Abstract
Integrin-ligand interactions are regulated in a complex manner by divalent cations, and previous studies have identified ligand-competent, stimulatory, and inhibitory cation-binding sites. In collagen-binding integrins, such as α2β1, ligand recognition takes place exclusively at the α subunit I domain. However, activation of the αI domain depends on its interaction with a structurally similar domain in the β subunit known as the I-like or βI domain. The top face of the βI domain contains three cation-binding sites: the metal-ion dependent adhesion site (MIDAS), the ADMIDAS (adjacent to MIDAS) and LIMBS (ligand-associated metal binding site). The role of these sites in controlling ligand binding to the αI domain has yet to be elucidated. Mutation of the MIDAS or LIMBS completely blocked collagen binding to α2β1; in contrast mutation of the ADMIDAS reduced ligand recognition but this effect could be overcome by the activating mAb TS2/16. Hence, the MIDAS and LIMBS appear to be essential for the interaction between αI and βI whereas occupancy of the ADMIDAS has an allosteric effect on the conformation of βI. An activating mutation in the α2 I domain partially restored ligand binding to the MIDAS and LIMBS mutants. Analysis of the effects of Ca2+, Mg2+ and Mn2+ on ligand binding to these mutants showed that the MIDAS is a ligand-competent site through which Mn2+ stimulates ligand binding, whereas the LIMBS is a stimulatory Ca2+-binding site, occupancy of which increases the affinity of Mg2+ for the MIDAS.
INTRODUCTION
Integrins are a large family of receptors for extracellular and cell surface proteins (1,2). Integrin-ligand binding is regulated by divalent ions and by conformational changes (3,4). The overall shape of an integrin is that of a large ligand-binding ‘head’ on two long ‘legs’ with flexible ‘knees’. Integrins can exist in low affinity (‘inactive’) and high affinity (primed or ‘active’) states; the balance between these states is regulated by both local and global conformational changes. The inactive form of the receptor has been shown to be bent with a closed headpiece, whereas the fully active form appears to be extended with an open headpiece (5,6). Integrins are heterodimers containing non-covalently associated α and β subunits. Eighteen α subunits (α1-α11, αV, αIIb, αD, αE, αM, αL, αX) and eight β subunits (β1-β8) have been identified in mammals. Two subfamilies can be identified depending on the presence or absence of an autonomous folding domain of approximately 200 amino acids, the inserted or ‘I’ domain in the α subunit (‘αI’1). The I domain comprises a central hydrophobic six-stranded β-sheet surrounded by seven α-helices, with the characteristic divalent cation-binding sequence motif DxSxS forming part of the metal-ion dependent adhesion site (MIDAS) on the top face of the domain (7). In each of the nine αI-containing integrins (α1, α2, α10, α11, αE, αD, αM, αL, αX), the MIDAS is the key site for ligand binding (8-10). I domains can exist in closed (inactive) or open (active) conformations, dependent upon a downward movement of the C-terminal α7 helix and an inward motion of the α1 helix (10,11). A domain sharing the same overall fold as the αI domain is present in all β subunits (the I-like or ‘βI’ domain); this domain forms a major part of the ligand binding pocket in integrins that lack αI (12-14).
Crystallographic studies of liganded, non-αI domain containing αVβ3 and αIIbβ3 integrin segments (13,14) have revealed the basis for the absolute dependence on divalent cations for ligand binding: a ligand carboxyl group coordinates to the cation present at the MIDAS of βI. The liganded β3 crystal structures also revealed two additional cation binding sites in βI, the ‘ADMIDAS’ site (adjacent to the MIDAS) and the ‘LIMBS’ (ligand-associated metal-binding site). The βI MIDAS cation shares coordination residues with both the ADMIDAS and LIMBS, hence the three sites form a metal ion cluster (15). The crystal structures of αIIbβ3 (14) also showed that the βI domain undergoes conformational shifts in its α1 and α7 helices that result in an outward swing of the underlying hybrid domain in the open (active) conformation of the ligand-binding integrin headpiece.
Although ligand binding in integrins that contain the αI domain does not directly involve βI, the C-terminal connecting loop between αI and the β-propeller contains a critical glutamate residue, which is proposed to bind to the βI MIDAS (16,17). Binding of this loop is proposed to pull downwards on the α7 helix of αI, holding it in an open (active) conformation (18). Hence, βI can potentially regulate ligand binding to αI. Through this mechanism, the binding of divalent cations or activating mAbs to the βI domain could stimulate ligand binding to αI (19).
The affinity of all integrin-ligand interactions is differentially regulated by Mn2+, Mg2+ and Ca2+. We have previously used the interaction between integrin α5β1 and fibronectin as a model system to identify and characterise cation binding sites that can support or modulate ligand recognition (20). Our results showed that several classes of cation binding sites exist. Occupancy of the first class by Mg2+ or Mn2+ is required for ligand binding (ligand-competent site). The second class is implicated in the allosteric inhibition of Mn2+-supported ligand binding by Ca2+ (inhibitory site). A third class of site also binds Ca2+ and is involved in the stimulation of Mg2+-supported ligand binding by increasing the affinity of Mg2+ for the ligand-competent site (stimulatory site). We subsequently provided evidence that in α5β1 the ligand-competent site for both Mg2+ and Mn2+ is the MIDAS of the βI domain (21). In contrast, the ADMIDAS was found to be a Ca2+-binding inhibitory site in α5β1 (22). Although the LIMBS is a candidate for a Ca2+-binding stimulatory site, it was not possible to test this suggestion for α5β1 since mutation of this site led to a complete loss of ligand recognition (22). Mutation of this site in β2 and β3 integrins also abrogates ligand binding (23-28). Hence, the identity of the stimulatory site is not fully resolved.
Previous studies on α4β7 have delineated separate functions for each of three βI cation-binding sites in this integrin (15,29). However, it is unclear if these findings are broadly applicable, e.g. to β1 integrins or to integrins that contain an αI domain. To study the regulation of ligand binding by divalent ions in an αI-containing integrin, we expressed the collagen receptor α2β1 in a recombinant soluble form. To elucidate the roles of the MIDAS, ADMIDAS and LIMBS we mutated critical residues for cation binding in the βI domain and tested for ligand binding ability and regulation by divalent cations in ELISA-type assays. In this system, an activating mutation in the α2 I domain was found to partially overcome the effect of LIMBS mutations, thereby allowing us to test whether the LIMBS is a stimulatory site. Our results demonstrate that the three sites have distinct roles in the regulation of collagen binding to the α2 I domain, and that the LIMBS is essential for the stimulation of Mg2+-supported ligand binding by Ca2+.
EXPERIMENTAL PROCEDURES
Monoclonal Antibodies
Mouse anti-human β1 mAbs 12G10 and 8E3 were produced as described (30,31). Mouse anti-human β1 mAb TS2/16 was a gift from Dr. F. Sánchez-Madrid (Hospital de la Princesa, Madrid, Spain). Mouse anti-human β1 mAb 15/7 was a gift from Dr. T. Yednock (Elan Pharmaceuticals, South San Francisco, CA). Mouse anti-human β1 mAbs 4B4 and HUTS-4 were purchased from Beckman Coulter (High Wycombe, UK) and Chemicon (Harrow, UK), respectively. Mouse anti-human α2 mAbs JA218 and JA212 were produced as previously described (32). Mouse anti-human α2 mAbs Gi9, P1E6 and 12F1 were from Beckman Coulter, Chemicon, and BD Biosciences (Oxford, UK), respectively. All mAbs were used as purified IgG.
Proteins
Integrin α2β1 was purified from HT-1080 cells as previously described (32). Type I collagen (from rat-tail tendon) was obtained from Sigma (Poole, U.K.). Collagen was biotinylated as described before (32) using sulfo-LC-NHS biotin (Perbio, Chester, U.K.). The α2 I domain-GST fusion protein was produced in E. coli as before (33).
Expression Vector Construction and Mutagenesis
Human α2 cDNA was obtained from D. Tuckwell (University of Manchester). C-terminally truncated human α2 constructs encoding residues 1-806 (TRα2), or 1-1129 (FLα2), and C-terminally truncated human β1 constructs encoding residues 1-455 (TRβ1) or 1-708 (FLβ1) were generated as essentially as previously described (34). Both α and β constructs were fused in frame to the hinge regions and CH2 and CH3 domains of human IgGγ1 (i.e., the Fc portion of the heavy chain). The sequence of the constructs was verified by DNA sequencing. To aid the formation of heterodimers, the CH3 domain of the α2 construct contained a “hole” mutation, while the CH3 domain of the β1 constructs carried a “knob” mutation as described (35). Mutagenesis of βI domain cation binding sites was carried out as before (6,22). Oligonucleotides were purchased from MWG Biotech (Southampton, U.K.). All constructs were verified by DNA sequencing.
Transfection of α2- and β1-Fc Constructs
Chinese hamster ovary cells L761h variant (34) were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 2 mM glutamine and 1% non-essential amino acids (growth medium). 75-cm2 flasks of sub-confluent CHO-L761h cells were transfected with 5μg of β1 construct, and 5 μg of α2 construct using LipofectaminePLUS reagent or Lipofectamine 2000 (Invitrogen, Paisley, Scotland) according to the manufacturer’s instructions. After 4 days, culture supernatants were harvested by centrifugation at 300x g for 5 min.
Reactivity of anti-α2 and anti-β1 mAbs with recombinant receptors
96-well plates (Costar ½-area EIA/RIA, Corning Science Products, High Wycombe, UK) were coated with goat anti-human Fc (Jackson ImmunoResearch, Cambridgeshire, UK) at a concentration of 2.6 μg/ml in Dulbecco’s PBS (50 μl/well) for 16 h. Wells were then blocked for 1 h with 200 μl of 5% (w/v) BSA, 150 mM NaCl, 0.05% (w/v) NaN3, 25 mM Tris-Cl, pH 7.4 (blocking buffer). The blocking solution was removed and the wells were then washed three times with 200 μl of 150 mM NaCl, 25 mM Tris-Cl, 1 mM MnCl2, pH 7.4, containing 1 mg/ml BSA (buffer A). Cell culture supernatants (50 μl/well) were added and the plate incubated at room temperature for 1 h. Wells were washed three times in buffer A and anti-α2 or anti-β1 mAbs (10 μg/ml in buffer A) were then added (50 μl/well). The plate was incubated for 2 h at room temperature and then washed 3 times in buffer A. Peroxidase-conjugated anti-mouse or anti-rat Fc secondary antibody (1:1000 dilution in buffer A; Jackson ImmunoResearch) was added (50 μl/well) for 30 min, the plate washed four times in buffer A, and color was developed using 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonic acid) substrate. Background binding of mAbs to wells incubated with supernatant from mock-transfected cells was subtracted from all measurements. Measurements obtained were the mean ± S.D. of three replicate wells.
Collagen Binding Assays
96-well plates were coated with goat anti-human Fc at a concentration of 2.6 μg/ml in Dulbecco’s PBS (50 μl/well) for 16 h. Wells were then blocked for 1–3 h with blocking buffer, and then washed three times with buffer A. Cell culture supernatants (50 μl/well) were added and the plate incubated at room temperature for 1 h. Wells were washed three times in buffer A and biotinylated collagen type I (0.03-10 μg/ml) in buffer A was added to the plate (50 μl/well), either alone, or in the presence of mAb TS2/16 (5 μg/ml). The plate was then incubated at room temperature for 2 h. Unbound ligand was removed and the wells washed three times with buffer A. Bound ligand was quantitated by addition of 1:500 dilution of ExtrAvidin peroxidase conjugate (Sigma) in buffer A for 20-30 min at room temperature (50 μl/well). Wells were then washed four times with buffer B, and color was developed using ABTS substrate (50 μl/well). Background binding of collagen to wells incubated with supernatant from mock-transfected cells was subtracted from all measurements. Measurements obtained were the mean ± S.D. of three replicate wells. In all assays comparing the binding of collagen to wild-type and mutant receptors, the level of ligand binding to mutant receptors was normalised relative to that of wild-type by measuring the binding of the anti-α2 mAb JA218 to a parallel set of three replicate wells for each receptor; this mAb only recognizes the heterodimeric state of the integrin.
Divalent cation regulation of collagen binding
96-well plates were coated with goat anti-human Fc at a concentration of 2.6 μg/ml in Dulbecco’s PBS (50 μl/well) for 16 h. Wells were then blocked for 1–3 h with blocking buffer, and then washed three times with buffer B (150 mM NaCl, 25 mM Tris-Cl, pH 7.4, containing 1 mg/ml BSA). Buffer B was treated with Chelex beads (BioRad, Hemel Hempstead, UK) to remove any small contaminating amounts of endogenous Ca2+ and Mg2+ ions. Cell culture supernatants (50 μl/well) were added and the plate incubated at room temperature for 1 h. Wells were washed three times in buffer B and biotinylated collagen (0.5 μg/ml) in buffer B was added to the plate (50 μl/well) in the presence of varying concentrations of Mn2+, Mg2+ and Ca2+, or varying concentrations of Ca2+ with Mg2+. The remainder of the assay was performed as described above.
Divalent cation regulation of collagen binding to α2 I domain
96-well plates were coated with recombinant α2 I-domain-GST fusion protein at a concentration of 10 μg/ml in Dulbecco’s PBS (50 μl/well) for 16 h. Wells were then blocked for 1–3 h with blocking buffer, and then washed three times with buffer B. Biotinylated collagen (0.5 μg/ml) in buffer B was added to the plate (50 μl/well) in the presence of varying concentrations of Mn2+, Mg2+ and Ca2+, or varying concentrations of Ca2+ with Mg2+. The remainder of the assay was performed as described above.
Calculation of Apparent Kd values
Binding of biotinylated collagen I to wild-type of mutant integrin was measured using solid phase assays as described above, over a range of collagen concentrations (0.1-10 μg/ml). Apparent affinity (App. Kd) of binding was estimated by non-linear regression analysis using SigmaPlot version 8 (Systat Software Inc.), assuming a single class of binding sites.
RESULTS
Expression of recombinant soluble α2β1-Fc integrins produces functional ‘full-length’ and truncated receptors
Recombinant integrins were produced in a CHO cell expression system by cloning the extracellular portions of α2 and β1 integrin cDNAs and replacing the transmembrane and cytoplasmic domains with modified Fc tags to favour heterodimerisation (34, 35). The soluble α2β1-Fc comprising the full-length extracellular domains of α2 and β1 is referred to as FLα2β1-Fc. In addition, α2 was modified by deleting 326 amino acids from its C-terminal end. The truncated subunit contains the headpiece region of α2, i.e. the β-propeller, αI domain, and the thigh domain. A parallel truncation in the β1 subunit (34) contains the PSI, βI, and hybrid domains. These two truncated subunits form a ‘minimised’ heterodimer referred to here as TRα2β1-Fc.
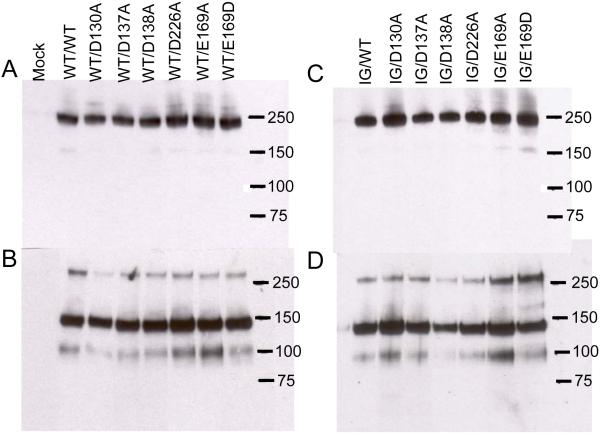
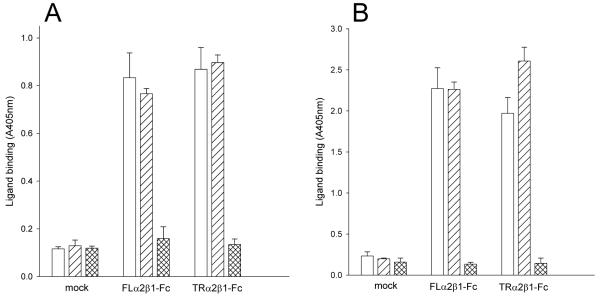
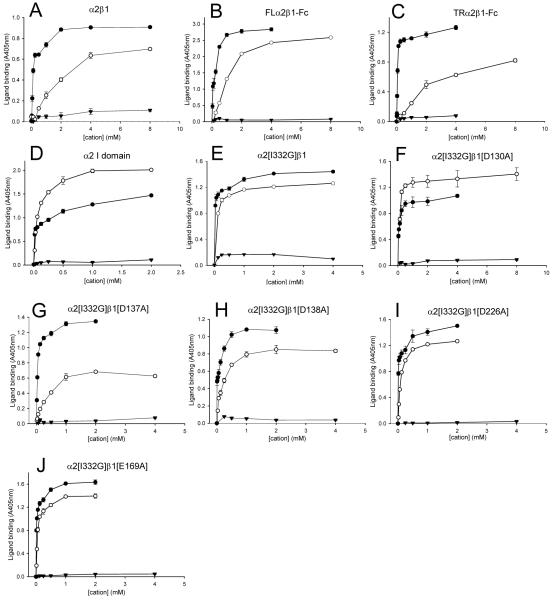
To assess whether the soluble integrins retained the native fold, anti-human α2 and β1 integrin monoclonal antibodies were examined for their ability to bind to the recombinant proteins in ELISA assays (Table I). Recombinant receptors were captured from CHO cell culture supernatants with anti-human Fc antibody onto microtiter plates and the binding of anti-integrin mAbs was evaluated. A panel of mAbs directed against α2 (12F1, JA218, 31H4, P1E6) and β1 (4B4, 8E3, 12G10, TS2/16) recognised the FL and the TRα2β1-Fc integrins to a similar extent as native α2β1 purified from HT-1080 cells (Table IA). The α2 mAbs directed against the αI domain (12F1, JA218 and P1E6) did not recognise the α2 constructs when these were transfected in the absence of a β1 subunit (data not shown). Western blotting of supernatants confirmed that heterodimer formation had taken place (Fig. 1A,C). Following confirmation of the correct protein folding and assembly, the soluble integrins were tested for collagen binding activity in solid phase assays (Fig. 2). The assays were performed in the presence either Mg2+ (Fig. 2A) or Mn2+ (Fig. 2B), using a concentration of the cation that supported maximal ligand binding. The FL and TRα2β1-Fc integrins bound collagen I to comparable extents in both Mg2+ and Mn2+, and specific collagen binding was cation-dependent as it was completely blocked by EDTA. In the presence of TS2/16, a stimulatory mAb that binds to the βI domain, collagen binding was unchanged for FLα2β1 and only slightly increased for the TRα2β1-Fc integrin. Hence, both FL and TRα2β1-Fc appear to be almost completely in an active form.
Table I.
Summary of mAb reactivity with α2β1 integrin variants. CHO L761h cells were transfected with α21-1129-Fc (FL), α21-806-Fc (TR) or α21-806[I332G] (TRα2[I332G]), and β11-708 (FL), wild-type β11-455 (TR) or mutated β11-455. Recombinant receptors were captured from cell culture supernatants with goat anti-human Fc antibody onto microtiter plates and were analysed for reactivity with 5μg/ml anti-α2 mAbs (JA218, 12F1, P1E6 or Gi9), or anti-β1 mAbs (12G10, TS2/16, 8E3, 4B4, 15/7 or HUTS-4) by ELISA. Results are expressed as a percentage of wild type full-length or truncated α2β1-Fc binding, normalised to the binding of anti-α2 JA128 mAb. Results shown are from one experiment, representative of at least three separate experiments.
| A | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| JA218 | 12F1 | P1E6 | Gi9 | 12G10 | TS2/16 | 8E3 | 4B4 | 15/7 | HUTS4 | |
| FL α2β1 | 107.96 | 53.39 | 109.63 | 77.19 | 115.89 | 116.97 | 57.16 | 50.53 | 83.18 | 86.42 |
| TRα2β1 | 100.00 | 54.25 | 100.30 | 67.90 | 101.05 | 108.20 | 55.80 | 50.25 | 45.10 | 65.25 |
| TRα2β1[D130A] | 89.21 | 43.18 | 89.43 | 51.16 | 97.86 | 98.17 | 52.41 | 42.28 | 55.53 | 73.64 |
| TRα2β1[D137A] | 103.10 | 54.65 | 128.62 | 66.71 | 101.51 | 110.32 | 61.81 | 49.44 | 24.02 | 51.66 |
| TRα2β1[D138A] | 100.73 | 57.87 | 112.22 | 71.67 | 98.21 | 107.11 | 60.19 | 65.48 | 2.42 | 24.88 |
| TRα2β1[D226A] | 95.77 | 56.08 | 101.76 | 69.63 | 103.48 | 98.55 | 62.73 | 53.35 | 55.26 | 75.04 |
| TRα2β1[E169A] | 98.92 | 59.35 | 106.93 | 64.99 | 109.41 | 110.94 | 58.86 | 64.30 | 60.34 | 91.35 |
| TRα2β1[E169D] | 99.18 | 56.33 | 101.41 | 67.49 | 103.59 | 109.94 | 49.39 | 51.37 | 71.65 | 88.86 |
| B | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| JA218 | 12F1 | P1E6 | Gi9 | 12G10 | TS2/16 | 8E3 | 4B4 | 15/7 | HUTS4 | |
| TRα2β1 | 100.00 | 83.15 | 102.00 | 88.75 | 68.30 | 64.65 | 63.20 | 52.55 | 93.15 | 81.90 |
| TRα2[I332G] β1 | 91.72 | 91.72 | 97.87 | 92.32 | 77.09 | 72.37 | 67.74 | 66.82 | 91.77 | 85.62 |
| TRα2[I332G] β1[D130A] | 93.82 | 83.17 | 96.49 | 84.44 | 63.70 | 64.88 | 71.87 | 66.42 | 90.21 | 85.80 |
| TRα2[I332G] β1[D137A] | 130.62 | 96.40 | 97.31 | 89.80 | 75.04 | 77.79 | 100.19 | 77.98 | 88.11 | 83.53 |
| TRα2[I332G] β1[D138A] | 108.93 | 88.83 | 88.50 | 85.78 | 76.79 | 82.95 | 85.29 | 80.93 | 88.99 | 85.13 |
| TRα2[I332G] β1[D226A] | 112.55 | 87.56 | 90.83 | 93.59 | 80.81 | 84.75 | 109.85 | 89.59 | 94.04 | 93.92 |
| TRα2[I332G] β1[E169A] | 96.69 | 93.06 | 98.29 | 95.09 | 83.59 | 79.91 | 93.11 | 85.96 | 94.37 | 94.37 |
| TRα2[I332G] β1[E169D] | 118.06 | 109.45 | 92.50 | 112.63 | 84.06 | 83.41 | 108.62 | 95.99 | 105.67 | 100.06 |
FIGURE 1. Western blotting of recombinant TRα2β1-Fc integrins.
Recombinant integrins were partially purified from cell culture supernatants using protein A Sepharose. Aliquots of purified proteins were run on 3-8 % SDS-PAGE gels under non-reducing (panels A, B) or reducing conditions (panels C, D), transferred to nitrocellulose and blotted with anti-human Fc conjugated with horseradish peroxidase. Panels A and C, culture supernatants from cells transfected with wild-type (WT) TRα2 and WT or cation-binding site mutant TRβ1. Panels B and D, culture supernatants from cells transfected with I332G mutant TRα2 (IG) and WT or cation-binding site mutant TRβ1. Migration positions of molecular weight markers are indicated (kDa). Under non-reducing conditions a predominant band at ~ 240 kDa is observed, close to the expected size for the TRα2β1-Fc heterodimer. Under reducing conditions disulfide bonds between the Fc hinge regions are broken and two predominant bands of ~ 140 kDa and ~ 100 kDa are observed, corresponding to the expected sizes of TRα2 and TRβ1 proteins, respectively. For unknown reasons the β subunit band reacts much more weakly with the anti-human Fc antibody than the α subunit band (31). The 240 and 100 kDa bands are reactive with the anti-β1 mAb TS2/16; the 240 and 140 kDa bands are reactive with anti-α2 mAb JA212 (not shown). These bands are not observed in proteins purified from the supernatants of mock-transfected cells.
FIGURE 2. Collagen binding to recombinant α2β1-Fc integrins.
Culture supernatants from CHO L761h cells. transfected with FLα2 and FLβ1 constructs (FLα2β1-Fc), TRα2 and TRβ1 constructs (TRα2β1-Fc) or no DNA (mock) were analysed by Fc-capture assays. Binding of collagen I (1 μg/ml) to captured proteins was measured in 5 mM Mg2+ (A) or 1 mM Mn2+ (B) in the absence (open bars) or presence of the activating anti-β1 mAb TS2/16 (diagonally-hatched bars) or 5 mM EDTA (cross-hatched bars). Supernatants from cells transfected with either α2 or β1 constructs alone did not show any collagen binding above that of supernatants from mock-transfected cells (data not shown). Absorbance values and error bars represent the mean and standard deviation of three replicate wells from one experiment, representative of four separate experiments.
Mutations in the βI cation-binding sites inactivate TRα2β1-Fc
To study the role of the β1 I domain cation-binding sites in regulation of α2β1 function, residues that form the sites (Table II) were selected for mutagenesis in the TRα2β1-Fc expression system. Each mutant contained an amino acid substitution of a single residue forming one of the three cation-binding sites. The β1 MIDAS substitution D130A and the two alternative alanine substitutions of residues Asp137 and Asp138 that coordinate the β1 ADMIDAS metal ion site have been shown to abrogate cation binding to the these sites in the non-αI domain-containing integrin α5β1 (21, 22). The β1 LIMBS contains residues Glu169, Asn224, Asp226, Glu229, and Pro228 (Table II). Of these residues Glu229 is also an integral residue of the MIDAS, and Pro228 contributes only a backbone carbonyl. The side chain of Asn224 is specific for the LIMBS but also participates in ligand binding (13). Thus, Glu169 and Asp226 were targeted as these are specific for the LIMBS and substitution of either Glu169 or Asp226 to alanine would be expected to abrogate cation binding to the LIMBS only. In parallel, a conservative mutation Glu169 to Asp, was examined as a control. This mutation is unlikely to perturb cation binding since an aspartate residue is present in place of glutamate at this position in almost all the other β subunits. The effect of these mutations was examined in the context of the TRα2β1-Fc protein. Expression of each recombinant integrin and protein folding were monitored using conformation-sensitive anti-human α2 and β1 integrin mAbs (Table IA). Since these antibodies recognised the wild-type and mutated receptors to similar extents, it was concluded that the mutations had no gross effect on the conformation.
Table II.
Cation-binding sites in the β1 and β3 A domains and their coordinating residues. Cation-binding site mutants made in β1 are shown in bold. Mutations were based on β3 structure as identified in the aVβ3 crystal structures. The cation-coordinating residues in β3 are shown alongside the corresponding residues in β1. Coordinating residues found only in the unliganded structure are marked with ‘*’ (12) and those found only in the liganded structure are marked with ‘**’ (13).
| Cation binding site |
Residues | |||||
|---|---|---|---|---|---|---|
| MIDAS | β3 | Asp119 | Ser121 | Ser123 | Glu220 | Asp251 |
| β1 | Asp130 | Ser132 | Ser134 | Glu229 | Asp259 | |
| mutation | D130A | |||||
| ADMIDAS | β3 | Ser123 | Asp126 | Asp127 | Met335* | Asp251** |
| β1 | Ser134 | Asp137 | Asp138 | Ala341 | Asp259 | |
| mutation | D137A | D138A | ||||
| LIMBS | β3 | Asp158 | Asn215 | Asp217 | Pro219 | Glu220 |
| β1 | Glu169 | Asn224 | Asp226 | Pro228 | Glu229 | |
| mutation |
E169A
E169D |
D226A | ||||
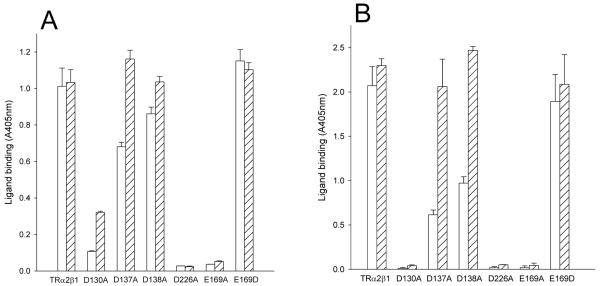
Next, we assessed the collagen-binding activity of the Fc-captured α2β1-Fc variants in solid phase assays (Fig. 3A, B). Of the six mutant integrins, only the control LIMBS E169D mutant bound collagen I to the same extent as wild-type TRα2β1. In contrast, the LIMBS D226A and E169A mutants did not support collagen binding under any experimental condition, with or without the stimulatory TS2/16 antibody. The β1 MIDAS D130A mutation also abolished ligand binding, although weak binding was observed in the presence of TS2/16 and Mg2+. In contrast, the ADMIDAS mutants D137A and D138A showed a moderate level of collagen binding, and addition of TS2/16 restored ligand binding to the same level as that for the wild-type integrin. Hence, the ADMIDAS mutants were partially active, whereas the LIMBS and MIDAS mutants were essentially inactive.
FIGURE 3. Collagen binding to TRα2β1-Fc cation-binding site mutants.
Culture supernatants from transfected CHO L761h cells with TRα2 and wild-type TRβ1 or TRβ1 mutated at the MIDAS (D130A), ADMIDAS (D137A, D138A) or LIMBS (D226A, E169A, E169D) were analysed in Fc-capture assays, performed in the presence of 5 mM Mg2+ (A) or 1 mM Mn2+ (B) in the absence (open bars) or presence of the activating mAb TS2/16 (diagonally-hatched bars). Absorbance values and error bars represent the mean and standard deviation of three replicate wells from one experiment, representative of three separate experiments. Non-specific collagen binding (as estimated from the binding to wells treated with supernatant from mock-transfected cells) has been subtracted in this and all subsequent figures.
To investigate further the activation state of the ADMIDAS mutants, we examined the binding of the activating mAbs HUTS-4 and 15/7 to α2β1D137A and α2β1D138A (Table IA). These mAbs recognise the swung-out conformation of the hybrid, and thereby report an open headpiece (6). The results showed that both ADMIDAS mutants had strongly decreased expression of the HUTS-4 and 15/7 epitopes, suggesting that these mutants mainly adopt the closed headpiece conformation. The expression of these epitopes on the ADMIDAS mutants was partially restored in the presence of TS2/16 (data not shown). Although the MIDAS and LIMBS mutations resulted in non-functional integrins these mutations did not lead to a reduction of HUTS-4 and 15/7 binding (Table IA). Hence, these mutations did not affect the activation state of the integrin; instead, it appears that the LIMBS and MIDAS are required for the interaction of the βI domain with the α2 I domain.
The α2[I332G] mutation promotes the active conformation of α2β1
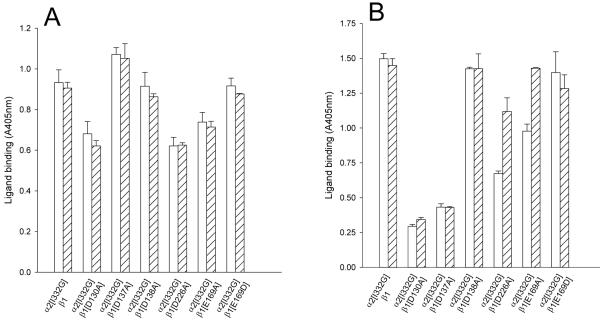
Since the α2β1-Fc LIMBS and MIDAS mutants were inactive for collagen binding, we investigated whether an activating mutation in the αI domain could restore ligand binding. Activation of αI-containing integrins by substitution of a conserved isoleucine residue in the α7 helix has been demonstrated by a number of studies (9,16,36). To test whether a similar mutation in α2 could rescue the ability of the βI domain mutants to bind to collagen, an Ile332 to Gly mutation (I332G) was introduced into the truncated α2 subunit. The TRα2[I332G]-Fc subunit was co-expressed with the wild-type or cation-binding site mutated β1-Fc subunits in CHO cells. The resulting seven mutants were examined for expression and correct folding in Fc-capture assays and Western blotting. The anti-α2 and the anti-β1 mAbs recognised the TRα2[I332G]β1-Fc mutants to a similar extent as the wild-type TRα2β1-Fc (Table IB), and each of the mutants formed a heterodimer (Fig. 1B,D). Thus, the α2[I332G]β1-Fc and all six double mutants assembled correctly and acquired a native fold.
The ability of the mutants to bind collagen I was then examined in solid-phase assays (Fig. 4A, B). The α2 activating mutation enhanced the low collagen binding levels of the D137A and D138A ADMIDAS mutants to levels close to those observed for the wild-type receptor in Mg2+, although the relative level of binding for the D137A mutant remained low in Mn2+. The activating mutation also restored 15/7 and HUTS-4 binding to these mutants to a level similar to that of wild-type integrin (Table IB). In addition, the MIDAS (D130A) mutant and both the LIMBS (D226A, E169A) mutants gained collagen-binding ability when co-expressed with the TRα2[I332G]-Fc mutant. The TS2/16 antibody caused no further increase in collagen binding, except for the LIMBS mutants in the presence of Mn2+ (Fig. 4B).
FIGURE 4. Collagen binding to TRα2[I332G]β1-Fc cation-binding site mutants.
Culture supernatants from transfected CHO L761h cells with TRα2 and wild-type TRβ1 or TRβ1 mutated at the MIDAS (D130A), ADMIDAS (D137A, D138A) or LIMBS (D226A, E169A, E169D) were analysed in Fc-capture assays, performed in the presence of 5mM Mg2+ (A) or 1 mM Mn2+ (B). The ligand-binding capacity of the α2β1-Fc mutants was detected with 0.5 μg/ml collagen I in the absence (open bars) or the presence of anti-β1 antibody TS2/16 (diagonally hatched bars). Absorbance values and error bars represent the mean and standard deviation from one experiment, representative of three separate experiments.
To estimate the apparent affinity (App. Kd) of the recombinant integrin-collagen I interaction, the binding of collagen was measured over a range of collagen concentrations (0.01 - 10 μg/ml) in the presence of 1 mM Mn2+. The App. Kd values for the binding of recombinant soluble integrins to collagen are summarised in Table III. The results showed that mutations which disrupted the βI cation-binding sites significantly decreased the apparent affinity of α2[I332G]β1 for collagen. Hence, the activating I332G mutation only partly overcomes the effects of mutations in the βI cation-binding sites, i.e., the βI domain still exerts some effect on ligand binding to αI in the double mutants.
Table III.
Apparent affinities of collagen I for α2β1-Fc integrins. Binding measurements were made in the presence of 1 mM Mn2+. Apparent affinity (App. Kd) values were estimated by non-linear regression analysis. Data shown are the mean ± S.D. from three experiments for each recombinant integrin.
| App. Kd (nM) | S.D. (nM) | |
|---|---|---|
| FLα2β1 | 0.92 | 0.16 |
| TRα2β1 | 1.52 | 0.23 |
| α2[I332G] β1 | 1.07 | 0.06 |
| α2[I332G]] β1[D130A] | 9.92 | 1.32 |
| α2[I332G] β1[D137A] | 8.17 | 2.14 |
| α2[I332G] β1[D138A] | 2.94 | 0.29 |
| α2[I332G] β1[D226A] | 6.22 | 1.25 |
| α2[I332G] β 1[E169A] | 4.87 | 0.69 |
| α2[I332G] β1 [E169D] | 1.17 | 0.06 |
The α2[I332G] and βI domain cation-binding site mutations result in changes in the relative abilities of Mn2+ and Mg2+ to support ligand binding
Next, we examined the effect of the β1 cation-binding site mutations on the specificity and requirement for cations in collagen binding, and compared it to that for purified α2β1 and for the solitary α2 I domain. As shown previously, the binding of purified α2β1 to collagen type I was supported strongly by Mn2+ and to a lesser extent by Mg2+, but not by Ca2+ (Fig. 5A). A similar profile of cation regulation was observed for both the FL and TRα2β1-Fc (Fig. 5B, C). The Mn2+ and Mg2+ concentrations required to support half-maximal collagen binding were very close to those observed for the purified α2β1. Thus, the results showed that ligand binding to the Fc-fusion proteins was modulated by divalent cations in a very similar manner to the purified integrin. The cation dependency of collagen binding to the solitary α2 I domain was also studied (Fig. 5D). The results showed that Mg2+ was slightly more effective than Mn2+ at supporting collagen binding. Hence, the observed stimulation of ligand binding by Mn2+ in the parental receptor derives from cation-binding sites outside of the αI domain.
FIGURE 5. Effect of divalent cations on the binding of collagen to purified α2β1 and recombinant proteins.
Binding of collagen I to purified α2β1 and to recombinant proteins was measured in the presence of varying concentrations of Mn2+ (●), Mg2+ (○), or Ca2+ (▲). Purified α2β1 (A), FLα2β1-Fc (B), TRα2β1-Fc (C), α2 I domain (D), TRα2[I332G]β1-Fc (E), TRα2[I332G]β1[D130A]-Fc (F), TRα2[I332G]β1[D137A]-Fc (G), TRα2[I332G]β1[D138A]-Fc (H), TRα2[I332G]β1[D226A]-Fc (I), and TRα2[I332G]β1[E169A]-Fc (J). Each experiment shown is representative of at least three separate experiments.
Next, the effect of Mn2+, Mg2+ and Ca2+ on collagen binding to the constitutively active mutant TRα2[I332G]β1-Fc and cation binding site double mutants was examined over a range of divalent cation concentrations (Fig. 5 E-J). For all the recombinant proteins, Mn2+ and Mg2+ but not Ca2+ ions support the binding of collagen. As observed for the wild-type integrins Mn2+ promoted higher maximal levels of collagen binding than Mg2+ to the ADMIDAS and LIMBS mutants (Fig. 5 G-J). In contrast, for the MIDAS mutant (Fig. 5F), Mg2+ was more effective than Mn2+ at promoting ligand binding (as observed for the solitary α2 I domain). Loss of Mn2+ stimulation for the MIDAS mutant indicates that the MIDAS is necessary for the ability of Mn2+ to promote ligand binding.
Mutation of the LIMBS or MIDAS leads to loss of Ca2+/Mg2+ synergy
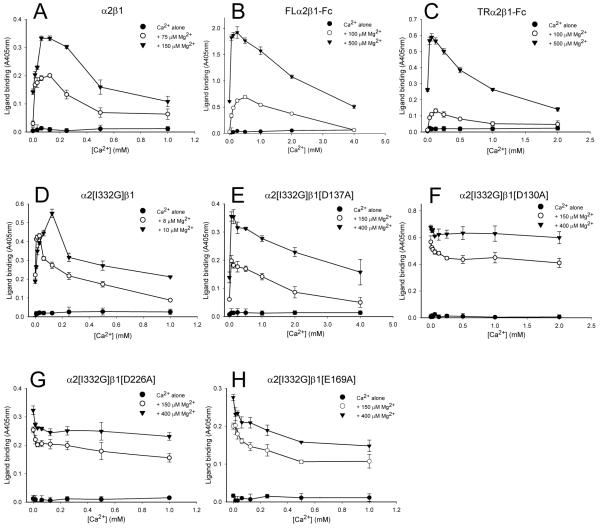
We next tested whether low concentrations of Ca2+ could stimulate collagen binding at sub-optimal Mg2+ concentrations for the wild-type and mutant receptors. In agreement with previous reports (37,38) low Ca2+ concentrations increased the Mg2+-dependent collagen binding to the purified α2β1 integrin, while Ca2+ ions at concentrations above 0.25 mM had an inhibitory effect on ligand binding (Fig. 5A). Similarly, Ca2+ concentrations below 0.25 mM reproducibly increased Mg2+-dependent collagen binding to both FL and TR recombinant α2β1-Fc integrins; however, increasing the Ca2+ concentration above 0.25 mM decreased collagen binding and strong inhibition of collagen binding occurred at 2 mM Ca2+ (Fig. 5B, C). Thus, in agreement to the reported regulatory effect of Ca2+ for the parental α2β1 integrin (37,38), low Ca2+ concentrations also stimulated the Mg2+-dependent collagen binding to the wild-type recombinant receptors, while Ca2+ at higher concentrations inhibited binding. As expected, no stimulatory effect of Ca2+ on Mg2+-supported ligand binding to the solitary α2 I domain was observed (data not shown).
We next tested the α2[I332G] cation-binding site mutants to determine if one of these sites was responsible for stimulation of ligand binding by Ca2+ at low Mg2+ concentrations. The effect of Ca2+ on Mg2+-dependent collagen binding to the TRα2[I332G]β1 integrin was examined first (Fig. 5D). In the presence of both Ca2+ and Mg2+ cations, collagen binding to the TRα2[I332G]β1-Fc had a biphasic pattern, similar to that for the TRα2β1-Fc integrin: Ca2+ at low concentrations stimulated Mg2+-dependent collagen binding, whereas higher concentrations of Ca2+ inhibited collagen binding. The α2[I332G]/β1[D137A]-Fc ADMIDAS mutant also bound collagen better in the presence of Ca2+ and Mg2+ than a low concentration of Mg2+ alone (Fig. 5E). Similar results were obtained for the I332G/D138A mutant (data not shown). Hence, Ca2+ ions retained the ability to stimulate Mg2+-supported collagen binding when the ADMIDAS was mutated.
In contrast, Ca2+ did not have a stimulatory effect on Mg2+-dependent collagen binding to the α2[I332G]/β1[D226A]-Fc and α2[I332G]/β1[E169A]-Fc LIMBS mutants (Fig. 5G, H). Micromolar concentrations of Ca2+ did not activate binding to collagen in the presence of sub-optimal (50 μM, data not shown) to optimal (0.15 - 0.5 mM) Mg2+ concentrations. Moreover, Ca2+ ions, even in the low μM range, inhibited collagen binding supported by Mg2+. Thus, Ca2+ failed to synergistically enhance the binding of collagen to either of the LIMBS mutants.
The stimulatory effect of Ca2+ was also lost in the α2[I332G]/β1[D130A] MIDAS mutant (Fig. 5F). Furthermore, it was also observed that Ca2+ concentrations in the range of 0.25–2 mM did not inhibit significantly collagen binding. Thus, Ca2+ ions were less effective in inhibiting collagen binding to the MIDAS mutant than to the ADMIDAS or LIMBS mutants.
DISCUSSION
In this study we have used a recombinant soluble α2β1-Fc integrin system to study the contribution of each of the βI domain cation-binding sites to the regulation of ligand binding to the αI domain. Our major findings are:
(i) Mutation of the βI MIDAS and LIMBS leads to a complete loss of ligand binding, suggesting that these sites are essential for the interaction between αI and βI.
(ii) The ADMIDAS is not essential for ligand recognition, however mutation of this site leads to a low activity state (closed headpiece).
(iii) An activating mutation in the α7 helix of the αI domain can partially overcome the effect of mutations in βI cation-binding sites.
(iv) Mutation of the βI MIDAS (in combination with the αI mutation) results in a loss of the capacity of Mn2+ to promote ligand binding (relative to Mg2+)
(v) Mutation of either the MIDAS or LIMBS (but not the ADMIDAS) leads to loss of synergy between low concentrations of Mg2+ and Ca2+ for supporting ligand binding.
A cell-free system was chosen to study the role of cation-binding sites in ligand recognition by α2β1 in order to avoid potential confounding effects due to the effects of divalent ions on cell signalling and interactions with other proteins. This is the first report of a functional αI domain-containing integrin preparation with a truncated extracellular domain. Our data show that the headpiece (β-propeller, I domain and thigh domain of α2, with PSI, hybrid and I domain of β1) of α2β1 is sufficient for native folding and ligand recognition. The TRα2β1-Fc recombinant proteins were also constitutively active for ligand binding; this is in contrast to the equivalent TRα5β1-Fc and TRα4β1-Fc proteins, which were found to be almost completely inactive (22,34,39). In addition, we found that the α2 I domain does not fold autonomously in the context of the α2 β-propeller domain; instead, association with the β1 subunit is necessary. This finding contrasts with a previous report that folding of the αM I domain can occur independently of association with the β2 subunit (40).
Identification of the MIDAS as the stimulatory Mn2+ binding site
Although the I332G mutation reduced the dependency of ligand binding on cation binding to the βI domain, Mn2+ remained more effective than Mg2+ for supporting ligand binding (whereas the converse is seen for the isolated α2 I domain). However, for the D130A MIDAS mutant Mg2+ was more effective than Mn2+, suggesting that regulation by the βI domain is absent in this mutant.
It has previously been suggested that the ADMIDAS is the site through which Mn2+ stimulates ligand binding, by displacing Ca2+ from this site (15). If Mn2+ activates integrins by binding to the ADMIDAS then mutation of the ADMIDAS should result in an integrin which is insensitive to stimulation of ligand binding by Mn2+ (relative to Mg2+). In contrast, we observed that the ADMIDAS mutants were more dependent on the presence of Mn2+ for ligand binding than the wild-type receptor, similar to the results obtained for α5β1 (22). Furthermore, we found that mutation of the AMIDAS did not alter the stimulatory effect of Mn2+ in the constitutively active (I332G) mutant, whereas mutation of the MIDAS abrogated this effect. Hence, our data support the alternative paradigm that the βI MIDAS is the site through which Mn2+ exerts its effects on ligand recognition (4). The differences between Mn2+ and Mg2+ in their abilities to stimulate ligand binding appears to be a consequence of the distinct capacities of these two ions to promote shape shifting in the βI domain (21). A possible explanation is that Mn2+ and Mg2+ have subtly different arrangements of coordinating residues at the βI MIDAS.
Mutation of the MIDAS decreased the propensity of Ca2+ to inhibit Mg2+-supported ligand binding at high Ca2+ concentrations. Hence the βI MIDAS also appears to be a low affinity Ca2+-binding site that contributes to the inhibition of ligand binding by Ca2+, presumably because occupancy of this site by Ca2+ does not support the interaction between βI and αI. Occupancy of cation-binding sites in the β-propeller may contribute to the inhibition of Mg2+-supported ligand binding observed at low Ca2+ concentrations, since these are high affinity Ca2+-binding sites (41).
Identification of the LIMBS as a stimulatory Ca2+-binding site
Mutation of the ADMIDAS does not lead to loss of the synergy between low concentrations of Ca2+ and Mg2+, either for α5β1(22) or α2β1 (this study), suggesting that the MIDAS and LIMBS are sufficient for synergy. However, it was not previously possible to test directly that mutation of the LIMBS leads to loss of Ca2+/Mg2+ synergy because LIMBS mutations abrogate ligand binding to α5β1. Although mutation of the LIMBS in wild-type α2β1 also led to a complete loss of ligand recognition, the α2[I332G] mutation partly restored ligand binding. As predicted, no synergy between Ca2+ and Mg2+ was seen in the LIMBS double mutants. This synergy was still present in the ADMIDAS mutants but was again lost in the MIDAS mutant. Hence, the MIDAS and LIMBS are both required (and sufficient) for synergism. Since the MIDAS preferentially binds Mg2+, the LIMBS must therefore be the stimulatory Ca2+ binding site.
Evidence to support the positive Ca2+-regulatory role of LIMBS has also been presented for cell-surface expressed α4β7 integrin (a non-αI domain-containing integrin) (13). The synergistic effects of low Ca2+ and Mg2+ concentrations were shown to require the LIMBS but not the ADMIDAS. In the same study, disruption of the LIMBS (by a mutation equivalent to the D226A mutation in β1) impaired the firm adhesion of cells on MAdCAM-1 coated surfaces, but did not block rolling. In contrast, mutation of the LIMBS in α2β1 (this study) and α5β1 abolishes ligand binding. Mutation in the LIMBS in β2 and β3 integrins has also been found to block ligand recognition (23-28,42). Mutation of the ADMIDAS resulted in a constitutively active (although bent) integrin in α4β7 (15,29,43) and αLβ2 (27), whereas for α2β1 (this study) and α5β1 (22) mutation of the ADMIDAS resulted in a constitutively inactive receptor. Subtle differences in the coordination of the βI cation binding sites among different integrins could account for these discrepancies.
Our results are in good agreement with data from the crystal structure of the ligand-occupied headpiece region of αIIbβ3 (14). In this structure the MIDAS was occupied by Mg2+, whereas the ADMIDAS and LIMBS were occupied by Ca2+. The higher propensity of Ca2+ than Mg2+ or Mn2+ to coordinate carbonyl oxygen atoms (44) may explain why Ca2+ will preferentially bind to the ADMIDAS and LIMBS rather than to the MIDAS, which lacks any carbonyl oxygen coordination (13,14). The LIMBS could act as the stimulatory Ca2+ site because occupancy of the LIMBS may reorient and stabilise the side chain carboxyl of E220 (E229 in β1), thereby favouring occupancy of the MIDAS, which is also coordinated by the same carboxyl group.
In summary, we have shown that the three βI cation-binding sites have distinct roles in ligand binding by α2β1. Our findings are likely to be broadly applicable to β1 integrins and to integrins that contain αI. The interaction of platelet α2β1 with collagen is a critical step during hemostasis and changes in the extracellular concentrations of Mg2+ and Ca2+ after wounding may regulate collagen binding in vivo (45). Similarly, alterations in the extracellular fluxes of these ions may alter tumor cell proliferation and migration (46). Our novel insights into the mechanisms of α2β1 regulation should assist in the development of α2β1 antagonists for therapeutic use (47,48).
FIGURE 6. Effect of Ca2+ on Mg2+-dependent collagen binding to purified α2β 1 and recombinant proteins.
Purified α2β1 (A), FLα2β1-Fc (B), TRα2β1-Fc (C), TRα2[I332G]β1-Fc (D), TRα2[I332G]β1[D137A]-Fc (E), TRα2[I332G]β1[D130A]-Fc (F), TRα2[I332G]β1[D226A]-Fc (G), TRα2[I332G]β1[E169A]-Fc( H). Binding of collagen I was measured in the presence of two constant concentrations of Mg2+ (as indicated in the panel legend) with varying concentrations of Ca2+. The level of ligand binding supported by Ca2+ alone is also shown. Each experiment shown is representative of at least three separate experiments.
Acknowledgments
We thank Stephanie Barton and Emlyn Symonds for the MIDAS and LIMBS β1 mutants.
We are grateful to T. Yednock, K. Yamada, and F. Sánchez-Madrid for mAbs, to D. Tuckwell for mAbs and cDNA constructs, and S. Craig for assistance with mAb production and purification.
Footnotes
These studies were supported by grants from the Wellcome Trust (to M. J. H.).
The abbreviations used are: αI, α subunit I-domain (also known as αA); βI, β subunit I-domain (also known as βA); MIDAS, metal-ion dependent adhesion site; ADMIDAS, adjacent to MIDAS; LIMBS, ligand-associated metal binding site; mAb, monoclonal antibody; PBS, phosphate-buffered saline; BSA, bovine serum albumin; ELISA, enzyme-linked immunosorbent assay; S.D., standard deviation; FLα2, α21-1129-Fc construct; TRα2, α21-806-Fc construct; FLβ1, β11-708-Fc construct; TRβ1, β11-455-Fc construct; FLα2β1-Fc, recombinant soluble integrin heterodimer containing the complete extracellular domains of α2 and β1 subunits fused to the CH2 and CH3 domains (Fc region) of human IgGγ1; TRα2β1-Fc, recombinant soluble integrin heterodimer containing C-terminally truncated α2 and β1 subunits fused to the CH2 and CH3 domains of human IgGγ1; CHO, Chinese hamster ovary
References
- 1.Hynes RO. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 2.Takada Y, Ye X, Simon S. Genome Biol. 2007;8:215. doi: 10.1186/gb-2007-8-5-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo BH, Carman CV, Springer TA. Annu. Rev. Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mould AP, Humphries MJ. Curr. Opin. Cell Biol. 2004;16:544–551. doi: 10.1016/j.ceb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Takagi J, Petre BM, Walz T, Springer TA. Cell. 2002;110:599–611. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 6.Mould AP, Barton SJ, Askari JA, McEwan PA, Buckley PA, Craig SE, Humphries MJ. J. Biol. Chem. 2003;278:17028–17035. doi: 10.1074/jbc.M304627200. [DOI] [PubMed] [Google Scholar]
- 7.Whittaker CA, Hynes RO. Mol. Biol. Cell. 2002;13:3369–3387. doi: 10.1091/mbc.E02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emsley J, Knight CG, Farndale RW, Barnes MJ. Cell. 2000;101:47–56. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- 9.Vorup-Jensen T, Ostermeier C, Shimaoka M, Hommel U, Springer TA. Proc. Natl. Acad. Sci. U S A. 2003;100:1873–1878. doi: 10.1073/pnas.0237387100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimaoka M, Xiao T, Liu JH, Yang Y, Dong Y, Jun CD, McCormack A, Zhang R, Joachimiak A, Takagi J, Wang JH, Springer TA. Cell. 2003;112:99–111. doi: 10.1016/s0092-8674(02)01257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JO, Bankston LA, Arnaout MA, Liddington RC. Structure. 1995;3:1333–1340. doi: 10.1016/s0969-2126(01)00271-4. [DOI] [PubMed] [Google Scholar]
- 12.Xiong J-P, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong J-P, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 14.Xiao T, Takagi J, Coller BS, Wang J-H, Springer TA. Nature (Lond.) 2004;432:59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Salas A, Springer TA. Nat. Struct. Biol. 2003;10:995–1001. doi: 10.1038/nsb1011. [DOI] [PubMed] [Google Scholar]
- 16.Huth JR, Olejniczak ET, Mendoza R, Liang H, Harris EA, Lupher ML, Jr., Wilson AE, Fesik SW, Staunton DE. Proc. Natl. Acad. Sci. USA. 2000;97:5231–5236. doi: 10.1073/pnas.97.10.5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alonso JL, Essafi M, Xiong JP, Stehle T, Arnaout MA. Curr. Biol. 2002;12:R340–R342. doi: 10.1016/s0960-9822(02)00852-7. [DOI] [PubMed] [Google Scholar]
- 18.Yang W, Shimaoka M, Salas A, Takagi J, Springer TA. Proc. Natl. Acad. Sci. USA. 2004;101:2906–2911. doi: 10.1073/pnas.0307340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu C, Ferzly M, Takagi J, Springer TA. J. Immunol. 2001;166:5629–5637. doi: 10.4049/jimmunol.166.9.5629. [DOI] [PubMed] [Google Scholar]
- 20.Mould AP, Akiyama SK, Humphries MJ. J. Biol. Chem. 1995;270:26270–26277. doi: 10.1074/jbc.270.44.26270. [DOI] [PubMed] [Google Scholar]
- 21.Mould AP, Askari JA, Barton S, Kline AD, McEwan PA, Craig SE, Humphries MJ. J. Biol. Chem. 2002;277:19800–19806. doi: 10.1074/jbc.M201571200. [DOI] [PubMed] [Google Scholar]
- 22.Mould AP, Barton SJ, Askari JA, Craig SE, Humphries MJ. J. Biol. Chem. 2003;278:51662–51629. doi: 10.1074/jbc.M306655200. [DOI] [PubMed] [Google Scholar]
- 23.Tozer EC, Liddington RC, Sutcliffe MJ, Smeeton AH, Loftus JC. J. Biol. Chem. 1996;271:21978–21984. doi: 10.1074/jbc.271.36.21978. [DOI] [PubMed] [Google Scholar]
- 24.Baker EK, Tozer EC, Pfaff M, Shattil SJ, Loftus JC, Ginsberg MH. Proc. Natl. Acad. Sci. USA. 1997;94:1973–1978. doi: 10.1073/pnas.94.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodman TG, Bajt ML. J. Biol. Chem. 1996;271:23729–23736. doi: 10.1074/jbc.271.39.23729. [DOI] [PubMed] [Google Scholar]
- 26.Yamanouchi J, Hato T, Tamura T, Fujita S. Thromb. Haemost. 2002;87:756–762. [PubMed] [Google Scholar]
- 27.Chen J, Yang W, Kim M, Carman CV, Springer TA. Proc. Natl. Acad. Sci. USA. 2006;103:13062–13067. doi: 10.1073/pnas.0605666103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng M, Foo SY, Shi ML, Tang RH, Kong LS, Law SK, Tan SM. J. Biol. Chem. 2007;282:18225–18232. doi: 10.1074/jbc.M701386200. [DOI] [PubMed] [Google Scholar]
- 29.Chen JF, Takagi J, Xie C, Xiao T, Luo B-H, Springer TA. J. Biol. Chem. 2004;279:55556–55561. doi: 10.1074/jbc.M407773200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mould AP, Garratt AN, Askari JA, Akiyama SK, Humphries MJ. FEBS Lett. 1995;363:118–122. doi: 10.1016/0014-5793(95)00301-o. [DOI] [PubMed] [Google Scholar]
- 31.Burrows L, Clark K, Mould AP, Humphries MJ. Biochem. J. 1999;344:527–533. [PMC free article] [PubMed] [Google Scholar]
- 32.Tuckwell DS, Smith L, Korda M, Askari JA, Santoso S, Barnes MJ, Farndale RW, Humphries MJ. Biochem. J. 2000;350:485–493. [PMC free article] [PubMed] [Google Scholar]
- 33.Tuckwell D, Calderwood DA, Green LJ, Humphries MJ. J. Cell Sci. 1995;108:1629–1637. doi: 10.1242/jcs.108.4.1629. [DOI] [PubMed] [Google Scholar]
- 34.Coe APF, Askari JA, Kline AD, Robinson MK, Kirby H, Stephens PE, Humphries MJ. J. Biol. Chem. 2001;276:35854–35866. doi: 10.1074/jbc.M103639200. [DOI] [PubMed] [Google Scholar]
- 35.Ridgway JB, Presta LG, Carter P. Protein Eng. 1996;9:617–621. doi: 10.1093/protein/9.7.617. [DOI] [PubMed] [Google Scholar]
- 36.Xiong JP, Li R, Essafi M, Stehle T, Arnaout MA. J. Biol. Chem. 2000;275:387623–8767. doi: 10.1074/jbc.C000563200. [DOI] [PubMed] [Google Scholar]
- 37.Staatz WD, Rajpara SM, Wayner EA, Carter WG, Santoro SA. J Cell Biol. 1989;108:1917–1924. doi: 10.1083/jcb.108.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grzesiak JJ, Davis GE, Kirchhofer D, Pierschbacher MD. J Cell Biol. 1992;117:1109–1117. doi: 10.1083/jcb.117.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barton SJ, Travis MA, Askari JA, Buckley PA, Craig SE, Humphries MJ, Mould AP. Biochem J. 2004;380:401–407. doi: 10.1042/BJ20031973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu C, Oxvig C, Springer TA. J. Biol. Chem. 1998;273:15138–15147. doi: 10.1074/jbc.273.24.15138. [DOI] [PubMed] [Google Scholar]
- 41.Oxvig C, Springer TA. Proc. Natl. Acad. Sci. USA. 1998;95:4870–4875. doi: 10.1073/pnas.95.9.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park EJ, Mora JR, Carman CV, Chen J, Sasaki Y, Cheng G, von Andrian UH, Shimaoka M. J. Clin. Invest. 2007;117:2526–2538. doi: 10.1172/JCI31570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murcia M, Jirouskova M, Li J, Coller BS, Filizola M. Proteins. 2008;71:1779–1791. doi: 10.1002/prot.21859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harding MM. Acta Crystallogr. D Biol. Crystallogr. 2001;57:401–411. doi: 10.1107/s0907444900019168. [DOI] [PubMed] [Google Scholar]
- 45.Grzesiak JJ, Pierschbacher MD. J. Clin. Invest. 1995;95:227–233. doi: 10.1172/JCI117644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grzesiak JJ, Bouvet M. Int. J. Cancer. 2008;122:2199–2209. doi: 10.1002/ijc.23368. [DOI] [PubMed] [Google Scholar]
- 47.Choi S, Vilaire G, Marcinkiewicz C, Winkler JD, Bennett JS, DeGrado WF. J. Med. Chem. 2007;50:5457–5462. doi: 10.1021/jm070252b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Käpylä J, Pentikäinen OT, Nyrönen T, Nissinen L, Lassander S, Jokinen J, Lahti M, Marjamäki A, Johnson MS, Heino J. J. Med. Chem. 2007;50:2742–2746. doi: 10.1021/jm070063t. [DOI] [PubMed] [Google Scholar]