
| BW8q1 | 1 | Body weight | D1mit354 | 22.1 ± 2.3 (57) | 25.0 ± 4.1 (68) | 24.3 ± 3.4 (132) | 5.45E-06 |
| W10q13 | 9 | Body weight | D9mit279 | 25.3 ± 3.5 (62) | 23.2 ± 3.4 (59) | 23.7 ± 3.5 (142) | 2.48E-03 |
| Wtmq9 | 19 | Body weight | D19mit90 | 22.7 ± 3.2 (64) | 24.8 ± 4.1 (69) | 24.2 ± 3.3 (131) | 1.94E-03 |
Measurements are expressed as means ± SD. The units for these measurements are: μm2/section for atherosclerotic lesions; mg/dl for glucose; and g for body weight. BB, homozygous for B6 alleles at the linked peak marker; CC, homozygous for BALB alleles; BC, heterozygous for B6 and BALB alleles at the peak marker. The number in the bracket represents the number of progeny with a specific genotype at a specific marker. ANOVA was used to determine the significance level (p value) of differences for a specific phenotype among progeny with three different genotypes at a specific marker.
Fasting glucose levels
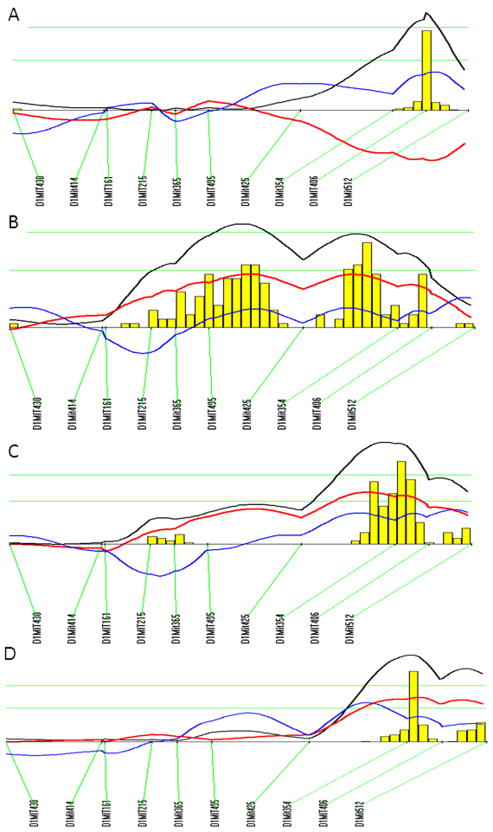
For fasting glucose on the chow diet, 4 significant QTLs on Chr1, Chr5, and Chr12, and two suggestive QTLs on Chr9 and Chr15 were identified (Figure 3). The significant locus on distal Chr1 and the 2 suggestive loci on Chr9 and Chr15 replicated the previously reported QTLs, Bglu3, a suggestive locus on Chr9 (now named Bglu14), and Fbg2, respectively (Table 1). The significant QTLs on the middle portion of Chr1, Chr5, and Chr12 were novel. The LOD score plot for Chr1 displayed two distinct peaks, located approximately 14 cM apart (Figure 3). The distal QTL peaked at 74.3 CM, overlapping with Bglu3, identified in a B6 x C3H Apoe−/− F2 cross 12. The proximal peak occurred at 60.3 cM with a LOD score reaching 3.94. We named this QTL Bglu12 to represent a significant mouse QTL for fasting glucose. The Chr5 locus had a highly significant LOD score of 6.72 and peaked at 47.3 cM. We named it Bglu13. The QTL on Chr9 had a suggestive LOD score of 3.06, and overlapped with a suggestive locus near D9mit229 (26.8 cM) for fasting glucose levels identified in a B6 x KK-Ay F2 cross 20. We designated this QTL as Bglu14 as it has not been named. The QTL on Chr12, named Bglu15, had a significant LOD score of 3.43 and peaked at 10 cM. Bglu15 is close to the centromere compared to Fbg-1, which is located in the middle portion of Chr12 between D12mit4 (35.5 cM) and D12mit227 (38.4 cM) 21. The suggestive QTL on Chr15 overlapped with Fbg-2 near D15Mit87 (16.7 cM), identified in (BALB x KK/Ta) x KK/Ta backcross 21. For fasting glucose on the Western diet, 2 significant QTLs on Chr1 and Chr5 and 1 suggestive QTL on Chr8 were identified (Figure 4). The Chr1 QTL replicated Bglu3, and the Chr5 replicated Bglu13. The Chr8 locus overlapped with Giq1, a locus with a strong influence on the late phase of glucose tolerance test identified in a B6×KK-A cross 22. Bglu13 affected fasting glucose levels on both chow and Western diets in a dominant mode of inheritance while all other QTLs exhibited an additive effect on the trait except for the QTL on Chr9 and Chr15 that affected glucose levels in a recessive and a heterosis mode, respectively (Table 2).
Figure 3.
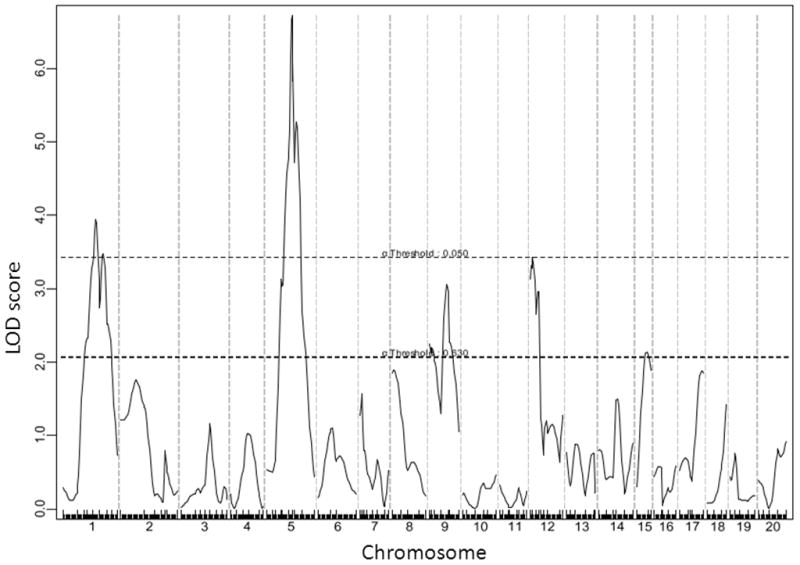
A genome-wide scan to search for loci for fasting plasma glucose levels when the F2 mice were fed the chow diet. Blood was collected from overnight fasted F2 mice the day before being started on the Western diet.
Figure 4.
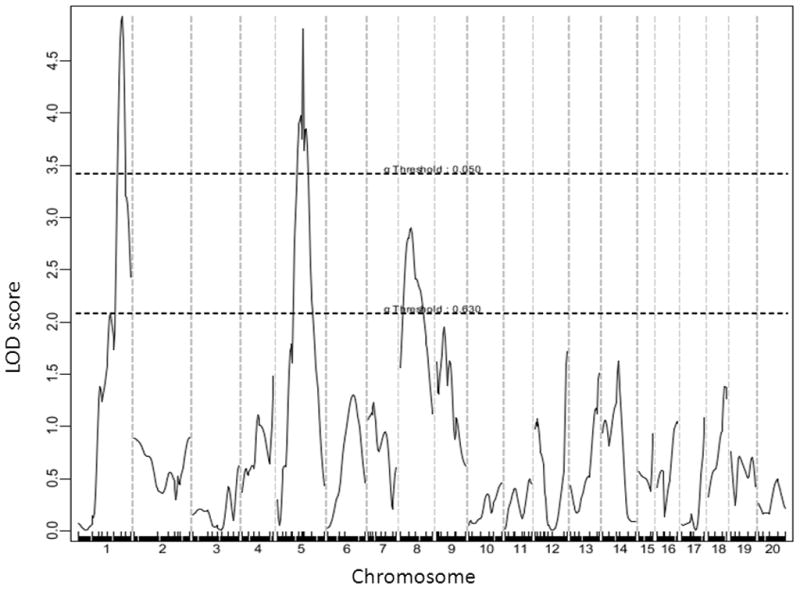
A genome-wide scan to search for loci for fasting plasma glucose levels on the Western diet. Mice were fed the high-fat diet for 12 weeks. Blood was collected from overnight fasted F2 mice the day before being euthanized.
Body weight
One significant QTL on distal Chr1 and 2 suggestive QTLs on Chr9 and Chr19 were identified for body weight (Figure 5). The Chr1 QTL overlaps with Wt3q2 and Wt6q2 for body weight mapped in two F2 populations created from a selection and an inbred mouse lines 23, Bw8q1 originally mapped in a B6×A/J intercross 24 and then replicated in a BxH Apoe−/− F2 cross12, and Nob3 for body fat body weight and blood glucose mapped in NZO×B6 F2 females 25. The Chr9 locus overlaps with W10q13 mapped in an M16i x L6 intercross 26, Do2 for dietary obese mapped in two crosses derived from AKR/J and SWR/J mice 27, and Obq18 for obesity mapped in B6 x 129 F2 females 28. The Chr19 locus corresponds to Wtmq9 mapped in a B6 x C3H Apoe−/− intercross 29, W3q14 from an M16i x L6 F2 intercross 26, and Abfw4 for abdominal fat mapped in DU6i x DBA/2 intercross 30.
Figure 5.
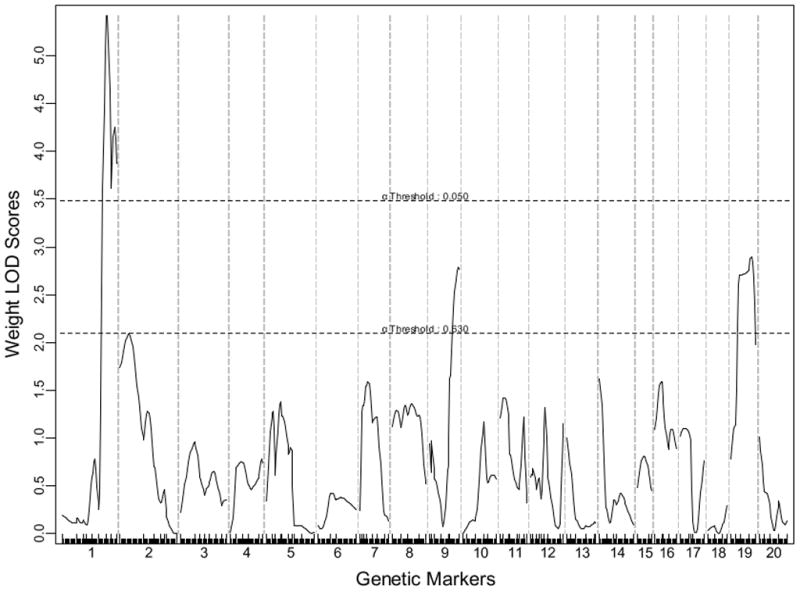
A genome-wide scan to search for loci for body weight. Mice were weighed the day being euthanized.
Coincident QTLs for atherosclerosis and fasting glucose
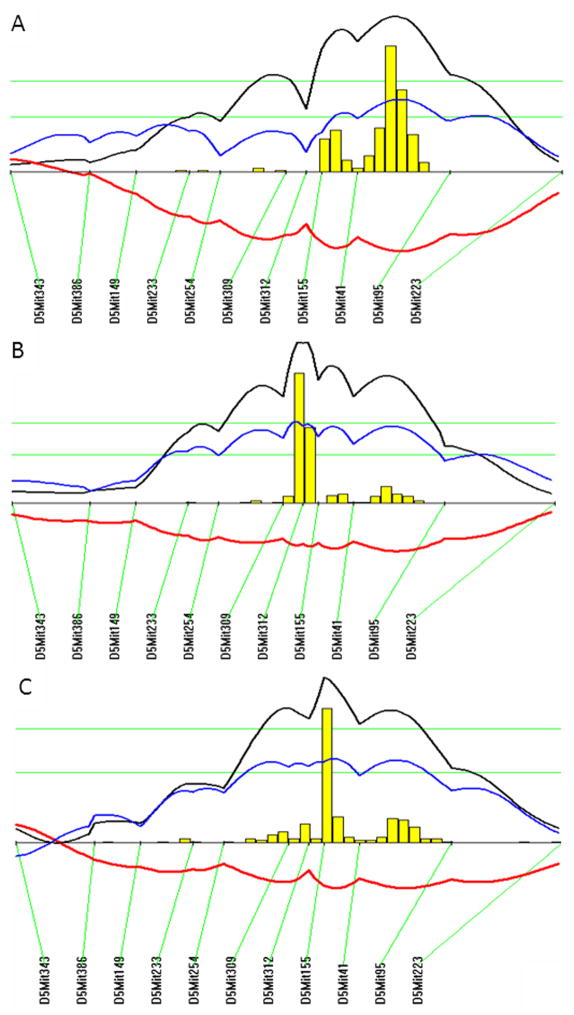
LOD score plots for chromosome 5 show that the QTLs for atherosclerosis (Ath42) coincided with the QTL for fasting glucose (Bglu13) in the confidence interval (Figure 6). Both loci exhibited a dominant effect from the B6 allele on atherosclerotic lesions or fasting glucose levels (Table 2). LOD score plots for chromosome 1 show partial overlapping of the QTL for atherosclerosis (Ath1) with the QTLs for fasting glucose (Bglu3) and body weight in the confidence interval (Figure 7). The B6 allele was associated with increased atherosclerotic lesions and decreased glucose levels and body weight while the BALB allele was associated with decreased lesions and increased glucose levels and body weight (Table 2). Ath29 was also partially overlapping with Bglu14 in the confidence interval on the Chr9 (Table 1).
Figure 6.
LOD score plots for atherosclerotic lesion size (A), fasting plasma glucose levels on the chow diet (B), and fasting plasma glucose levels on the Western diet (C) on chromosome 5. Plots were created with the interval mapping function of Map Manager QTX, including a bootstrap test shown as a histogram estimating the confidence interval for the QTL. Two green horizontal lines represent genome-wide significance thresholds for suggestive or significant peaks (P=0.63 and P=0.05, respectively). Black plots reflect the LOD calculated at 1-cM intervals. The blue plot represents the effect of the B6 allele, and the red plot represents the effect of the BALB allele.
Figure 7.
LOD score plots for atherosclerotic lesion size (A), fasting plasma glucose levels on the chow diet (B), fasting plasma glucose levels on the Western diet (C), and body weight (D) on chromosome 1. Plots were created with the interval mapping function of Map Manager QTXb20, as stated above. The histogram denotes the confidence interval of the QTL.
Discussion
In this study, we have identified five loci contributing to the development of atherosclerosis, six loci to fasting glucose levels on the chow diet, three loci contributing to fasting glucose levels on the Western diet, and three loci for body weight in an intercross between B6 and BALB Apoe−/− mouse strains. Moreover, we have observed the colocalization of QTLs for atherosclerotic lesions and for plasma glucose levels on chromosomes 1, 5, and 9.
B6 and BALB are prototype mouse strains for genetic studies of atherosclerosis. In pioneering studies of recombinant inbred strains derived from the two strains, as well as from B6 and C3H/HeJ, Paigen et al 18 identified the first atherosclerosis susceptibility locus, Ath-1. Subsequent studies of female F2 and N2 progeny derived from the B6 and BALB strains demonstrates the segregation of Ath-1 with HDL cholesterol levels 31. However, there are several limitations in those studies: First, the number rather than the size of atherosclerotic lesions was measured. Thus, atherosclerotic lesions were treated as a “qualitative trait” rather than a “quantitative trait”. Second, the mapping was performed using a rather small numbers of animals, especially the recombinant inbred strains, thus the power for detecting susceptibility loci was low. Third, there were fewer polymorphic markers available at the time when the studies were conducted. Lastly, the diet-induced mouse model of atherosclerosis develops only small fatty streak lesions that are largely limited to the aortic root 32. In contrast, Apoe-deficient mice develop all phases of atherosclerotic lesions in large and medium sized arteries seen in humans 11. Our present work has extended the prior studies by finding five atherosclerosis QTLs, including Ath1. Among the five QTLs, Ath1, Athsq1, and Ath29, have been previously reported 15,18,33. Tnfsf4 has been identified to be the causal gene of Ath1 34. We recently have identified Rcn2, a calcium-binding protein in the endoplasmic reticulum, as a key regulator in oxidized phospholipid-induced cytokine production and a probable candidate gene of Ath29 35. The confidence interval of Athsq1 is corresponding to human chromosome 9p21, a region that is associated with coronary heart disease 36, 37,38.
The QTL on chromosome 2, named Ath41, is close to Athla1, an atherosclerosis susceptibility locus mapped in a (PERA×B6-Ldlr−/−)×B6-Ldlr−/− N2 backcross mice 39. Athla1 is located in a more distal region (69 cM), and it increases lesion size only when homozygous for the B6 allele. Candidate genes for Ath41 include Dab2ip, Tfpi, and Slc38a11, which have been shown to be associated with coronary heart disease in humans 40,41,42.
We identified a major locus on chromosome 5, approximately between 40 and 60 cM, which affected both atherosclerotic lesion size and fasting plasma glucose levels. We named it Ath42 for atherosclerotic lesions and Bglu13 for fasting glucose. As the two loci overlap significantly in the confidence interval, it is plausible to postulate that they share the same underlying causal gene. The present observation that both QTLs exhibited the same dominant B6 allele effect on the two different traits supports this speculation. Nevertheless, it is also likely that the two phenotypes are affected by two linked but unique genes residing in the QTL interval. The region from 40 to 60 cM on chromosome 5 in the mouse corresponds to chromosomal regions of 4q13, 4q21, and 12q24 in humans. The 4q13 region has been shown to be associated with variations in metabolic traits, including blood glucose 43,44, and the 12q24 region is associated with coronary heart disease 45,46, metabolic syndrome 47,48, type 1 and type 2 diabetes 49,50,51. One promising candidate gene in the region is Hnf1a, which encodes hepatocyte nuclear factor 1α. One A/G SNP in exon 9 between B6 and BALB leads to amino acid substitution (P580R) in the Hnf1a protein. In humans, Hnf1a mutations are the most common cause of maturity-onset diabetes of the young (MODY) 52. Polymorphisms in the Hnf1a gene are associated with risk for T2DM and coronary heart disease 49, 53.
The QTLs for plasma glucose and body weight on distal chromosome 1 have been reported previously in three separate mouse intercrosses, including two B6×C3H crosses deficient in Apoe 12, 29, and a cross between New Zealand obese (NZO) and B6 mice 25. The confidence interval of the QTLs overlaps with a region of linkage to type 2 diabetes found in multiple human populations that has been extensively examined by the International Chromosome 1q Type 2 Diabetes Consortium 54. In the current cross, we have observed two distinct peaks of the linkage curve for plasma glucose on the chow diet with the distal peak at 74.3 cM and the proximal peak at 60.3 cM. The bootstrap test, an effective statistical method for defining the confidence interval of QTLs using simulation 55, also indicated the existence of two QTLs for the trait on chromosome 1. We have named the proximal QTL Bglu12 to represent a new locus for fasting glucose in the mouse.
The QTLs for atherosclerosis (Ath1), fasting glucose (Bglu3), and body weight (BW8q1) overlap in the confidence interval on the distal chromosome 1 region. The B6 allele was associated with increased atherosclerosis but decreased glucose levels and body weight. Apoa2 is a major gene in the region that may contribute to variations in the traits. The QTL effect on body weight disappeared when the influence from the Apoa2 allele was eliminated 56. On the other hand, transgene expression of Apoa2 in mice results in several phenotypes observed in T2DM, including glucose intolerance, insulin resistance, hypertriglyceridemia, and obesity 57,58. Apoa2 is also a major gene in the mouse that has a dramatic influence on plasma HDL cholesterol levels 59. BALB mice have an Apoa2b allele that elevates HDL cholesterol levels and B6 mice have an Apoa2a allele that decreases HDL cholesterol levels 60. High HDL cholesterol levels protect against atherosclerosis. Apcs, encoding serum amyloid P (SAP), is another candidate in the distal chromosome 1 region that may contribute to T2DM and atherosclerosis. Plasma SAP levels, which are primarily regulated by the Apcs gene, are correlated with blood glucose and body weight in a segregating F2 population derived from B6 and C3H Apoe−/− mice12. In humans SAP is significantly correlated with obesity, blood pressure, lipids, common and internal carotid wall thickness, and ankle-brachial index 61. Cxcr4, Pask, Cntnap5a, Lct, and Pik3c2b are positional candidate genes for Bglu12. Variants of these genes have been found to be associated with susceptibility to T2DM, fasting glucose, or insulin resistance in humans 62, 63,64, 65, 66, 67.
The QTL for fasting glucose on chromosome 9 was partially overlapping with the QTL for atherosclerosis. Sorl1, Rcn2, and Apoc3 are potential candidate genes in the region that may affect both atherosclerosis and T2DM 35, 68, 69. For Bglu15, Adam17, encoding a disintegrin and metallopeptidase domain 17, and Ahr, encoding aryl-hydrocarbon receptor, are two likely candidate genes. Adam17 is involved in the shedding of the extracellular domain of cytokines, growth factors, receptors or adhesion molecules 70. Ahr signaling affects molecular clock genes associated with glucose metabolism, and Ahr deficiency enhances insulin sensitivity and reduces PPAR-α pathway activity 71. In the present study, we have found some QTLs, such as Bglu3 and Bglu13, that influenced glucose levels when mice had normoglycemia on a chow diet also affected glucose levels when mice developed hyperglycemia on a high-fat diet. However, some other QTLs, such as Bglu12, Bglu14, and Bglu15 only exerted effect under a specific condition (the chow diet). Six QTLs were found for fasting plasma glucose when mice were fed the chow diet while only three QTLs were detected when fed the Western diet. The reasons for the discrepancy in the results are unknown. One probable explanation is that the Western diet has a significant influence on plasma glucose levels, which might overwhelm the influence from genetic factors on the trait. In addition, the western diet induces some metabolic changes that suppress gene expression. We recently have found that there are much more differentially expressed genes in the aorta of two strains when fed a chow diet than a Western diet 72.
In summary, we have identified multiple QTLs contributing to the development of atherosclerosis and glucose homeostasis in a segregating F2 population. The finding on the colocalization of QTLs for atherosclerosis and glucose has laid the basis for further study to determine whether they are controlled by the same genes or different unique genes in the QTL intervals.
Acknowledgments
Funding Sources: This work was supported by NIH grant HL82881.
Footnotes
Conflict of Interest Disclosures: None.
References
- 1.Bloomgarden ZT. Cardiovascular disease in diabetes. Diabetes Care. 2008;31:1260–1266. doi: 10.2337/dc08-zb06. [DOI] [PubMed] [Google Scholar]
- 2.Mani A, Radhakrishnan J, Wang H, Mani A, Mani MA, Nelson-Williams C, et al. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278–1282. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodzioch M, Orso E, Klucken J, Langmann T, Bottcher A, Diederich W, et al. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat Genet. 1999;22:347–351. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- 4.Saleheen D, Nazir A, Khanum S, Haider SR, Frossard PM. R1615P: a novel mutation in ABCA1 associated with low levels of HDL and type II diabetes mellitus. Int J Cardiol. 2006;110:259–260. doi: 10.1016/j.ijcard.2005.06.059. [DOI] [PubMed] [Google Scholar]
- 5.Pulai JI, Latour MA, Kwok PY, Schonfeld G. Diabetes mellitus in a new kindred with familial hypobetalipoproteinemia and an apolipoprotein B truncation (apoB-55) Atherosclerosis. 1998;136:289–295. doi: 10.1016/s0021-9150(97)00222-0. [DOI] [PubMed] [Google Scholar]
- 6.Pfister R, Barnes D, Luben RN, Khaw KT, Wareham NJ, Langenberg C. Individual and cumulative effect of type 2 diabetes genetic susceptibility variants on risk of coronary heart disease. Diabetologia. 2011;54:2283–2287. doi: 10.1007/s00125-011-2206-5. [DOI] [PubMed] [Google Scholar]
- 7.Ridker PM, Cook NR, Cheng S, Erlich HA, Lindpaintner K, Plutzky J, et al. Alanine for proline substitution in the peroxisome proliferator-activated receptor gamma-2 (PPARG2) gene and the risk of incident myocardial infarction. Arterioscler Thromb Vasc Biol. 2003;23:859–863. doi: 10.1161/01.ATV.0000068680.19521.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paynter NP, Chasman DI, Pare G, Buring JE, Cook NR, Miletich JP, et al. Association between a literature-based genetic risk score and cardiovascular events in women. JAMA. 2010;303:631–637. doi: 10.1001/jama.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bielinski SJ, Pankow JS, Folsom AR, North KE, Boerwinkle E. TCF7L2 single nucleotide polymorphisms, cardiovascular disease and all-cause mortality: the Atherosclerosis Risk in Communities (ARIC) study. Diabetologia. 2008;51:968–970. doi: 10.1007/s00125-008-1004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarzani R, Salvi F, Bordicchia M, Guerra F, Battistoni I, Pagliariccio G, et al. Carotid artery atherosclerosis in hypertensive patients with a functional LDL receptor-related protein 6 gene variant. Nutr Metab Cardiovasc Dis. 2011;21:150–156. doi: 10.1016/j.numecd.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Nakashima Y, Plump AS, Raines EW, Breslow JL, Ross R. ApoE-deficient mice develop lesions of all phases of atherosclerosis throughout the arterial tree. Arterioscler Thromb. 1994;14:133–140. doi: 10.1161/01.atv.14.1.133. [DOI] [PubMed] [Google Scholar]
- 12.Su Z, Li Y, James JC, Matsumoto AH, Helm GA, Lusis AJ, et al. Genetic linkage of hyperglycemia, body weight and serum amyloid-P in an intercross between C57BL/6 and C3H apolipoprotein E-deficient mice. Hum Mol Genet. 2006;15:1650–1658. doi: 10.1093/hmg/ddl088. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Wang Q, Chai W, Chen MH, Liu Z, Shi W. Hyperglycemia in apolipoprotein E-deficient mouse strains with different atherosclerosis susceptibility. Cardiovasc Diabetol. 2011;10:117. doi: 10.1186/1475-2840-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian J, Pei H, James JC, Li Y, Matsumoto AH, Helm GA, et al. Circulating adhesion molecules in apoE-deficient mouse strains with different atherosclerosis susceptibility. Biochem Biophys Res Commun. 2005;329:1102–1107. doi: 10.1016/j.bbrc.2005.02.090. [DOI] [PubMed] [Google Scholar]
- 15.Su Z, Li Y, James JC, McDuffie M, Matsumoto AH, Helm GA, et al. Quantitative trait locus analysis of atherosclerosis in an intercross between C57BL/6 and C3H mice carrying the mutant apolipoprotein E gene. Genetics. 2006;172:1799–1807. doi: 10.1534/genetics.105.051912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan Z, Pei H, Roberts DJ, Zhang Z, Rowlan JS, Matsumoto AH, et al. Quantitative trait locus analysis of neointimal formation in an intercross between C57BL/6 and C3H/HeJ apolipoprotein E-deficient mice. Circ Cardiovasc Genet. 2009;2:220–228. doi: 10.1161/CIRCGENETICS.108.792499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abiola O, Angel JM, Avner P, Bachmanov AA, Belknap JK, Bennett B, et al. The nature and identification of quantitative trait loci: a community's view. Nat Rev Genet. 2003;4:911–916. doi: 10.1038/nrg1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paigen B, Mitchell D, Reue K, Morrow A, Lusis AJ, LeBoeuf RC. Ath-1, a gene determining atherosclerosis susceptibility and high density lipoprotein levels in mice. Proc Natl Acad Sci U S A. 1987;84:3763–3767. doi: 10.1073/pnas.84.11.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welch CL, Bretschger S, Latib N, Bezouevski M, Guo Y, Pleskac N, et al. Localization of atherosclerosis susceptibility loci to chromosomes 4 and 6 using the Ldlr knockout mouse model. Proc Natl Acad Sci U S A. 2001;98:7946–7951. doi: 10.1073/pnas.141239098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suto J, Matsuura S, Imamura K, Yamanaka H, Sekikawa K. Genetic analysis of non-insulin-dependent diabetes mellitus in KK and KK-Ay mice. Eur J Endocrinol. 1998;139:654–661. doi: 10.1530/eje.0.1390654. [DOI] [PubMed] [Google Scholar]
- 21.Shike T, Hirose S, Kobayashi M, Funabiki K, Shirai T, Tomino Y. Susceptibility and negative epistatic loci contributing to type 2 diabetes and related phenotypes in a KK/Ta mouse model. Diabetes. 2001;50:1943–1948. doi: 10.2337/diabetes.50.8.1943. [DOI] [PubMed] [Google Scholar]
- 22.Suto J, Sekikawa K. A quantitative trait locus that accounts for glucose intolerance maps to chromosome 8 in hereditary obese KK-A(y) mice. Int J Obes Relat Metab Disord. 2002;26:1517–1519. doi: 10.1038/sj.ijo.0802152. [DOI] [PubMed] [Google Scholar]
- 23.Moody DE, Pomp D, Nielsen MK, Van Vleck LD. Identification of quantitative trait loci influencing traits related to energy balance in selection and inbred lines of mice. Genetics. 1999;152:699–711. doi: 10.1093/genetics/152.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang S, Gershenfeld HK. Genetic contributions to body weight in mice: relationship of exploratory behavior to weight. Obes Res. 2003;11:828–838. doi: 10.1038/oby.2003.114. [DOI] [PubMed] [Google Scholar]
- 25.Vogel H, Nestler M, Ruschendorf F, Block MD, Tischer S, Kluge R, et al. Characterization of Nob3, a major quantitative trait locus for obesity and hyperglycemia on mouse chromosome 1. Physiol Genomics. 2009;38:226–232. doi: 10.1152/physiolgenomics.00011.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rocha JL, Eisen EJ, Van Vleck LD, Pomp D. A large-sample QTL study in mice: I. Growth. Mamm Genome. 2004;15:83–99. doi: 10.1007/s00335-003-2312-x. [DOI] [PubMed] [Google Scholar]
- 27.West DB, Goudey-Lefevre J, York B, Truett GE. Dietary obesity linked to genetic loci on chromosomes 9 and 15 in a polygenic mouse model. J Clin Invest. 1994;94:1410–1416. doi: 10.1172/JCI117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishimori N, Li R, Kelmenson PM, Korstanje R, Walsh KA, Churchill GA, et al. Quantitative trait loci that determine plasma lipids and obesity in C57BL/6J and 129S1/SvImJ inbred mice. J Lipid Res. 2004;45:1624–1632. doi: 10.1194/jlr.M400098-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Ghazalpour A, Doss S, Zhang B, Wang S, Plaisier C, Castellanos R, et al. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet. 2006;2:e130. doi: 10.1371/journal.pgen.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlborg O, Brockmann GA, Haley CS. Simultaneous mapping of epistatic QTL in DU6i x DBA/2 mice. Mamm Genome. 2005;16:481–494. doi: 10.1007/s00335-004-2425-4. [DOI] [PubMed] [Google Scholar]
- 31.Paigen B, Mitchell D, Holmes PA, Albee D. Genetic analysis of strains C57BL/6J and BALB/cJ for Ath-1, a gene determining atherosclerosis susceptibility in mice. Biochem Genet. 1987;25:881–892. doi: 10.1007/BF00502607. [DOI] [PubMed] [Google Scholar]
- 32.Paigen B, Morrow A, Holmes PA, Mitchell D, Williams RA. Quantitative assessment of atherosclerotic lesions in mice. Atherosclerosis. 1987;68:231–240. doi: 10.1016/0021-9150(87)90202-4. [DOI] [PubMed] [Google Scholar]
- 33.Seidelmann SB, Kuo C, Pleskac N, Molina J, Sayers S, Li R, et al. Athsq1 is an atherosclerosis modifier locus with dramatic effects on lesion area and prominent accumulation of versican. Arterioscler Thromb Vasc Biol. 2008;28:2180–2186. doi: 10.1161/ATVBAHA.108.176800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Ria M, Kelmenson PM, Eriksson P, Higgins DC, Samnegard A, et al. Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nat Genet. 2005;37:365–372. doi: 10.1038/ng1524. [DOI] [PubMed] [Google Scholar]
- 35.Manichaikul A, Wang Q, Shi YL, Zhang Z, Leitinger N, Shi W. Characterization of Ath29, a major mouse atherosclerosis susceptibility locus, and identification of Rcn2 as a novel regulator of cytokine expression. Am J Physiol Heart Circ Physiol. 2011;301:H1056–61. doi: 10.1152/ajpheart.00366.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wahlstrand B, Orho-Melander M, Delling L, Kjeldsen S, Narkiewicz K, Almgren P, et al. The myocardial infarction associated CDKN2A/CDKN2B locus on chromosome 9p21 is associated with stroke independently of coronary events in patients with hypertension. J Hypertens. 2009;27:769–773. doi: 10.1097/HJH.0b013e328326f7eb. [DOI] [PubMed] [Google Scholar]
- 38.Helgadottir A, Thorleifsson G, Magnusson KP, Gretarsdottir S, Steinthorsdottir V, Manolescu A, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 39.Seidelmann SB, De Luca C, Leibel RL, Breslow JL, Tall AR, Welch CL. Quantitative trait locus mapping of genetic modifiers of metabolic syndrome and atherosclerosis in low-density lipoprotein receptor-deficient mice: identification of a locus for metabolic syndrome and increased atherosclerosis on chromosome 4. Arterioscler Thromb Vasc Biol. 2005;25:204–210. doi: 10.1161/01.ATV.0000149146.32385.1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrison SC, Cooper JA, Li K, Talmud PJ, Sofat R, Stephens JW, et al. Association of a sequence variant in DAB2IP with coronary heart disease. Eur Heart J. 2011 doi: 10.1093/eurheartj/ehr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 42.Lettre G, Palmer CD, Young T, Ejebe KG, Allayee H, Benjamin EJ, et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet. 2011;7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabatti C, Service SK, Hartikainen AL, Pouta A, Ripatti S, Brodsky J, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gieger C, Geistlinger L, Altmaier E, Hrabe de Angelis M, Kronenberg F, Meitinger T, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Erdmann J, Grosshennig A, Braund PS, Konig IR, Hengstenberg C, Hall AS, et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zabaneh D, Balding DJ. A genome-wide association study of the metabolic syndrome in Indian Asian men. PLoS One. 2010;5:e11961. doi: 10.1371/journal.pone.0011961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Illig T, Gieger C, Zhai G, Romisch-Margl W, Wang-Sattler R, Prehn C, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42:137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shepherd M, Ellis I, Ahmad AM, Todd PJ, Bowen-Jones D, Mannion G, et al. Predictive genetic testing in maturity-onset diabetes of the young (MODY) Diabet Med. 2001;18:417–421. doi: 10.1046/j.1464-5491.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 53.Reiner AP, Gross MD, Carlson CS, Bielinski SJ, Lange LA, Fornage M, et al. Common coding variants of the HNF1A gene are associated with multiple cardiovascular risk phenotypes in community-based samples of younger and older European-American adults: the Coronary Artery Risk Development in Young Adults Study and The Cardiovascular Health Study. Circ Cardiovasc Genet. 2009;2:244–254. doi: 10.1161/CIRCGENETICS.108.839506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das SK, Elbein SC. The search for type 2 diabetes susceptibility loci: the chromosome 1q story. Curr Diab Rep. 2007;7:154–164. doi: 10.1007/s11892-007-0025-3. [DOI] [PubMed] [Google Scholar]
- 55.Visscher PM, Thompson R, Haley CS. Confidence intervals in QTL mapping by bootstrapping. Genetics. 1996;143:1013–1020. doi: 10.1093/genetics/143.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suto J. Quantitative trait locus analysis of plasma cholesterol levels and body weight by controlling the effects of the Apoa2 allele in mice. J Vet Med Sci. 2007;69:385–392. doi: 10.1292/jvms.69.385. [DOI] [PubMed] [Google Scholar]
- 57.Castellani LW, Goto AM, Lusis AJ. Studies with apolipoprotein A-II transgenic mice indicate a role for HDLs in adiposity and insulin resistance. Diabetes. 2001;50:643–651. doi: 10.2337/diabetes.50.3.643. [DOI] [PubMed] [Google Scholar]
- 58.Castellani LW, Gargalovic P, Febbraio M, Charugundla S, Jien ML, Lusis AJ. Mechanisms mediating insulin resistance in transgenic mice overexpressing mouse apolipoprotein A-II. J Lipid Res. 2004;45:2377–2387. doi: 10.1194/jlr.M400345-JLR200. [DOI] [PubMed] [Google Scholar]
- 59.Welch CL, Bretschger S, Wen PZ, Mehrabian M, Latib N, Fruchart-Najib J, et al. Novel QTLs for HDL levels identified in mice by controlling for Apoa2 allelic effects: confirmation of a chromosome 6 locus in a congenic strain. Physiol Genomics. 2004;17:48–59. doi: 10.1152/physiolgenomics.00124.2003. [DOI] [PubMed] [Google Scholar]
- 60.Wang X, Korstanje R, Higgins D, Paigen B. Haplotype analysis in multiple crosses to identify a QTL gene. Genome Res. 2004;14:1767–1772. doi: 10.1101/gr.2668204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jenny NS, Arnold AM, Kuller LH, Tracy RP, Psaty BM. Serum amyloid P and cardiovascular disease in older men and women: results from the Cardiovascular Health Study. Arterioscler Thromb Vasc Biol. 2007;27:352–358. doi: 10.1161/01.ATV.0000254150.97741.fe. [DOI] [PubMed] [Google Scholar]
- 62.Gupta V, Vinay DG, Rafiq S, Kranthikumar MV, Janipalli CS, Giambartolomei C, et al. for the Indian Migration Study Group. Association analysis of 31 common polymorphisms with type 2 diabetes and its related traits in Indian sib pairs. Diabetologia. 2011 doi: 10.1007/s00125-011-2355-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.da Silva Xavier G, Farhan H, Kim H, Caxaria S, Johnson P, Hughes S, et al. Per-arnt-sim (PAS) domain-containing protein kinase is downregulated in human islets in type 2 diabetes and regulates glucagon secretion. Diabetologia. 2011;54:819–827. doi: 10.1007/s00125-010-2010-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berisha SZ, Serre D, Schauer P, Kashyap SR, Smith JD. Changes in whole blood gene expression in obese subjects with type 2 diabetes following bariatric surgery: a pilot study. PLoS One. 2011;6:e16729. doi: 10.1371/journal.pone.0016729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Enattah NS, Forsblom C, Rasinpera H, Tuomi T, Groop PH, Jarvela I FinnDiane Study Group. The genetic variant of lactase persistence C (-13910) T as a risk factor for type I and II diabetes in the Finnish population. Eur J Clin Nutr. 2004;58:1319–1322. doi: 10.1038/sj.ejcn.1601971. [DOI] [PubMed] [Google Scholar]
- 66.Arkan MC, Hevener AL, Greten FR, Maeda S, Li ZW, Long JM, et al. IKK-beta links inflammation to obesity-induced insulin resistance. Nat Med. 2005;11:191–198. doi: 10.1038/nm1185. [DOI] [PubMed] [Google Scholar]
- 67.Le Stunff C, Dechartres A, Mariot V, Lotton C, Trainor C, Miraglia Del Giudice E, et al. Association analysis indicates that a variant GATA-binding site in the PIK3CB promoter is a Cis-acting expression quantitative trait locus for this gene and attenuates insulin resistance in obese children. Diabetes. 2008;57:494–502. doi: 10.2337/db07-1273. [DOI] [PubMed] [Google Scholar]
- 68.Lane RF, Raines SM, Steele JW, Ehrlich ME, Lah JA, Small SA, et al. Diabetes-associated SorCS1 regulates Alzheimer's amyloid-beta metabolism: evidence for involvement of SorL1 and the retromer complex. J Neurosci. 2010;30:13110–13115. doi: 10.1523/JNEUROSCI.3872-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Onat A, Erginel-Unaltuna N, Coban N, Cicek G, Yuksel H. APOC3 -482C>T polymorphism, circulating apolipoprotein C-III and smoking: interrelation and roles in predicting type-2 diabetes and coronary disease. Clin Biochem. 2011;44:391–396. doi: 10.1016/j.clinbiochem.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 70.Peiretti F, Canault M, Morange P, Alessi MC, Nalbone G. The two sides of ADAM17 in inflammation: implications in atherosclerosis and obesity. Med Sci (Paris) 2009;25:45–50. doi: 10.1051/medsci/200925145. [DOI] [PubMed] [Google Scholar]
- 71.Wang C, Xu CX, Krager SL, Bottum KM, Liao DF, Tischkau SA. Aryl Hydrocarbon Receptor Deficiency Enhances Insulin Sensitivity and Reduces PPAR-alpha Pathway Activity in Mice. Environ Health Perspect. 2011;119:1739–1744. doi: 10.1289/ehp.1103593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuan Z, Miyoshi T, Bao Y, Sheehan JP, Matsumoto AH, Shi W. Microarray analysis of gene expression in mouse aorta reveals role of the calcium signaling pathway in control of atherosclerosis susceptibility. Am J Physiol Heart Circ Physiol. 2009;296:H1336–43. doi: 10.1152/ajpheart.01095.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]