Summary
Integrin α5β1 is a key receptor for the extracellular matrix protein fibronectin. Antagonists of human α5β1 have therapeutic potential as anti-angiogenic agents in cancer and diseases of the eye. However, the structure of the integrin is unsolved and the atomic basis of fibronectin and antagonist binding by α5β1 is poorly understood. Here we demonstrate that zebrafish α5β1 integrins do not interact with human fibronectin or the human α5β1 antagonists JSM6427 and cyclic peptide CRRETAWAC. Zebrafish α5β1 integrins do bind zebrafish fibronectin-1, and mutagenesis of residues on the upper surface and side of the zebrafish α5 subunit β-propeller domain shows that these residues are important for the recognition of RGD and synergy sites in fibronectin. Using a gain-of-function analysis involving swapping regions of the zebrafish α5 subunit with the corresponding regions of human α5 we show that blades 1-4 of the β-propeller are required for human fibronectin recognition, suggesting that fibronectin binding involves a broad interface on the side and upper face of the β-propeller domain. We find that the loop connecting blades 2 and 3 of the β-propeller (D3-A3 loop) contains residues critical for antagonist recognition, with a minor role played by residues in neighbouring loops. A new homology model of human α5β1 supports an important function for D3-A3 loop residues Trp-157 and Ala-158 in the binding of antagonists. These results will aid the development of reagents that block α5β1 functions in vivo.
Keywords: integrin, fibronectin, RGD, synergy region, antagonist, interactions, gain of function
INTRODUCTION
The interactions of integrin receptors with extracellular-matrix macromolecules are critical for development, responses to injury and normal tissue homeostasis [1-3]. Integrin α5β1 is a fibronectin receptor found on a wide variety of cell types. Two distinct sites in fibronectin are involved in binding to α5β1; the first lies in the tenth type III repeat (3Fn10) and encompasses the Arg-Gly-Asp (RGD) sequence [4], while a second, weaker, interaction site is found in the ninth type III repeat (3Fn9) and includes the sequence Pro-His-Ser-Arg-Asn (PHSRN; the so-called ‘synergy’ sequence) [5-7]. These two sequences are separated by 30-40 Å in the tertiary structure of this region of fibronectin [8].
α5β1 is also highly expressed in pathological situations, for example on many tumour cell types and on activated endothelial cells during the formation of tumour vasculature [9-13]. Moreover, there is evidence that α5β1-fibronectin interactions play a key role in diseases of the eye that involve neovascularisation [14-18]. Hence, there is a great deal of interest in the development of antagonists of α5β1 for therapeutic use [19,20].
Presently, no high resolution structural information has been obtained for α5β1-ligand interactions. However, crystal structures of the closely related integrins αVβ3 and αIIbβ3 [21-23] show that the RGD sequence binds in a pocket formed by loops on top of the α subunit β-propeller domain and the A-domain of the β subunit (known as βA or βI). Small molecule antagonists bind in the same pocket [22] thereby preventing ligand recognition. Homology models of α5β1 have been constructed using the structure of αVβ3 bound to the antagonist cilengitide [24, 25] as a template; these models have aided the rational design of high affinity α5β1 antagonists such as JSM6427 [19]. Phage display technology has also been used to identify high affinity peptide ligands for α5β1 such as the disulphide-bridged, cyclic peptides Cys-Arg-Arg-Glu-Thr-Ala-Trp-Ala-Cys (CRRETAWAC), Cys-Arg-Gly-Asp-Gly-Phe-Cys (CRGDGFC), and Cys-Arg-Gly-Asp-Gly-Trp-Cys (CRGDGWC) [26-27]. These peptides have been shown to interact with loops on the upper face of the α5 subunit β-propeller domain [28-30]. However, few data are currently available concerning the precise mode of antagonist binding by α5β1. In addition, there is controversy concerning the mechanism of fibronectin recognition, specifically whether the synergy site binds directly to the α5 subunit [31], or indirectly supports the binding of RGD to the integrin [32]. Hence, there is a pressing need to define further the mechanisms of ligand binding by α5β1.
Previously, we identified two close homologues of the mammalian β1 subunit in zebrafish: β1-1 and β1-2 (also known as β1a and β1b) [33]. Here we show that the zebrafish α5 subunit can form functional heterodimers with both zebrafish β1 subunits or with human β1; however, all of these zebrafish α5 integrins show no binding to human fibronectin or human α5β1 antagonists. This lack of ligand binding enabled us to use a gain-of-function approach to identify the regions of the α5 subunit required for interactions with human fibronectin and antagonists. We demonstrate that a loop region between the second and third blades of the β-propeller (D3-A3 loop) plays a key role in antagonist binding, but a much more extensive region of the β-propeller is necessary for recognition of the whole fibronectin ligand. A new homology model of α5β1, based on the αIIbβ3-tirofiban structure [22], supports a prominent function for residues at the apex of the D3-A3 loop in the binding of antagonists.
EXPERIMENTAL
Materials
Peptides GACRRETAWACGA (CRRETAWAC), GACRRETADACGA (CCRETADAC) and GCRGDSPCG (cyclic-RGD) were purchased from Peptide 2.0 Inc. (Chantilly, VA, USA). Peptides were cyclized by oxidation as previously described [28]. Small molecules JSM6427 and JSM6406 were provided by Jerini AG.
Cloning and mutagenesis
Full-length zebrafish α5 cDNA clone was a gift from S. Koshida (Okazaki, Japan). Full-length cDNA clones for two zebrafish β1 subunits β1-1 and β1-2 were obtained as previously described [33]. Recombinant soluble versions of human, zebrafish and chimeric integrins fused to the hinge region and CH2 and CH3 domains of the human IgGγ1 chain were made essentially as before [34], using the complete extracellular domain of the α5 subunit and the headpiece of the β1 subunit (PSI, hybrid and βI domains). For cloning into the pEE12.2Fc vector [34], an internal Hind III site in zebrafish α5 was altered from AAGCTT to AAACTT by overlap extension PCR, creating a silent mutation. Constructs were verified by DNA sequencing. The amino acid sequence of the α5 construct was identical to that predicted by B3DJJ0: UniProtKB/TrEMBL (although note that the signal peptide should be 32, not 14, amino acid residues in length) with the exception that Ser488 in the mature sequence was substituted by Asn. This substitution lies outside the β-propeller domain and so is unlikely to affect function. The sequences of β1-1 and β1-2 were identical to those previously reported (DQ149101 and DQ149102: GenBank). Constructs containing the head region of the β1-1 or β1-2 (equivalent to the previously reported β1TR construct [34]) were made by cloning a PCR product containing residues Gln-21–Pro-475 of β1-1 (Gln-1–Pro-455 in the mature sequence) or Gln-18–Pro-472 (Gln-1–Pro-455 in the mature sequence) of β1-2 fused with a murine antibody leader sequence [34,39] into the pV.16hFc vector. The corresponding human α5- and β1TR-Fc clones [34] were a gift from J. Askari (University of Manchester). Blade swapping chimeras were created by overlap extension PCR using human α5 and zebrafish α5-Fc constructs as templates. W1-3 blade swap contains residues Phe-1–Asp-228 of human α5, W1-4 swap contains residues Phe-1–Met-281 of human α5, W2-4 swap contains residues Ile-70–Met-281 of human α5. Loop-swapping mutations in zebrafish α5 were created using overlap extension PCR. To determine the sequences for swapping, alignment of zebrafish and human sequences was performed using ClustalW (www.ebi.ac.uk/Tools/clustalw2/index.html). Site-directed mutagenesis was carried out using the QuikChange II XL kit (Stratagene, Dorchester) or by overlap extension PCR. Oligonucleotides for cloning and mutagenesis were supplied by Eurogentec (Seraing, Belgium), oligonucleotides for alanine scanning mutations were a gift from S. Holley (Yale University, CT, USA) or from Eurogentec. Wild-type human α5-Fc and human β1TR-Fc constructs were prepared as previously described [34].
A cDNA encoding a cell binding fragment of zebrafish FN-1 (3Fn6-10 including the constitutively included EIIIB domain [35]) was obtained by reverse transcription of RNA from 3-day old zebrafish embryos [33]. The sequence of the cDNA was identical to that predicted by B3DGZ1 (UniProtKB/TrEMBL database; residues Thr1093–Thr1632) but differed from the published sequence [35] at the 3Fn8/9 boundary. The cDNA was cloned into the pCEP-PU expression vector, which encoded a C-terminal FLAG-tag.
Transient transfection of CHO cells
Chinese hamster ovary cells L761h variant [34] were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% foetal calf serum, 2 mM glutamine and 1% non-essential amino acids (Invitrogen, Paisley). 6-well plates of sub-confluent CHO-L761h cells were transfected with 1 μg of β1 construct and 1 μg of α5 construct/well using LipofectaminePLUS reagent (Invitrogen) according to the manufacturer’s instructions. After 6 days, culture supernatants were harvested by centrifugation at 1000x g for 5 min.
Western Blotting
All the reagents for SDS-PAGE were from Invitrogen except for the protein markers that were obtained from Bio-Rad Laboratories (Hemel Hempstead). Aliquots of cell culture supernatants were run on 3-8 % NuPAGE™ Tris-Acetate gels (Invitrogen) under non-reducing conditions, transferred to nitrocellulose and blotted with anti-human Fc peroxidase conjugate (Stratech Scientific, New Market). Bands were visualized using UptiLight enhanced chemiluminescence reagent (Cheshire Biosciences, Chester).
Solid-phase ligand-binding assays
The 50 kDa cell binding domain of human fibronectin (3Fn6-10; ‘50K’) was produced in E. coli as before [6]. The 70 kDa cell binding fragment of zebrafish FN-1 (‘70K’) was produced by transient transfection of 293-EBNA cells using LipofectAMINE PLUS reagent according to the manufacturer’s instructions. Cells were cultured in DMEM/F12 with Glutamax containing 0.1 unit/ml penicillin, and 10 μg/ml streptomycin for 7 days. 70K was purified from the cell culture medium using anti-FLAG M2 affinity gel (Sigma-Aldrich, Poole) chromatography.
For receptor–ligand binding assays, the binding of biotinylated 50K or 70K fibronectin fragments to recombinant receptors (captured from cell culture supernatants using goat polyclonal anti-human Fc) was measured in the presence of 1mM Mn2+ at room temperature as previously described [36]. Measurements obtained were the mean ± S.D. of four replicate wells. Background binding to wells coated with BSA alone was subtracted from all measurements.
In all assays comparing the binding of fibronectin fragments to wild-type and mutant receptors, the level of ligand binding to mutant receptors was normalised relative to that of wild-type zfα5β1-1 by measuring the binding of the anti-human Fc peroxidase conjugated antibody to a parallel set of replicate wells for each receptor [34]. Each experiment shown is representative of at least three separate experiments. Binding or inhibition curves were fitted using global optimization by simulated annealing (GOSA-fit, www.bio-log.biz).
Apparent KI values for inhibitors were calculated using the formula: KI = IC50/(1 + [ligand]/KD), where IC50= concentration of inhibitor for 50% inhibition of binding and KD= apparent affinity of ligand binding to α5β1. In inhibition assays the concentration of ligand was 2–3 nM and inhibitor concentrations were 0.1–100 μg/ml for CRRETAWAC and 0.1–300 nM for JSM6427 (prepared by serial dilution).
Homology modelling
Alignment of α5 with αV and αIIb, and β1 with β2 and β3 was performed using Clustal W2 (www.ebi.ac.uk). This alignment was identical to that of Xiong and co-workers [21] within the β-propeller and βI/A domains. Homology modelling of the α5 β-propeller domain, and β1 head region, was carried out using Modeller 9v6 [37] (http://salilab.org/modeller/release.html) using the 2vdr structure of αIIbβ3 [22] as a template, except for residues Arg-220–Tyr-233 of α5 where the 1L5G structure of αV [21] was used as a template. Lowest energy models were chosen, and selected loops were refined using Modeller. Final quality of the model was assessed using PROCHECK [38] (http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/upload.html): 87.8% of side chains were in the most favoured regions, 10.9% in additional allowed regions, 0.5% in generously allowed, and 0.8% in disallowed regions of the Ramachandran plot. Residues in disallowed or generously allowed regions of the plot did not contribute to the ligand-binding pocket. DS Vizualizer 2.0 (Accelerys Software Inc.) was used to visualize the model. Docking of a fragment of JSM6427 into the ligand-binding pocket was performed with the constraints of placing the 2-amino group in a suitable position for hydrogen bonding to the carboxylate group of Asp-227 on the α5 subunit and orientation of the carboxylate group of JSM6427 towards the MIDAS cation on the β subunit. The program PyMOL (version 0.99) (http://www.pymol.org) was used to display the model.
Results
Characterization of zebrafish α5 integrins
Constructs containing the complete extracellular domain of the zebrafish α5 subunit and the head region of the β1-1 and β1-2 subunits fused to the hinge region and CH2 and CH3 domains of the Fc region of the human IgGγ1 chain were created as described in ‘Experimental’. These constructs were chosen because expression of the head and leg regions of the α subunit together with the head region of the β subunit has previously been shown to be sufficient to create a constitutively active integrin [40]. The zebrafish α5 construct (zfα5-Fc) was transiently co-expressed with β1-1, β1-2 or human β1TR [34] (huβ1) Fc constructs in CHOL761h cells. Recombinant integrins were partially purified from the cell culture medium and the formation of heterodimers was examined using Western blotting. The results (Figure 1A) showed that in each case a predominant band at ~240 kDa was observed, corresponding to the expected mass of an α,β-Fc heterodimer. Under reducing conditions two predominant bands were observed at ~ 50 and ~ 100 kDa, concordant with the expected sizes of α5 light chain-Fc and β1-Fc subunits, respectively (not shown). Together, these data show that zebrafish α5 can form heterodimers not only with zebrafish β1-1 and β1-2 but also with human β1.
Figure 1. Preparation and characterization of recombinant soluble α5 integrins.

(A) Western blotting of recombinant-soluble zebrafish (zf) α5β1-Fc integrins. The expected migration positions of α,β heterodimer and α,α homodimers are indicated by arrows. In each case a predominant band at ~ 240 kDa is observed, at the expected size for the zfα5β1-Fc heterodimer. (B) Western blotting of recombinant-soluble human (hu) α5β1-Fc integrins. A predominant band at ~ 260 kDa is observed, at the expected size for the huα5β1-Fc heterodimer. These bands are not observed in proteins purified from the supernatants of mock-transfected cells.
Zebrafish α5 integrins bind to zebrafish but not human fibronectin
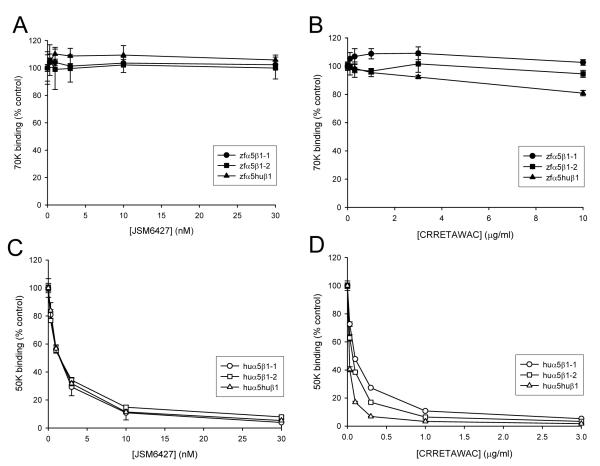
A recombinant fragment of zebrafish fibronectin-1 (FN-1) containing the α5β1 binding sites (type III repeats 6-10) was produced in mammalian cells as described in ‘Experimental’. Since this fragment has an apparent molecular mass of approx. 70kDa by SDS-PAGE, we refer to this protein as ‘70K’. The 70K protein has a similar synergy region to human fibronectin (PPSRS in place of PHSRN) and an identical RGD-containing loop (Supplementary Figure S1). To test whether the recombinant integrins were functional, binding of biotin-labelled 70K to recombinant zebrafish α5 integrins was examined in solid-phase assays. Ligand binding was measured in the presence of 1mM Mn2+ in order to fully activate the receptors [36]. The results (Figure 2A) showed that each receptor bound strongly to 70K in a concentration-dependent and saturable manner. 70K binding could be inhibited by EDTA or a cyclic RGD peptide, showing that the interaction was specific (Supplementary Figure S2). Apparent affinities could be calculated from the binding curves (Supplementary Table 1) and these data showed that the affinity of zfα5β1-1 for FN-1 was about 3-fold higher than that of zfα5β1-2. Surprisingly, zfα5huβ1 bound 70K with the highest affinity. We also examined whether the zebrafish integrins could bind to an equivalent recombinant fragment of human fibronectin (‘50K’ [6]). The data (Figure 2B) showed that there was essentially no binding of zfα5β1-1 or zfα5β1-2, and only very low binding of zfα5huβ1 to 50K.
Figure 2. Fibronectin binding to recombinant integrins in solid phase assays.
(A) Binding of 70K fragment of zebrafish fibronectin to zfα5β1-1, zfα5β1-2 and zfα5huβ1. (B) Binding of 50K fragment of human fibronectin to zfα5β1-1, zfα5β1-2 and zfα5huβ1. (C) Binding of 50K to huα5β1-1, huα5β1-2 and huα5huβ1. (D) Binding of 70K to huα5β1-1, huα5β1-2 and huα5huβ1.
Human α5 can form functional heterodimers with zebrafish β1 subunits
As described above, we found that zebrafish α5 could associate with human β1. We next tested if the human α5 subunit could form heterodimers with the two zebrafish β1 subunits. The human α5-Fc construct (huα5-Fc) was co-transfected with huβ1, β1-1, or β1-2-Fc constructs in CHO L761h cells. Cell culture supernatants were analysed for the formation of heterodimers as described above (Figure 1B). The results showed that a predominant band of ~260 kDa in each case, the expected size of the heterodimer. The apparent molecular mass of the human α5 heterodimers was observed to be slightly greater than that of the corresponding zebrafish receptors (Figure 1B), probably due to the presence of additional glycosylation sites in human α5 [41]. Heterodimer formation was also assessed using mAbs recognizing the β-propeller domain of the α5 subunit (Supplementary Figure S3). The epitopes of these mAbs are only expressed when the β-propeller assumes its native fold, and this folding is dependent on the association of α5 with the βI-domain [34]. We found that anti-α5 β-propeller epitopes were expressed to a similar level for huα5β1-1, huα5β1-2 and huα5huβ1 receptors. No binding of these antibodies was observed when cells were transfected with the α5 subunit alone. Collectively, these data demonstrate that the human α5 subunit can form heterodimers with zebrafish β1-1 and β1-2.
We next examined if the human α5 integrins could bind to the human 50K fibronectin fragment. The results (Figure 2C and Supplementary Table 1) showed that all three receptors bound with high affinity to 50K. This binding was specific since it could be fully inhibited by EDTA or cyclic RGD peptide (Supplementary Figure S2B). We then tested if the human receptors could bind to the zebrafish 70K protein (Figure 2D). We found that the human α5 integrins could interact with zebrafish fibronectin fragment but with markedly lower affinity than with the human protein (Supplementary Table 1).
In summary, we found that zebrafish α5 integrins interacted strongly with the FN-1 fragment but not with the human protein, whereas the human α5 integrins bound preferentially to human fibronectin fragment. To further support these findings we investigated if zfα5β1-1-Fc could bind to purified human plasma fibronectin (huFN) in an ‘inverted’ assay in which fibronectin fragments or whole fibronectin are coated onto a 96-well plate and the integrin is in the solution phase. The results (Supplementary Figure S4A) showed that zfα5β1-1-Fc bound well to 70K but not to 50K or huFN. In contrast, huα5β1-1-Fc bound poorly to 70K but strongly to 50K or huFN (Supplementary Figure S4B). We also found that zebrafish fibroblasts, which express zfα5β1-1, spread very well on the 70K zebrafish protein but poorly on the 50K human protein (Supplementary Figure S5), suggesting that our in vitro data is relevant to the function of cell-surface integrins.
Alanine-scanning mutagenesis identifies residues in loops on the side and upper surface of the zebrafish α5 subunit β-propeller involved in the binding of fibronectin
We next examined whether the mechanism of fibronectin binding to zebrafish α5 integrins was similar to that previously reported for human α5β1. In human α5, residues around Phe-187 and Asp-227 are predicted to be important for the binding of the RGD sequence [21,42], while Tyr-208 and Ile-210 appear to play a role in recognition of the synergy region [31]. Based on an alignment with human α5 (Supplementary Figure S7) alanine mutations were made in zebrafish α5 residues in either the RGD (F183A, D224A, D225A) or proposed synergy region (Y204A, L206A) binding sites [31]. These residues lie in loops on the upper face or side of the β-propeller domain (the corresponding residues in human α5 are Phe-187, Asp-227, Asp-228, Tyr-208 and Ile-210). CHO cells were transfected with wild-type or α5 mutants together with β1-1-Fc and recombinant integrins were tested for binding to 70K. The results (Supplementary Figure S6, Table 1) showed that the Tyr-204 or Leu-206 putative synergy region binding site mutations reduced the affinity of ligand binding approx. 30-fold. A previously described mutation of Tyr-204 to asparagine (Y204N) that causes a loss-of-function phenotype in vivo [43] had a more severe effect on ligand recognition than the Y204A mutation. The F183A and D224A putative RGD binding site mutations essentially abrogated ligand recognition. The D225A mutation had an intermediate effect (Supplementary Figure S6, Table 1) and a control mutation K154A in a neighbouring loop had no effect on ligand binding (not shown). In summary, the same residues appear to be important for recognition of fibronectin by both zebrafish and human α5 integrins.
Table 1.
Apparent affinities of binding of wild-type and mutant integrins to 70K and 50K fibronectin fragments
| integrin | 70K | 50K |
|---|---|---|
|
| ||
| huα5β1-1 | 25 ± 2 | 1.3 ± 0.2 |
| zfα5β1-1 | 2.8 ± 0.2 | ND |
| zfα5β1-1(D225A) | 42 ± 14 | ND |
| zfα5β1-1(Y204A) | 79 ± 7 | ND |
| zfα5β1-1(L206A) | 106 ± 16 | ND |
| zfα5(huW1-4)β1-1 | 14 ± 1 | 1.3 ± 0.1 |
| zfα5(EPEK)β1-1 | 4.6 ± 0.1 | ND |
| zfα5(SWAA)β1-1 | 1.7 ± 0.1 | ND |
| zfα5(ASSIY)β1-1 | 8.3 ± 0.3 | ND |
| Triple swap | 5.8 ± 0.4 | ND |
Apparent KD values (nM) for binding to 70K fragment of zebrafish fibronectin or 50K fragment of human fibronectin. Values are mean ± S.D.; n=3-5 experiments. ND: not determined
Zebrafish α5 integrins are not inhibited by human α5 antagonists
We next investigated whether there were any differences between the human and zebrafish integrins in their sensitivity to human α5β1 antagonists. For these assays we used the small molecule JSM6427 (Supplementary Figure S8 [19]) and the cyclic peptide CRRETAWAC [26]. The ability of these compounds to inhibit binding of 50K to human α5 integrins or 70K to zebrafish integrins was tested. The data (Figure 3 A-D) showed that all the human α5 receptors were potently inhibited by JSM6427 and CRRETAWAC but these reagents had little or no effect on the zebrafish heterodimers. In control experiments, the compound JSM6406 (an inactive derivative of JSM6427) and the peptide CRRETADAC in which the essential tryptophan residue is replaced by aspartate were ineffective for blocking 50K binding to huα5β1-1 (Supplementary Figure S9). Apparent KI values calculated from the inhibition curves for JSM6427 and CRRETAWAC (Supplementary Table 2) showed that the affinity of JSM6427 binding was unaffected by the β subunit partner of human α5; similarly, only small differences were observed between the apparent KI values of CRRETAWAC for huα5β1-1, huα5β1-2 and huα5huβ1. Similar KI values were obtained when JSM6427 or CRRETAWAC were used to inhibit binding of the human α5 integrins to 70K (Supplementary Table 2). These findings show that the α subunit, not the β, determines the specificity and affinity of antagonist binding. Since zebrafish α5 integrins do not recognise human fibronectin or human α5 antagonists, these data suggested that a gain-of-function approach could be employed to define the regions within the α5 subunit that confer ligand-binding specificity.
Figure 3. Effect of human α5β1 antagonists on fibronectin binding to recombinant integrins.
(A) Effect of JSM6427 on binding of 70K fragment of zebrafish fibronectin to zfα5β1-1, zfα5β1-2 and zfα5huβ1. (B) Effect of CRRETAWAC on binding of 70K to zfα5β1-1, zfα5β1-2 and zfα5huβ1. (C) Effect of JSM6427 on binding of 50K fragment of human fibronectin to huα5β1-1, huα5β1-2 and huα5huβ1. (D) Effect of CRRETAWAC on binding of 50K to huα5β1-1, huα5β1-2 and huα5huβ1. Binding is expressed as a percentage of 70K or 50K binding in absence of inhibitor.
Swapping blades 1-4 of the zebrafish α5 subunit β-propeller domain results in a complete gain of function for binding to human fibronectin and human α5β1 antagonists
To identify regions of the human α5 subunit that are essential for ligand recognition portions of the zebrafish α5 subunit were exchanged with the corresponding regions of the human subunit (Supplementary Figure S7) and the binding of fibronectin fragments or antagonists tested in solid-phase assays. Previously, we have used a similar approach to show that a region of the human α5 subunit β-propeller encompassing blades 2 and 3 contains key sites involved in ligand recognition [29]. As a first step, we swapped blades 2 and 3 of the zebrafish α5 β-propeller with the human α5 sequence and co-expressed the chimeric α5 subunit with β1-1-Fc subunit. We found that this exchange was not sufficient to create a receptor that bound to human fibronectin (not shown); however, exchange of a larger section of the propeller (blades 1 to 3; W1-3) gave weak gain of 50K binding, and exchange of blades 1 to 4 (W1-W4) resulted in a near-complete gain of binding (Figure 4A). Exchange of blades 2 to 4 (W2-W4) only was not sufficient for full gain of binding, suggesting that all the first four blades are important in forming the ligand-binding pocket for fibronectin. The W1-W4 chimera recognized the 50K human and 70K zebrafish fibronectin fragments with very similar apparent affinities as the wild-type huα5β1-1Fc (Figures 4A and 4B, Table 1). The W1-4 chimera also expressed the epitopes of human anti-α5 mAbs to a similar extent as huα5β1-1 (Supplementary Figure S10).
Figure 4. Characterization of chimaeric zebrafish/human α5β1-1 blade swap integrins.
Binding of wild-type huα5β1-1, and zfα5(huW1-3)β1-1, zfα5(huW2-4)β1-1 and zfα5(huW1-4)β1-1 blade swap receptors to (A) 50K fragment of human fibronectin and (B) 70K fragment of zebrafish fibronectin. No significant binding of wild-type zfα5β1-1 to 50K was observed in the same assay (not shown).
We next tested whether the zfα5(huW1-W4)β1-1Fc chimeric integrin could recognise the human α5 antagonists. The results (Table 2) showed that the chimera bound these antagonists with essentially the same apparent KI values as the wild-type huα5β1-1. Hence, blades 1-4 of the β-propeller contain all the sequences necessary for fibronectin and antagonist binding.
Table 2.
Apparent KI values of JSM6427 and CRRETAWAC for wild-type and chimaeric α5β1-1 integrins
| JSM6427 | CRRETAWAC | |||
|---|---|---|---|---|
| integrin | 70K | 50K | 70K | 50K |
| huα5β1-1 | ND | 0.65 ± 0.15 | ND | 47 ± 9 |
| zfα5(huW1-W4)β1-1 | ND | 0.75 ± 0.04 | ND | 31 ± 12 |
| zfα5(SWAA)β1-1 | 7.2 ± 2.4 | ND | 126 ± 43 | ND |
| Triple swap | 1.21 ± 0.04 | ND | 76 ± 20 | ND |
Apparent KI values (nM) for 70K fragment of zebrafish fibronectin or 50K fragment of human fibronectin binding to recombinant integrins. Values are mean ± S.D.; n=3-6 experiments. ND: not determined.
Swapping a loop region in blade 3 of the β-propeller confers a partial gain of function for binding of human α5β1 antagonists
Although swapping a large region of the β-propeller was necessary for gain of fibronectin binding we reasoned that, since the binding interface of low molecular weight antagonists is likely to cover a much reduced area compared to that of fibronectin, it may be possible to swap only small regions of the β-propeller to gain antagonist binding. We chose to swap predicted loop regions on the top face of the β-propeller in repeats 2 to 4, since previously we have shown that these loops are involved in the binding of RGD and CRRETAWAC [28-30]. Three loop swap mutations were made: KEDTPH to EKEPL (B2-C2 loop), RGRK to SWAA (D3-A3 loop) and RKEIRF to ASSIY (D4-A4 loop) (Supplementary Figure S7). No changes were made to the B3-C3 loop, since it is identical in zebrafish and human α5. These mutations only had a small effect on the affinity of 70K binding (Figure 5A, Table 1). We then examined whether JSM6427 or CRRETAWAC could inhibit 70K binding. The results (Figure 5B,C) showed that the SWAA loop swap mutant was strongly inhibited by the antagonists, whereas these compounds had little or no effect on the EKEPL or ASSIY loop swap mutants. Hence, the D3-A3 loop contains residues that are essential for antagonist recognition. To determine which of these four residues were critical for antagonist binding we made mutants in which one (RGRA), two (RGAA) or three (SGAA) residues were swapped. JSM6427 inhibited the binding of 70K to the SGAA and RGAA mutants but not the RGRA mutant, and more strongly inhibited 70K binding to the SWAA mutant (Figure 6A, Figure 6 legend). In contrast, CRRETAWAC only inhibited the interaction of 70K with the SWAA mutant (Figure 6B). These results indicate that both the tryptophan residue and the first alanine residue in the SWAA sequence play important roles in JSM6427 binding but only the tryptophan residue is critical for gain of CRRETAWAC binding. The serine and the second alanine residues do not appear to be necessary for the gain of JSM6427 binding.
Figure 5. Characterization of chimaeric zebrafish/human α5β1-1 loop swap integrins.
(A) Binding of wild-type zfα5β1-1, zfα5(EPEK)β1-1, zfα5(SWAA)β1-1 and zfα5(ASSIY)β1-1 loop swap receptors to 70K fragment of zebrafish fibronectin. (B) Effect of JSM6427 on binding of loop swap receptors to 70K. (C) Effect of CRRETAWAC on binding of loop swap receptors to 70K. In panels B and C 70K binding is expressed as a percentage of binding in absence of inhibitor.
Figure 6. Effect of human α5β1 antagonists on fibronectin binding to D3-A3 swap mutants.
Effect of JSM6427 (A) and CRRETAWAC (B) on binding of zfα5(SGAA)β1-1 swap zfα5(RGAA)β1-1 swap, zfα5(RGRA)β1-1 swap, zfα5(SWAA)β1-1 loop swap and wild-type zfα5β1-1 to 70K fragment of zebrafish fibronectin. One experiment, representative of three separate experiments. In this set of experiments, apparent KI values for JSM6427 were calculated as 25 ± 4, 29 ± 6, and 4.2 ± 0.2 nM (mean ± SD) for zfα5(SGAA)β1-1, zfα5(RGAA)β1-1, and zfα5(SWAA)β1-1, respectively.
Apparent KI values for the SWAA mutant (Table 2) showed that the apparent affinity of CRRETAWAC binding was approx 3-fold lower, and that for JSM6427 was approx 10-fold lower, than for wild-type human α5 integrin. Hence, although the D3-A3 loop contains residues that are essential for antagonist recognition other residues outside of this loop may be necessary for a full gain of function.
A triple loop swap results in a complete gain of function for binding of human α5β1 antagonists but only a minor gain of binding to human fibronectin
To investigate whether residues other than those in the SWAA sequence are involved in the binding of the antagonists, a construct with a triple loop swap KEDTPH to EKEPL, RGRK to SWAA and RKEIRF to ASSIY (Supplementary Figure S7) was created. The triple swap only had a minor effect on the affinity of 70K binding (Table 1). We then examined the effects of JSM6427 and CRRETAWAC on 70K binding to this mutant. The results (Supplementary Figure 11A,B) showed that 70K binding to the triple swap mutant was inhibited more potently by the antagonists than the single SWAA swap. Apparent KI values for the triple swap mutant were very close to those of wild-type huα5β1-1 integrin (Table 2). Hence, the exchanged residues in the B2-C2 and D4-A4 loops also appear to play a role in antagonist binding.
Although the triple loop swap mutant fully restored antagonist binding, this receptor demonstrated only a small increase of binding to 50K (Supplementary Figure 11C). Hence, the triple swap was not sufficient to restore recognition of human fibronectin.
A new homology model of the α5β1 ligand-binding pocket supports a prominent role for D3-A3 loop residues in binding of human α5 antagonists
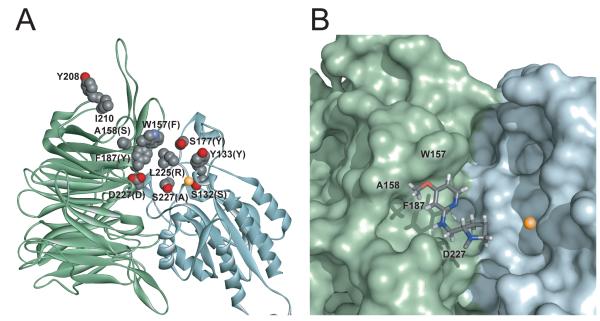
Homology modelling of human α5β1 has guided the development of antagonists [19,24,25]. However, these models were based on the medium resolution αVβ3-cilengitide structure [21]. The αIIβ3-γ chain peptide structure, 2vdr [23], was chosen as a template for homology modelling of α5β1 rather than the αVβ3-cilengitide structure (1L5G) for three reasons: (i) the 2vdr structure is of higher resolution than the 1L5G structure (2.4 versus 3.2 Å), (ii) the 2vdr structure is a high affinity open conformation of the integrin, whereas the 1L5G structure represents a lower affinity closed conformation, (iii) the αV subunit has an aspartate residue in the D3-A3 loop (Asp-150) that interacts with the arginyl side chain of the ligand; there is unlikely to be an equivalent interaction in α5, instead the D3-A3 loop of α5 contains mainly hydrophobic residues and is more similar to αIIb in the same region. However, we used the 1L5G structure for modelling the D4-A4 loop of α5 because the αV subunit has an Asp residue at the same position as Asp-227 of α5 whereas αIIb has a Phe residue. Homology modelling was carried out as described in ‘Experimental’. The final model (Figure 7A, Supplementary Figure 12) shows residues that have been proposed to be critical for ligand binding in α5 ([31,42] and this study) or β1 [22,24]. On the α subunit side, residues in the D3-A3 and D4-A4 loops (Trp-157, Ala-158 and Asp-227) form walls on either side of the binding pocket for antagonists, while Phe-187 in the B3-C3 loop lines the base of the pocket. Potentially, other residues in these loops and the nearby B2-C2 loop could also participate in antagonist binding. On the β subunit side, residues in the β1-α1 loop of the βI domain (Ser-132 and Tyr-133) form another wall of the pocket and nearby residues Leu-225, Ser-227 and Ser-172 are also predicted to have essential roles [22,24]. Recognition of the synergy site in fibronectin involves residues Tyr-208 and Ile-210, situated in the C3-D4 loop on the side of the β-propeller [31]. A possible binding mode for the guanidine-mimetic region of JSM6427 is shown in Figure 7B.
Figure 7. Homology model of the ligand-binding region of human α5β1.
A. Ribbon diagram of the α subunit β-propeller domain (green) and β subunit I/A domain (pale cyan). The structure of αIIbβ3 [22] was used as a template for modelling, except for the D4-A4 loop where the structure of αVβ3 [21] was used. Model shows side chains of residues likely to be critical for ligand recognition [22,24,41,30,31, and this study]. Tyr-208 and Ile-210 on the side of the β-propeller contribute to binding of the synergy region of fibronectin, while the other labelled residues contribute to the binding of the RGD sequence in fibronectin and/or α5β1 antagonists. Side chain atoms are depicted as spheres with carbon, nitrogen and oxygen atoms coloured in grey, blue and red, respectively. Residues in the RGD-binding pocket of the corresponding αIIb (or αV) or β3 subunits are indicated in parentheses. The MIDAS cation is depicted by the gold sphere. B. Possible binding mode of JSM6427. A fragment of JSM6427 containing the 4-methoxy-2-aminopyridine ring (guanidine mimetic region) is shown docked into the binding pocket of α5β1. The integrin is shown as a Connolly surface with α5 subunit in green and β1 subunit in pale cyan. Fragment of JSM6427 is depicted as sticks with carbon, nitrogen, oxygen and hydrogen atoms coloured in grey, blue, red and white, respectively. The program PyMOL (version 1.1) was used to display the model.
DISCUSSION
The major findings of this report are the following: (i) the zebrafish α5 subunit can form heterodimers with zebrafish β1-1, β1-2 or human β1 subunits but these receptors do not recognise human fibronectin or human α5β1 antagonists, (ii) swapping the first four blades of zebrafish α5 β-propeller domain with those of human α5 endows the chimeric receptor with the ligand binding properties of human α5β1, (iii) exchange of the D3-A3 loop region in zebrafish α5 with the corresponding region of human α5 (residues Ser-156–Ala-159) confers zebrafish α5 integrins with the capacity to recognise human α5β1 antagonists, (iv) residues Trp-157 and Ala-158 are important for the binding of small molecule antagonists, (v) swapping of three loop regions (B2-C2, D3-A3 and D4-A4 loops) is sufficient for complete gain of antagonist binding but not of fibronectin binding (vi) homology modelling of the α5β1 ligand-binding pocket supports a prominent role for residues in the D3-A3 loop in binding of human α5β1-specific antagonists.
We found that the zebrafish α5 subunit could form heterodimers with human β1 in addition to the two zebrafish β subunits. Similarly, human α5 could form heterodimers with β1-1 and β1-2 as well as the human β1 subunit. Formation of heterodimers is known to be dependent upon an association of the β subunit I domain with the upper face of the α subunit β-propeller. One of the most important interactions that promotes heterodimer assembly is the binding of an arginine/lysine residue in the βD-βD’ loop of the βI domain to a hydrophobic ‘cage’ at the centre of the β-propeller [22,44-45]. Significantly, the aromatic residues that form the cage for the Arg/Lys side chain are conserved between human and zebrafish α5. In addition the sequence of the βD-βD’ loop is almost identical in β1-1, β1-2 and human β1. These features may explain why it is possible to form cross-species heterodimers despite the very long evolutionary separation of humans and fish (~350 My). The high level of identity between the I/A domains of β1-1, β1-2 and human β1 [33] may also explain why the specificity and affinity of ligand binding was influenced mainly by the α5 subunit and not by the β.
We do not currently have a clear explanation for why zebrafish α5 integrins are unable to interact with human fibronectin. However, an intriguing feature of zebrafish FN-1 is that a highly conserved threonine residue in the putative integrin-binding face of the molecule [9] is replaced with a much less bulky glycine residue (Supplementary Figure 13). Hence it is possible that steric clashes may interfere with human fibronectin binding to zebrafish α5β1. We found that it was necessary to swap a large section of the β-propeller (repeats 1-4) in order to gain binding to human fibronectin, suggesting that the site of fibronectin binding involves a broad interface. However, it is also possible that a relatively small interface is involved in ligand recognition but the tertiary structure of the ligand-binding loops is dependent upon the folding of adjacent loops throughout blades 1-4. The key residues involved in fibronectin recognition include Phe-187, Tyr-208, Ile-210, and Asp-227 in blades 3 and 4 of the human α5 subunit [42,33,21], and further studies will be necessary to pinpoint additional essential residues. Previously, it has been shown that the fibrinogen binding site on αIIb involves many residues in blades 2-4, covering a large portion of the upper face and side of the β-propeller [46]. Nearly all these residues are now known to lie in a specialised sub-region of the β-propeller known as the ‘cap’ [22].
Although zebrafish α5 integrins were unable to bind to human fibronectin, alanine mutagenesis suggested that the same residues involved in the interaction of human fibronectin with the human receptor also participate in zebrafish α5–FN-1 binding. For example, mutation of F183A and D224A putative RGD binding site residues fully abrogated ligand recognition. In addition, we found that mutation of residues Tyr-204 and Leu-206 in the C3-D4 loop, equivalent to Tyr-208 and Ile-210 in human α5, strongly perturbed ligand recognition. Mutation of either Tyr-204 or Leu-206 to alanine reduced the apparent affinity of fibronectin binding approx. 30-fold, whereas mutation of the corresponding residues in human α5 reduced the affinity of fibronectin binding by 5- and 200-fold, respectively [31]. These data are consistent with previous reports that loss of interaction with the synergy region leads to ~100-fold lower affinity of fibronectin binding [5,6]. A model of the interaction between α5β1 and fibronectin based on X-ray scattering data [31] predicts that the Tyr and Ile/Leu residues on the side of the β-propeller are located 30-40 Å away from the site of RGD binding, which is consistent with the spacing of the synergy region from RGD in fibronectin [8]. In agreement with our in vitro data, mutation of Tyr-204 to Asn results in a loss-of-function mutation of α5 in zebrafish [43]. The C3-D4 loop of αIIb contains residues essential for fibrinogen binding by αIIbβ3 [46], and alternative splicing of this loop can regulate the specificity of ligand interactions in the α7 and αPS subunits [47-49]. In addition this loop contains the binding site of function-blocking mAbs in αIIb and α5 [22,50]. Hence, the C3-D4 loop may form an important secondary site of ligand recognition in many integrins.
Peptides containing the sequence RGDW or RGDF are potent inhibitors of β3 integrins. The crystal structure of αIIbβ3 bound to the cyclic hexapeptide eptifibatide shows that the GDW sequence has a tight reverse turn following the Asp residue, and that this bend allows the Trp side chain to occupy a hydrophobic pocket formed by Phe-160 and Tyr190 on the αIIb subunit [22]. In contrast, RGDW and RGDF peptides are poor inhibitors of α5β1 but peptides containing the sequences RGDGW or RGDGF are very good α5β1 inhibitors [26,27]. The presence of the Gly residue immediately following the Asp residue may favour a more open (less tight) turn in these peptides when binding to α5β1. As noted previously [24,25], replacement of bulky β3 residues Tyr-166, Arg-214 and Arg-216 by Ser-177, Gly-223 and Leu-225, respectively, in β1 opens up more space adjacent to the RGD binding pocket. This additional space may allow for the more extended turn in RGDGW/F peptides. In previous homology models of α5β1 [24,25,31], which used the αVβ3 structure [21] as the template, the side chain of Trp-157 was pointing away from the pocket. In our new model, Trp-157 and Phe-187 of α5 occupy similar positions to Phe-160 and Tyr-190 in αIIb [22]. The side chains of Trp-157 and Phe-187 are suitably positioned to allow stacking interactions with the aromatic moieties of peptide antagonists, such as the phenyl group of Phe in RGDGF peptides and the indole ring of Trp in RGDGW and RRETAWA. Hence, our model helps to explain why these moieties are critical for high affinity binding of these antagonists.
We found that the specificity of antagonist binding appears to derive mainly from the α subunit. In our model (Figure 7A), residues in human β1 predicted to be important for ligand recognition by α5β1 [24] are completely conserved in zebrafish β1-1 and β1-2 [33]. Ligand binding pocket residues in human α5 are also conserved in zebrafish α5, with the important exceptions that Trp-157 is substituted by Gly and Ala-158 by Arg. We have previously shown that Trp-157 in the D3-A3 loop is involved in binding of human α5β1-specific and α5β1-selective antagonists such as CRRETAWAC and RGDGW [28-30]. Here we demonstrate that residues in this loop (SWAA) are also important for the binding of small molecule antagonists. Swapping only two loop residues (RGAA mutant) gave a partial gain of function for binding of JSM6427 but no gain of binding for CRRETAWAC. Hence, although the tryptophan residue is essential for high affinity binding of CRRETAWAC, this residue plays a more minor role in the interaction of α5β1 with JSM6427. Instead, our data suggest that Ala-158 has an important function in the binding of this compound either by direct interaction or gaining space for other interactions. The 4-methoxy substituent of the 2-aminopyridine ring of JSM6427 is involved in high affinity binding to α5β1 [19], and our modelling (Figure 7B) suggests that this moiety could interact with residues in the D3-A3 loop region.
JSM6427 is undergoing phase I clinical trials for treatment of age-related macular degeneration. Our findings will aid in the rational design of more potent and specific α5β1 antagonists. Finally, animal models such as zebrafish provide a valuable means of drug screening [51]. However, due to sequence differences in the D3-A3 loop, antagonists of human integrins often do not cross react with integrins of other species [52,53, this study]. Our results suggest that introduction of the SWAA loop swap into the zebrafish α5 gene could create an in-vivo model suitable for drug screening or discovery [54].
Supplementary Material
Acknowledgements
We thank Drs. Adam Hurlstone, Scott Holley and Jordi Bella for helpful discussions, Simon Williams, Patrick Buckley, and Guillaume Jacquemet for assistance with homology modelling, Scott Holley for gift of oligonucleotides, S. Koshida for zebrafish α5 cDNA, and Janet Askari for human α5 and β1-Fc constructs. We are grateful to Elizabeth Lord, Jennifer Hamilton and Simon Mathers for technical assistance, and Amanda Kelly and Paul Walker for PAC2 cell line and cDNA. We thank Roland Stragies (Jerini AG) for synthesis of JSM6427 and JSM6406, and Ulf Reimer (Jerini AG) and Patrick Bryant for helpful discussion of homology modelling.
Funding These studies were supported by grants 045225 and 074941 from The Wellcome Trust to MJH.
Abbreviations used are
- FN-1
fibronectin-1 (zebrafish)
- MIDAS
metal-ion dependent adhesion site
- mAb
monoclonal antibody
- PBS
phosphate-buffered saline
- BSA
bovine serum albumin
- S.D.
standard deviation
- α5β1-Fc
recombinant soluble integrin heterodimer containing the extracellular domain of the α5 subunit and a truncated β1 subunit, fused to the Fc region of human IgGγ1
- CHO
Chinese hamster ovary
- ABTS
2,2′-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid)
- EM
electron microscopy
Footnotes
Declarations of interest GZ is an employee of Jerini AG
REFERENCES
- 1.Askari A, Buckley PA, Mould AP, Humphries MJ. Linking integrin conformation to function. J. Cell Sci. 2009;122:165–170. doi: 10.1242/jcs.018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hynes RO. Integrins: bidirectional, allosteric signalling machines. Cell (Cambridge, Mass.) 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 3.Streuli CH, Akhtar N. Signal co-operation between integrins and other receptor systems. Biochem. J. 2009;418:491–506. doi: 10.1042/BJ20081948. [DOI] [PubMed] [Google Scholar]
- 4.Pierschbacher MD, Ruoslahti E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature. 1984;309:30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 5.Aota S, Nomizu M, Yamada KM. The short amino acid sequence Pro-His-Ser-Arg-Asn in human fibronectin enhances cell-adhesive function. J. Biol. Chem. 1994;269:24756–24761. [PubMed] [Google Scholar]
- 6.Mould AP, Askari JA, Aota S, Yamada KM, Irie A, Takada Y, Mardon HJ, Humphries MJ. Defining the topology of integrin α5β1-fibronectin interactions using inhibitory anti-α5 and anti-β1 monoclonal antibodies - Evidence that the synergy sequence of fibronectin is recognized by the amino-terminal repeats of the α5 subunit. J. Biol. Chem. 1997;272:17283–17292. doi: 10.1074/jbc.272.28.17283. [DOI] [PubMed] [Google Scholar]
- 7.Redick SD, Settles DL, Briscoe G, Erickson HP. Defining fibronectin’s cell adhesion synergy site by site-directed mutagenesis. J. Cell Biol. 2000;149:521–527. doi: 10.1083/jcb.149.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leahy DJ, Aukhil I, Erickson HP. 2.0 Å crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- 9.Martin S, Cosset EC, Terrand J, Maglott A, Takeda K, Dontenwill M. Caveolin-1 regulates glioblastoma aggressiveness through the control of α5β1 integrin expression and modulates glioblastoma responsiveness to SJ749, an α5β1 integrin antagonist. Biochim Biophys Acta. 2009;1793:354–367. doi: 10.1016/j.bbamcr.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 10.Färber K, Synowitz M, Zahn G, Vossmeyer D, Stragies R, van Rooijen N, Kettenmann H. An α5β1 integrin inhibitor attenuates glioma growth. Mol. Cell. Neurosci. 2008;39:579–585. doi: 10.1016/j.mcn.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Yamada SD, Lengyel E. Loss of E-cadherin promotes ovarian cancer metastasis via α5 integrin, which is a therapeutic target. Cancer Res. 2008;68:2329–2339. doi: 10.1158/0008-5472.CAN-07-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S, Bell K, Mousa SA, Varner JA. Regulation of angiogenesis in vivo by ligation of integrin α5β1 with the central cell-binding domain of fibronectin. Am. J. Pathol. 2000;156:1345–1362. doi: 10.1016/s0002-9440(10)65005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsons-Wingerter P, Kasman IM, Norberg S, Magnussen A, Zanivan S, Rissone A, Baluk P, Favre CJ, Jeffry U, Murray R, McDonald DM. Uniform overexpression and rapid accessibility of α5β1 integrin on blood vessels in tumors. Am. J. Pathol. 2005;167:193–211. doi: 10.1016/s0002-9440(10)62965-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umeda N, Kachi S, Akiyama H, Zahn G, Vossmeyer D, Stragies R, Campochiaro PA. Suppression and Regression of Choroidal Neovascularization by Systemic Administration of an α5β1 Integrin Antagonist. Molecular Pharmacology. 2006;69:1820–1828. doi: 10.1124/mol.105.020941. [DOI] [PubMed] [Google Scholar]
- 15.Maier AK, Kociok N, Zahn G, Vossmeyer D, Stragies R, Muether PS, Joussen AM. Modulation of hypoxia induced neovascularization by JSM6427, an integrin α5β1 inhibiting molecule. Curr. Eye Res. 2007;32:801–812. doi: 10.1080/02713680701553052. [DOI] [PubMed] [Google Scholar]
- 16.Muether PS, Dell S, Kociok N, Zahn G, Stragies R, Vossmeyer D, Joussen AM. The role of integrin α5β1 in the regulation of corneal neovascularization. Exp. Eye Res. 2007;85:355–356. doi: 10.1016/j.exer.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Dietrich T, Onderka J, Bock F, Kruse FE, Vossmeyer D, Stragies R, Zahn G, Cursiefen C. Selective inhibition of lymphangiogenesis by integrin α5 blockade – Functional role of integrins in lymphangiogenesis. Am. J. Path. 2007;171:361–372. doi: 10.2353/ajpath.2007.060896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zahn G, Vossmeyer D, Stragies R, Wills M, Wong CG, Loeffler KU, Adamis AP, Knolle J. Preclinical Evaluation of the Novel Small Molecule Integrin α5β1 Inhibitor JSM6427 in Monkey and Rabbit Models of Choroidal Neovascularization. 2009. in press AO. [DOI] [PubMed]
- 19.Stragies R, Osterkamp F, Zischinsky G, Vossmeyer D, Kalkhof H, Reimer U, Zahn G. Design and synthesis of a new class of selective integrin α5β1 antagonists. J. Med. Chem. 2007;50:3786–3794. doi: 10.1021/jm070002v. [DOI] [PubMed] [Google Scholar]
- 20.Ricart AD, Tolcher AW, Liu G, Holen K, Schwartz G, Albertini M, Weiss G, Yazji S, Ng C, Wilding G. Volociximab, a chimeric monoclonal antibody that specifically binds α5β1 integrin: a phase I, pharmacokinetic, and biological correlative study. Clin. Cancer Res. 2008;14:7924–7929. doi: 10.1158/1078-0432.CCR-08-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiong J-P, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman S, Arnaout MA. Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg-Gly-Asp ligand. Science (Washington, D.C.) 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 22.Xiao T, Takagi J, Coller BS, Wang JH, Springer TA. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 2004;432:59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Springer TA, Zhu J, Xiao T. Structural basis for distinctive recognition of fibrinogen γC peptide by the platelet integrin αIIbβ3. J. Cell Biol. 2008;182:791–800. doi: 10.1083/jcb.200801146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heckmann D, Meyer A, Marinelli L, Zahn G, Stragies R, Kessler H. Probing integrin selectivity: Rational design of highly active and selective ligands for the α5β1 and αvβ3 integrin receptor. Angew Chem. Int. Ed. Engl. 2007;46:3571–357. doi: 10.1002/anie.200700008. [DOI] [PubMed] [Google Scholar]
- 25.Marinelli L, Meyer A, Heckmann D, Lavecchia A, Novellino E, Kessler H. Ligand binding analysis for human α5β1 integrin: Strategies for designing new α5β1 integrin antagonists. J. Med. Chem. 2005;48:4204–4207. doi: 10.1021/jm040224i. [DOI] [PubMed] [Google Scholar]
- 26.Koivunen E, Wang B, Ruoslahti E. Isolation of a highly specific ligand for the α5β1 integrin from a phage display library. J. Cell Biol. 1994;124:373–380. doi: 10.1083/jcb.124.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koivunen E, Wang B, Ruoslahti E. Phage libraries displaying cyclic peptides with different ring sizes: ligand specificities of the RGD-directed integrins. Biotechnology. 1995;13:265–270. doi: 10.1038/nbt0395-265. [DOI] [PubMed] [Google Scholar]
- 28.Mould AP, Burrows L, Humphries MJ. Identification of amino acid residues that form part of the ligand-binding pocket of integrin α5β1. J. Biol. Chem. 1998;273:25664–25672. doi: 10.1074/jbc.273.40.25664. [DOI] [PubMed] [Google Scholar]
- 29.Mould AP, Askari JA, Humphries MJ. Molecular Basis of Ligand Recognition by Integrin α5β1: I. Specificity of ligand binding is determined by amino acid sequences in the second and third NH2-terminal repeats of the α subunit. J. Biol. Chem. 2000;275:20324–20336. doi: 10.1074/jbc.M000572200. [DOI] [PubMed] [Google Scholar]
- 30.Humphries JD, Askari JA, Zhang X-P, Takada Y, Humphries MJ, Mould AP. Molecular Basis of Ligand Recognition by Integrin α5β1: II. Specificity of Arg-Gly-Asp binding is determined by Trp157 of the α subunit. J. Biol. Chem. 2000;275:20337–20345. doi: 10.1074/jbc.M000568200. [DOI] [PubMed] [Google Scholar]
- 31.Mould AP, Symonds EJ, Buckley PA, Grossmann JG, McEwan PA, Barton SJ, Askari JA, Craig SE, Bella J, Humphries MJ. Structure of an integrin-ligand complex deduced from solution x-ray scattering and site-directed mutagenesis. J. Biol. Chem. 2003;278:39993–39999. doi: 10.1074/jbc.M304627200. [DOI] [PubMed] [Google Scholar]
- 32.Takagi J, Strokovich K, Springer TA, Walz T. Structure of integrin α5β1 in complex with fibronectin. EMBO J. 2003;22:4607–4615. doi: 10.1093/emboj/cdg445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mould AP, McLeish JA, Huxley-Jones J, Goonesinghe AC, Hurlstone AF, Boot-Handford RP, Humphries MJ. Identification of multiple integrin β1 homologs in zebrafish (Danio rerio) BMC Cell Biol. 2006;7:24. doi: 10.1186/1471-2121-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coe APF, Askari JA, Kline AD, Robinson MK, Kirby H, Stephens PE, Humphries MJ. Generation of a minimal α5β1 integrin-Fc fragment. J. Biol. Chem. 2001;276:35854–35866. doi: 10.1074/jbc.M103639200. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Q, Liu X, Collodi P. Identification and characterization of a novel fibronectin in zebrafish. Exp. Cell Res. 2001;268:211–219. doi: 10.1006/excr.2001.5291. [DOI] [PubMed] [Google Scholar]
- 36.Mould AP, Barton SJ, Askari JA, McEwan PA, Buckley PA, Craig SE, Humphries MJ. Conformational changes in the integrin βA domain provide a mechanism for signal transduction via hybrid domain movement. J. Biol. Chem. 2003;278:17028–17035. doi: 10.1074/jbc.M213139200. [DOI] [PubMed] [Google Scholar]
- 37.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2008;426:145–149. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 39.Kabat EA, Wu TT, Reid-Miller M, Perry HM, Gottesman KS. Sequences of Proteins of Immunological Interest. 4th Ed United States Department of Health and Human Services; Washington, D.C.: 1987. [Google Scholar]
- 40.Mould AP, Travis MA, Barton SJ, Hamilton JA, Askari JA, Craig SE, Macdonald PR, Kammerer RA, Buckley PA, Humphries MJ. Evidence that monoclonal antibodies directed against the integrin β subunit plexin/semaphorin/integrin domain stimulate function by inducing receptor extension. J. Biol. Chem. 2005;280:4238–4246. doi: 10.1074/jbc.M412240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isaji T, Sato Y, Zhao Y, Miyoshi E, Wada Y, Taniguchi N, Gu J. N-glycosylation of the β-propeller domain of the integrin α5 subunit is essential for α5β1 heterodimerization, expression on the cell surface, and its biological function. J. Biol. Chem. 2006;281:33258–33267. doi: 10.1074/jbc.M607771200. [DOI] [PubMed] [Google Scholar]
- 42.Irie A, Kamata T, Puzon-McLaughlin W, Takada Y. Critical amino acid residues for ligand binding are clustered in a predicted β-turn of the third N-terminal repeat in the integrin α4 and α5 subunits. EMBO J. 1995;15:5550–5556. doi: 10.1002/j.1460-2075.1995.tb00242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crump JG, Swartz ME, Kimmel CB. An integrin-dependent role of pouch endoderm in hyoid cartilage development. PLoS Biol. 2004;2:E244. doi: 10.1371/journal.pbio.0020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin αVβ3. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta V, Alonso JL, Sugimori T, Essafi M, Xiong JP, Arnaout MA. Role of the β-subunit arginine/lysine finger in integrin heterodimer formation and function. J. Immunol. 2008;180:1713–1718. doi: 10.4049/jimmunol.180.3.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamata T, Tieu KK, Irie A, Springer TA, Takada Y. Amino acid residues in the αIIb subunit that are critical for ligand binding to integrin αIIbβ3 are clustered in the β-propeller model. J. Biol. Chem. 2001;276:44275–44283. doi: 10.1074/jbc.M107021200. [DOI] [PubMed] [Google Scholar]
- 47.Graner MW, Bunch TA, Baumgartner S, Kerschen A, Brower DL. Splice variants of the Drosophila PS2 integrins differentially interact with RGD-containing fragments of the extracellular proteins tiggrin, ten-m, and D-laminin 2. J. Biol. Chem. 1998;273:18235–18241. doi: 10.1074/jbc.273.29.18235. [DOI] [PubMed] [Google Scholar]
- 48.Bunch TA, Kendall TL, Shakalya K, Mahadevan D, Brower DL. Modulation of ligand binding by alternative splicing of the αPS2 integrin subunit. J. Cell. Biochem. 2007;102:211–23. doi: 10.1002/jcb.21288. [DOI] [PubMed] [Google Scholar]
- 49.von der Mark H, Pöschl E, Lanig H, Sasaki T, Deutzman R, von der Mark K. Distinct acidic clusters and hydrophobic residues in the alternative splice domains X1 and X2 of α7 integrins define specificity for laminin isoforms. J. Mol. Biol. 2007;371:1188–1203. doi: 10.1016/j.jmb.2007.05.074. [DOI] [PubMed] [Google Scholar]
- 50.Burrows L, Clark K, Mould AP, Humphries MJ. Fine mapping of inhibitory anti-α5 monoclonal antibody epitopes that differentially affect integrin-ligand binding. Biochem. J. 1999;344:527–533. [PMC free article] [PubMed] [Google Scholar]
- 51.Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat. Rev. Drug Discov. 2005;4:35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
- 52.Basani RB, Zhu H, Thornton MA, Soto CS, Degrado WF, Kowalska MA, Bennett JS, Poncz M. Species differences in small molecule binding to αIIbβ3 are the result of sequence differences in 2 loops of the αIIb β propeller. Blood. 2009;113:902–910. doi: 10.1182/blood-2008-09-177337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blue R, Kowalska MA, Hirsch J, Murcia M, Janczak CA, Harrington A, Jirouskova M, Li J, Fuentes R, Thornton MA, Filizola M, Poncz M, Coller BS. Structural and therapeutic insights from the species specificity and in vivo antithrombotic activity of a novel αIIb-specific αIIbβ3 antagonist. Blood. 2009 May 4; doi: 10.1182/blood-2008-08-169243. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogt A, Cholewinski A, Shen X, Nelson SG, Lazo JS, Tsang M, Hukriede NA. Automated image-based phenotypic analysis in zebrafish embryos. Dev. Dyn. 2009;238:656–663. doi: 10.1002/dvdy.21892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.