Abstract
An ETS family member, ETS Related Gene (ERG) is involved in the Ewing family of tumors as well as leukemias. Rearrangement of the ERG gene with the TMPRSS2 gene has been identified in the majority of prostate cancer patients. Additionally, overexpression of ERG is associated with un- favorable prognosis in prostate cancer patients similar to leukemia patients. Histone acetyltransferases (HATs) and histone deacetylases (HDACs) regulate transcription as well as epigenetic status of genes through acetylation of both histones and transcription factors. Deregulation of HATs and HDACs is frequently seen in various cancers, including prostate cancer. Many cellular oncogenes as well as tumor viral proteins are known to target either or both HATs and HDACs. Several studies have demonstrated that there are alterations of HDAC activity in prostate cancer cells. Recently, we found that ERG binds and inhibits HATs, which suggests that ERG is involved in deregulation of protein acetylation. Additionally, it has been shown that ERG is associated with a higher expression of HDACs. In this study, we tested the effect of the HDAC inhibitors valproic acid (VPA) and trichostatin-A (TSA) on ERG-positive prostate cancer cells (VCaP). We found that VPA and TSA induce apoptosis, upregulate p21/Waf1/CIP1, repress TMPRSS2-ERG expression and affect acetylation status of p53 in VCaP cells. These results suggest that HDAC inhibitors might restore HAT activity through two different ways: by inhibiting HDAC activity and by repressing HAT targeting oncoproteins such as ERG.
Keywords: valproic acid, trichostatin A, histone deacetylase, histone acetyltransferase, CBP/p300, p53, ETS related gene, prostate cancer
Introduction
ERG (Ets Related Gene) belongs to the ETS family of transcriptional factors, which shares a homologous 84 amino acids DNA binding domain (1,2). ERG gene codes for sequence specific DNA binding proteins that function as transcriptional activators (3,4). ERG gene is rearranged in Ewing family of tumors and also in leukemias (5–9). ERG gene and its fusion genes (EWS-ERG and TLS/FUS-ERG) code for sequence specific transcriptional activators (7,8). It appears that fusion of RNA binding proteins (EWS and TLS/FUS) with DNA binding proteins (ERG and Fli-1) leads to human cancers (7,8,10,11). These fusion proteins were shown to inhibit apoptosis, which may be one of the reasons for the activation of the ERG gene in leukemias and human solid tumors (12).
Rearrangement of ERG is also observed in prostate cancer (13). Among genetic alterations associated with prostate cancer, the rearrangement between the androgen responsive trans-membrane protease serine 2 (TMPRSS2) and transcriptional factor ERG has been detected in approximately 50% of patients (13). This rearrangement results in overexpression of ERG (14–17). TMPRSS2 also fuses with other ETS genes, namely ETV1 (ER81), ETV4 (PEA3) and ETV5; however, they are less common in prostate cancers (15,18,19). ERG is involved in numerous roles in the developmental processes of neural crest, angiogenesis/vasculogenesis, hematogenesis, and chondrogenesis (20–24). Overexpression of normal or aberrant ERG is associated with transformation, antiapoptotic property, and invasiveness (12,25–28). In prostate cancer, the majority of TMPRSS2-ERG rearrangements produce truncated ERG proteins (29,30). The expression of the chimeric protein, TMPRSS2-ERG, in which the first five amino acids of TMPRSS2 are fused to truncated ERG is associated with early PSA recurrences and seminal vesicle invasion. Additionally, chimeric TMPRSS2-ERG mRNA which translates into full length ERG proteins, is associated with aggressive phenotypes (29). Several studies suggest that an overexpression of ERG alone is responsible for inducing epithelial hyperplasia and prostatic intraepithelial neoplasia (PIN) in mouse; however, it requires PTEN loss or forced expression of activated Akt for full carcinoma progression (31–34). Thus, it is possible that ERG is a potential therapeutic target in prostate cancer.
Covalent modification of amino-terminal tails of core histones (H2A, H2B, H3, and H4) plays an important role in transcriptional regulation and epigenetic control. Acetylation/deacetylation of core histones represent just one of the complex modulations on nucleosome (35). In general, acetylation of histone H3 and H4 following recruitment of histone acetyl-transferases (HATs), are attributed to transcriptional activation. On the other hand, recruitment of histone deacetylases (HDACs) represses transcription by reversing acetylation. The substrates of HATs as well as HDACs are not limited to histones, but expanded to transcriptional factors such as p53, E2F, and NF-κB (36).
Several lines of evidence suggest that disruption of acetylation/deacetylation activity contributes to tumorigenesis. The individuals diagnosed with Rubinstein-Tybi syndrome exhibit hemizygosity at CBP locus show higher incidence of cancer of neural crest origin (37). Chromosomal translocation of CBP/p300 with MOZ, MORF, and MLL has been documented in several hematological malignancies (28–40). Viral onco-proteins such as E1A of adenovirus, E6 of human Papilloma virus, Tax of HTLV-1, and large T of SV40 are known to target CBP/p300 and repress its cofactor activity (41–44). Cellular chimeric fusion proteins AML1-ETO and E2A-PbX-1 also target p300/CBP (45,46). We have shown that EWS-ATF-1 from malignant melanoma of soft parts repress p53 transcriptional activation through binding to CBP (47). Additionally, we and others found that EWS-Fli-1 in Ewing’s sarcoma and normal Fli-1, which is the closest member of ERG in ETS family, also targets CBP/p300 and repress its HAT activity of transcriptional cofactor (48,49). Recently, we also found that truncated ERG specific to prostate cancer can bind to CBP and repress its cofactor activity for a nuclear receptor, which requires CBP/p300 (unpublished data). These studies suggest that alteration of HAT activity is a contributing factor for pathogenesis of these malignancies. In terms of HDAC, AML1-ETO recruits a co-repressor complex that contains HDAC1 activity (50,51). Administration of HDAC inhibitors subvert ETO-mediated repression and induce differentiation. PML-RARα in acute promyelocytic leukemia (APL) associates N-CoR/Sin3/HDAC1 complex with higher affinity than normal RARα and its dissociation requires a pharmacological dose of all-trans retinoic acid (52). These studies suggest that modulating HAT/HDAC activity by oncoproteins plays an important role in tumorigenesis.
Recently HDAC inhibitors have begun emerging as a new class of chemotherapeutic agents for various cancers including prostate cancer. Histone deacetylase (HDAC) family including 18 members are subdivided into four classes based on homology of the catalytic domain, organization of the domain, and requirement for cofactor (53). Class I and class II HDAC subfamilies are yeast Rpd3 and Hda1 homologues, respectively. Class III is comprised of NAD-dependent deacetylases (sirtuins) and shares homology with yeast Sir2. Class IV contains only HDAC11 and its characterization remains for further study. There are several lines of evidence suggesting upregulation of HDACs in prostate cancer. Higher expression of class I HDAC (HDAC1, 2, 3) was observed in aggressive prostate cancer (54,55). High HDAC2 expression correlates with shorter survival time (55). Nuclear localization of class II HDAC4, which is shuttling between nucleus and cytoplasm in normal cells, was detected in hormone refractory prostate cancer (56).
Valproic acid (2-propylpentanoic acid) (VPA) is a short-chain fatty acid class of histone deacetylase inhibitor (HDACI). VPA is a well-tolerated, established drug for epilepsy and bi- polar disorder (57). VPA inhibits class I and class II HDACs except HDAC6 or HDAC10 (58). It is suggested that VPA inhibits HDAC activity by binding to the HDAC catalytic site (59). Both Valproic acid and Trichostatin-A serve as a basis for newly-developing HDACI inhibitors. Because VPA is one of the least toxic among HDAC inhibitors, it is now subjected to late clinical trials; however, little is known as to how it affects gene network in prostate cancer.
In this report, we show that VPA and TSA induce apoptosis, upregulate p21/Waf1/CIP1 expression, repress TMPRSS2-ERG expression, and affect acetylation status of p53 in ERG-positive prostate cancer cells.
Materials and methods
Tissue culture and cell viability assay
The human VCaP prostate cancer cell line was obtained from the American Type Culture Collection. VCaP cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 15% fetal bovine serum and 1% penicillin/streptomycin at 37°C with 5% CO2 atmosphere. Valproic acid (VPA) and trichostatin-A (TSA) were purchased from Sigma-Aldrich. For viability assay, cells were seeded at 15,000 cells/well in opaque-walled 96-well plates for 24 h before the administration of drugs. TSA, VPA, or vehicle control was added to the cells at the indicated concentrations. Cells were allowed to incubate at 37°C for 24, 48, or 72 h. The cell viability was measured using CellTiter-Glo reagent according to the manufacturer’s instructions (Promega). The luminescent signal was measured using Fluoroskan Ascent FL and data were analyzed with Ascent software version 2.6 (Thermo Electron Corp.). IC50 was calculated using the Hill-Slope model.
In situ cell death detection (TUNEL) assay
Cells were seeded at 200,000 cells per chamber on glass slides and incubated overnight. Medium was replaced with fresh medium containing TSA or VPA for 24 h. Cells were washed with PBS, then allowed to air dry. Cells were fixed using freshly prepared 4% formaldehyde in PBS and incubated at room temperature for 1 h. Cells were washed with PBS and then incubated with permeabilization solution (0.1% Triton X-100 in 0.1% sodium citrate) for 2 min on ice. Cell death was detected using In Situ Cell Death Detection kit, Fluorescein (Roche) according to the manufacturer’s instructions. Subsequently cells were stained using DAPI (Santa Cruz). Micrographs were taken using Olympus IX-71 fluorescence microscope.
Caspase 3/7 activity assay
Cells were seeded at 15,000 cells/well in 96-well plates 24 h before the addition of TSA or VPA. Cells were treated with TSA (5–1000 nM) or VPA (0.05–20 mM) or control (DMSO) for 12 or 24 h. Caspase 3/7 activity was measured using Caspase-Glo 3/7 assay reagent (Promega) according to manufacturer’s protocol. The luminescent signals were measured using Fluoroskan Ascent FL (Thermo Electron Corp.).
Western blot analysis
VCaP cells were seeded at 10 millions per 100 mm plate overnight. The next day TSA or VPA was added to cells at the indicated concentrations for 12 or 24 h. Total cell lysate was prepared using the lysis buffer (50 mM Tris-HCl (pH 7.4), 1% NP-40, 0.125 % sodium deoxcholate, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, 100 μM NaF, protease inhibitor tablet (Roche). Protein concentration was determined using Bradford method (Bio-Rad). Cells were then lysed and protein complexes were separated on 4–20% gradient SDS-PAGE gel. Membrane was incubated with ERG (C-20, Santa Cruz), p21 (12D1, Cell Signaling), acetylated p53 (Lys373, Upstate), total p53 (Ab-1, Calbiochem) and β-actin (C-4, Santa Cruz) primary antibodies. Proteins were detected using ECL detection kit (GE Healthcare).
Quantitative RT-PCR studies
Total RNA was isolated from VCaP cells treated with 1 μM TSA or 10 mM valproic acid for 24 h using RNAspin Mini kit (GE Healthcare) and immediately treated with RNase inhibitor (Roche). First strand cDNA was synthesized with 1 μg of total RNA using Advantage RT-for-PCR kit (Clontech). SYBR Green (Qiagen) was used to detect PCR products using Bio-Rad Mini Opticon real-time PCR system and data were analyzed with Opticon Monitor 3 software (Bio-Rad). GAPDH was used to normalize samples. The primers used were as follows: ERG (forward: 5′-CGCCTACAAGTTCG ACTTCC-3′, reverse: 5′-CCCAGTTGGTGAATTCCAGT-3′), p21 (forward: 5′-CCTCATCCCGTGTTCTCCTTT-3′, reverse: 5′-GTACCACCCAGCGGACAAGT-3′), and GAPDH (forward: 5′-AAGGTGAAGGTCGGAGTCAA-3′, reverse: 5′-AATGAA GGGGTCATTGATGG-3′).
Statistical analysis
One way analysis of variance was performed to detect overall difference among the samples. Then the Student-Newman-Keuls (SNK) test was applied to determine the significant values among the samples. Also student’s t-test was used wherever applicable.
Results
Induction of apoptosis by TSA and VPA in ERG-positive prostate cancer cells
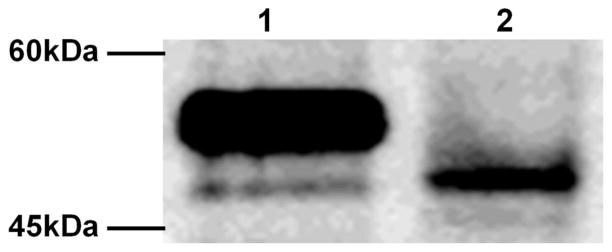
VCaP cell line was established from a vertebral metastasis of hormone refractory prostate cancer (60). VCaP cells have an androgen-responsive, AR-positive and PSA-positive phenotype. Recently, the expression of fusion genes by rearrangements of TMPRSS2 and ERG on chromosome 21 was found in VCaP cells. In consequence of this rearrangement, amino-terminal 39 amino acids of ERG are deleted and the open reading frame starts from the first in-frame ATG of ERG resulting in shorter 423 aa protein. We confirmed the expression of truncated ERG proteins in VCaP cells by Western blot analysis as a major band shown in Fig. 1. There also exists minor lower bands possibly reflecting alternative splicing variants in VCaP cells suggested in other studies (29).
Figure 1.
Truncated-ERG is expressed in a prostate cancer cell line. Western blot analysis was performed with either COS-1 transfected with full-length ERG-2 expression vector (lane 1) or VCaP cell lysate (lane 2).
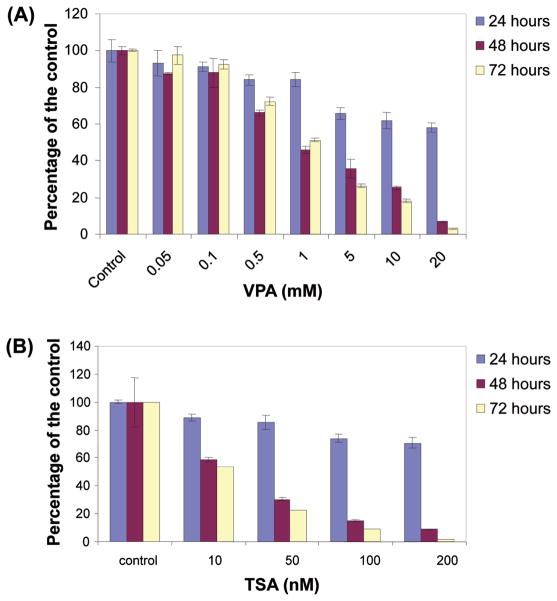
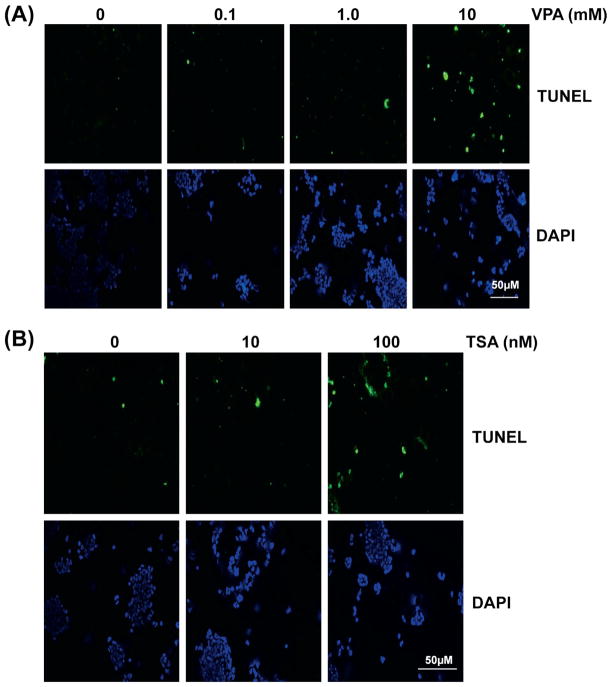
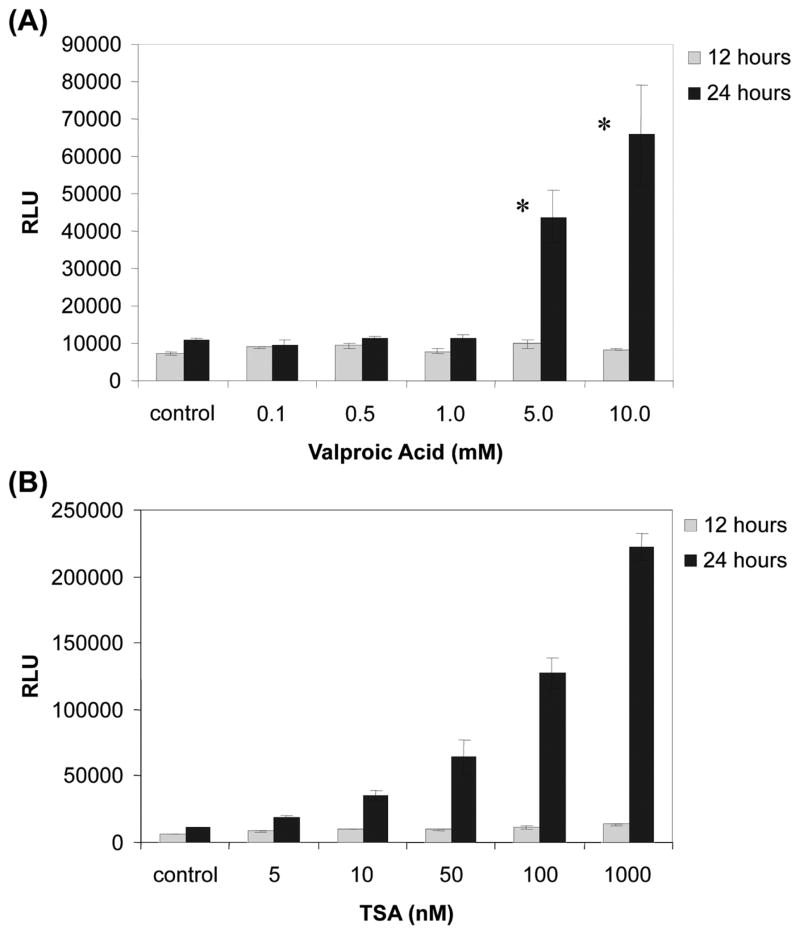
Previous studies suggest that Valproic acid is effective on ERG-negative prostate cancer cell lines, therefore we examined the effect on TMPRSS2-ERG positive VCaP cells (61). First we tested whether VPA or TSA would have any effect on VCaP cell growth. Cell viability assays were performed using VPA or TSA at various concentrations and time intervals. Our results indicate that VPA and TSA inhibit cell growth in a dose- and time- dependent manner (Fig. 2). More than 80% decrease in viability was observed at the concentration of 10 mM VPA after 72 h incubation, or 100 nM TSA after 72 h incubation, respectively. The IC50 (95% CI) values after 72 h treatment were 1.3 mM (1.0 mM–1.5 mM) for VPA and 10 nM (6.4 nM–14 nM) for TSA, respectively. These declines of viability are comparable to other TMPRSS2-ERG-negative prostate cancer cell lines (61). We observed that morphological changes characteristics to apoptosis such as membrane blebbing and nuclear condensation appeared within 24 h with either 100 nM TSA or 10 mM VPA (data not shown). These observations were confirmed by terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assay. As shown in Fig. 3, the number of apoptotic cells labeled with fluorescein-dUTP increased in dose-dependent manner in both VPA (Fig. 3A) and TSA (Fig. 3B) while vehicle treated control cells showed no significant sign apoptosis. The induction of apoptosis was further confirmed by caspase activities. As shown in Fig. 4A and B, caspase 3/7 activity was dramatically increased over 5 mM VPA or 100 nM TSA after 24 h incubation. Taken together these data suggest that these drugs induce apoptosis in VCaP cells. The effect of VPA for apoptosis was consistent with VPA effect on ERG- negative prostate cancer cell lines (61).
Figure 2.
Both TSA and valproic acid impair cell viability of VCaP cells. (A) Cells were treated with variable amount of valproic acid (0.05–20 mM) or vehicle control for 24, 48, or 72 h. (B) Cells were treated with variable amount of trichostatin-A (10–200 nM) or vehicle control for 24, 48, or 72 h. Results are shown as the mean of percentage of control of three independent experiments with standard deviations.
Figure 3.
Both TSA and valproic acid induce apoptosis in VCaP cells. Apoptosis of VCaP cells were analyzed using TUNEL reaction. (A) Cells were treated with variable amount of valproic acid (0.1–10 mM) or vehicle control for 24 h. (B) Cells were treated with variable amount of TSA (10–00 nM) or vehicle control for 24 h. Fluorescent signals after labeling with fluorescein-dUTP are shown in the top panels. Bottom panels show DAPI staining for nucleus. Scale bar, 50 μm.
Figure 4.
Both valproic acid and TSA increase caspase 3/7 activity. (A) Cells were treated with valproic Acid (0.1–10 mM) or vehicle control for 12 or 24 h before the measurement of caspase 3/7 activity. (B) Cells were treated with TSA (5–1000 nM) or vehicle control (DMSO) for 12 or 24 h before the measurement of caspase 3/7 activity. Caspase 3/7 activity was determined using Caspase-Glo 3/7 assay reagent (Promega) according to manufacturer’s protocol. Values represent the mean of luminescence signals of three independent experiments with standard deviations. *P<0.01
The effect of TSA and VPA on TMPRSS2-ERG and tumor suppressor genes
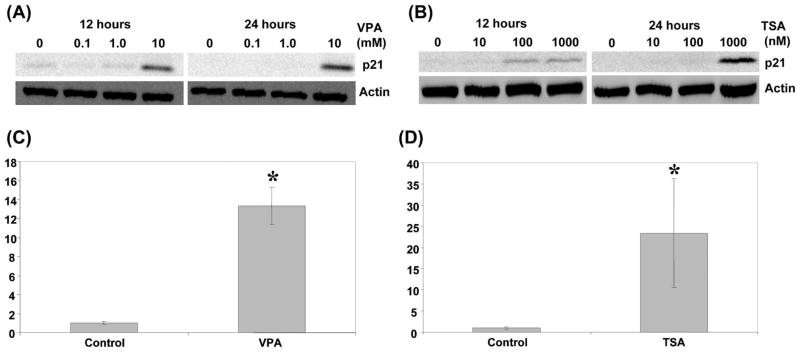
Next we tested the effect of TSA and VPA on p21Waf1/CIP1 (CDKN1A) expression. Western blot assay revealed that p21 was induced at 100 nM TSA at 12 h (Fig. 5A and B). Similar induction was observed at 10 mM VPA for 12 h, however, a strong induction occurred after 24 h incubation (Fig. 5B). The induction of p21 was further confirmed with quantitative RT-PCR. Fourteen-fold induction of p21 mRNA at 10 mM VPA (Fig. 5C) and 25-fold induction at 1 μM TSA (Fig. 5D) were observed after 24 h treatment, suggesting that both VPA and TSA activate p21 transcription. These results are consistent with the induction of p21 by HDAC inhibitors in various cancer cells including prostate cancer (62–64).
Figure 5.
Effect of TSA and valproic acid on p21 expression. (A) Western blot assay was performed after 12 or 24 h valproic acid treatments at the indicated concentrations. The blot with anti-p21 antibody is shown at the top. β-actin used as a loading control is shown at the bottom. (B) Western blot assay was performed after 12 or 24 h TSA treatments at the indicated concentrations. The blot with anti-p21 antibody is shown at the top. β-actin used as a loading control is shown at the bottom. (C) The relative expression level of p21 mRNA was determined with quantitative RT-PCR after 24 h treatment with 10 mM valproic acid. (D) The relative expression level of p21 mRNA was determined with quantitative RT-PCR after 24 h treatment with 1 μM TSA. In both cases, GAPDH was used to normalize samples. The results represent fold activation over control of three independent experiments with standard deviations. *P<0.05
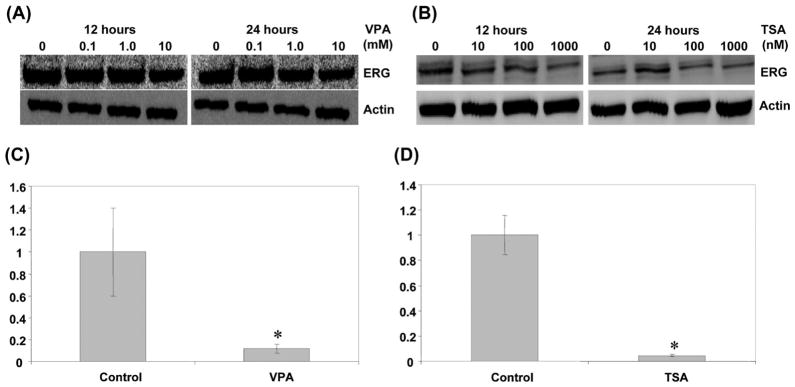
Previous studies have shown that HDAC inhibitors repress ERG expression (64,65). Therefore, we investigated the effect of TSA and VPA on TMPRSS2-ERG expression. Western blot analysis showed that expression of TMPRSS2-ERG decreased at 1 μM TSA treatment or 10 mM VPA treatment at as early as 12 h of incubation (Fig. 6). Similar reduction was observed after 24 h of incubation (Fig. 6). These results were confirmed with quantitative RT-PCR. VCaP cells were treated with either TSA or VPA and the relative expression level of mRNA was measured. As shown in Fig. 6C, the mRNA levels were significantly decreased after 24 h incubation with 10 mM VPA. Similar repression was observed in 1 μM of TSA (Fig. 6D). These results suggest that VPA and TSA may interfere with TMPRSS2-ERG expression in prostate cancer.
Figure 6.
Effect of TSA and valproic acid on TMPRSS2-ERG expression. (A) Western blot assay was performed after 12 or 24 h valproic acid treatments at the indicated concentrations. The blot with anti-ERG antibody is shown at the top. β-actin used as a loading control is shown at the bottom. (B) Western blot assay was performed after 12 or 24 h TSA treatments at the indicated concentrations. The blot with anti-ERG antibody is shown at the top. β-actin used as a loading control is shown at the bottom. (C) The relative expression level of TMPRSS2-ERG mRNA was determined with quantitative RT-PCR after 24 h treatment with 10 mM valproic acid. (D) The relative expression level of TMPRSS2-ERG mRNA was determined with quantitative RT-PCR after 24 h treatment with 1 μM TSA. In both cases, GAPDH was used to normalize samples. The results represent relative values against control (arbitrarily set to 1) of three independent experiments with standard deviations. * P<0.05
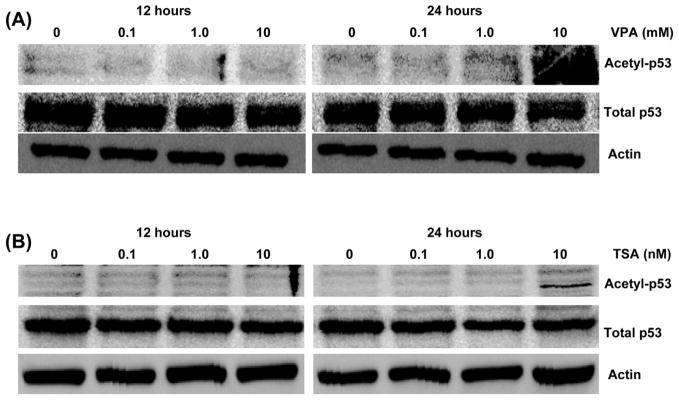
Histone acetyltransferase CBP/p300 acetylates p53 at Lys-373/Lys382 which affects DNA binding activity, and transcriptional activation of target genes such as p21 and Bax (66). Viral oncoproteins are known to inhibit this acetylation by targeting CBP/p300 (41–44). We found that both normal and truncated ERG also bind to CBP and repress its activity (unpublished data). Since VPA and TSA repress ERG expression in VCaP cells, we postulated that the downregulation of expression of ERG by HDAC inhibitor leads to the restoration of CBP/p300 histone acetylase activity subsequently allowing HATs to acetylate its target transcriptional factors, such as p53. As shown in Fig. 7, acetylation of p53 at Lys-373 was greatly enhanced at 10 mM VPA and 1 μM TSA after 24 h incubation. These treatments do not appear to have any effect on total p53 protein levels. These results suggest that VPA and TSA affect acetylation status of p53 in prostate cancer.
Figure 7.
Both TSA and valproic acid induce acetylation of p53 in VCaP cells. (A) Western blot assays were performed on VCaP cells incubated for 12 or 24 h at the indicated concentrations of valproic acid. Blot with anti-acetylated p53-Lys 373 (top), anti-p53 (middle), or anti-β-actin (bottom) are shown. (B) Western blots were performed on VCaP cells incubated for 12 or 24 h at the indicated concentrations of TSA. Blot with anti-acetylated p53-Lys 373 (top), anti-p53 (middle), or anti-β-actin (bottom) are shown.
Discussion
In this study, we found that VPA induces apoptosis in ERG rearranged prostate cancer cells, affects status of tumor suppressor genes, and represses expression of TMPRSS2-ERG. TMPRSS2-ERG expression is one of the most frequent genetic alterations known to occur in prostate cancer patients (13). The majority of TMPRSS2-ERG transcripts seen in patients skip Exon 3, which contain native ATG and result in a truncated ERG protein (29). Also, several studies indicate that ERG expression in prostate cancer contributes to the oncogenic properties of cell proliferation, invasion, metastasis, and cell motility (28,31,67). The higher expression of ERG in prostate cancer correlates with unfavorable prognosis (14). Therefore, targeting ERG expression by HDAC inhibitor may have an impact on developing new therapeutic strategy.
Deregulation of HDAC activities are observed in a variety of cancers. Administration of HDAC inhibitors to prostate cancer cells demonstrate suppression of growth, differentiation, and induce apoptosis, which suggests that HDAC activity is essential for maintaining tumor status (61,64,68). However how these HDAC inhibitors affect transcriptional network that sustain tumor properties is poorly understood. HDAC inhibitors are thought to modulate selectively the expression of fraction of genes. Among these genes, p21/Waf1/CIP1 has emerged as a common target of HDAC inhibitors for upregulation (62–64). It appears that the repression of negatively regulating factor is required for upregulation of p21. For example, p21 is repressed by c-Myc through the interaction of Sp1/Sp3 or Miz-1 (69). Sp1/Sp2 on p21 promoter has an important role in both activation and repression of HDAC1 (63). EWS-Fli-1, which is closely related to ERG at DNA-binding domain binds to p21 promoter and downregulates p21 expression through inhibition of p300 acetyltransferase activity. This repression of EWS-Fli-1 was relieved by either overexpression of p300 or by the administration of sodium-butylate (48). Downregulation of EWS-Fli-1 by antisense dramatically increased p21 expression (70). We and others showed that EWS-Fli-1 and normal ERG share the same target sequences (3,4,7,10). Therefore, it might be possible that TMPRSS2-ERG is involved in p21 repression through binding to its promoter. The association between ERG and HDAC has been demonstrated in several studies. ERG recruits HDAC and represses collagen promoter activity and TSA relieves this repression (71). ESET, which was cloned as interacting protein with ERG, recruits HDAC1/mSin3 (72). One might think administration of VPA and TSA could reduce this inhibition at least in part by repressing TMPRSS2-ERG expression.
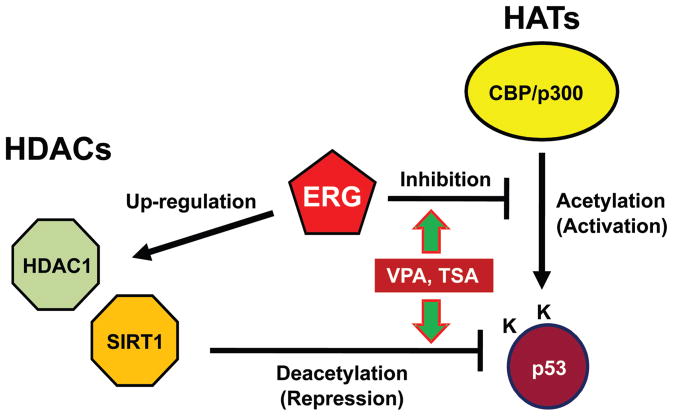
Acetylation of p53 by CBP/p300 increases DNA binding and transcriptional activity on p21 promoter (66,73). Viral oncoproteins frequently target histone acetylase CBP/p300 and repress its activity (41–44). Recently, we found that truncated ERG binds and interferes with CBP mediated transcriptional activation of a nuclear receptor. Since VPA and TSA repress TMPRSS2-ERG expression, it could be possible that these HDAC inhibitors rescue CBP/p300 by subverting the inhibitory effect of ERG that restore p53 acetylation under certain situations where the HDACs are inactivated by VPA or TSA. In this regard, acetylation of p53 is not evident until 24 h after administration of TSA or VPA while the repression of TMPRSS2-ERG precedes acetylation by at least 12 h. The acetylation also requires the amount of 1 μM TSA and 10 mM VPA that is enough to repress TMPRSS2-ERG. The reason that the effective concentration of VPA to affect p53 is relatively high in this study might reside in its ineffectiveness to repression of TMPRSS2-ERG even at higher concentration or HDAC selectivity of VPA (59). It might be possible that the release of CBP/p300 from blockade of TMPRSS2-ERG following the repression of TMPRSS2-ERG expression restores acetylation of p53 in the absence of the activity of HDACs. The TMPRSS2-ERG fusion gene was associated with upregulation of HDAC1 (74). Recently, we found that ectopic expression of truncated-ERG increases SIRT1 activity (data not shown). Given that the re-arrangement of ERG is supposedly an early event of tumorigenesis, it is hypothesized that TMPRSS2-ERG insulates p53 from acetylation through both interference with CBP/p300 HAT activity and inducing HDAC expression and therefore maintains p53 inactive (Fig. 8). In this model, HDAC inhibitors reactivate tumor suppressor genes not only by repressing HDAC activities, but also by repressing the expression of HAT targeting oncogenes such as ERG.
Figure 8.
ERG interferes with acetylation of p53 in two ways. ERG binds CBP/p300 histone acetyltransferase and inhibits its activity of acetylation on p53. ERG also induces expression of HDACs which deacetylate p53.
We showed the dual role of HDAC inhibitors in repressing TMPRSS2-ERG expression and affecting p53 acetylation status, therefore, valproic acid and other HDAC inhibitors may be useful to prevent further advance of early stage prostate cancer. In this regard, a combination of different HDAC inhibitors might help in order to retain the therapeutic dose of each inhibitor in the moderate range and to inhibit all of the HDACs involved in repressing major tumor suppressor genes.
Acknowledgments
We thank other colleagues of the Reddy and Rao laboratories for their kind cooperation. This study was funded in part by GCC Distinguished Cancer Scholar award to E.S.P. Reddy and V.N. Rao, DOD grants W81XWH-09-1-0236, W81XWH-08-1-0628 to E.S.P. Reddy and U56/U54 MSM/UAB/TU partnership grants 2U54CA118948, U54CA118638-05S1 to E.S.P. Reddy and G.K. Bhat. We thank K. Aysola for help with the Figures.
References
- 1.Rao VN, Papas TS, Reddy ES. erg, a human ets-related gene on chromosome 21: alternative splicing, polyadenylation, and translation. Science. 1987;237:635–639. doi: 10.1126/science.3299708. [DOI] [PubMed] [Google Scholar]
- 2.Reddy ES, Rao VN, Papas TS. The erg gene: a human gene related to the ets oncogene. Proc Natl Acad Sci USA. 1987;84:6131–6135. doi: 10.1073/pnas.84.17.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reddy ES, Rao VN. erg, an ets-related gene, codes for sequence-specific transcriptional activators. Oncogene. 1991;6:2285–2289. [PubMed] [Google Scholar]
- 4.Siddique HR, Rao VN, Lee L, Reddy ES. Characterization of the DNA binding and transcriptional activation domains of the erg protein. Oncogene. 1993;8:1751–1755. [PubMed] [Google Scholar]
- 5.Erkizan HV, Uversky VN, Toretsky JA. Oncogenic partnerships: EWS-FLI1 protein interactions initiate key pathways of Ewing’s sarcoma. Clin Cancer Res. 2010;16:4077–4083. doi: 10.1158/1078-0432.CCR-09-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ichikawa H, Shimizu K, Hayashi Y, Ohki M. An RNA-binding protein gene, TLS/FUS, is fused to ERG in human myeloid leukemia with t(16;21) chromosomal translocation. Cancer Res. 1994;54:2865–2868. [PubMed] [Google Scholar]
- 7.Ohno T, Ouchida M, Lee L, Gatalica Z, Rao VN, Reddy ES. The EWS gene, involved in Ewing family of tumors, malignant melanoma of soft parts and desmoplastic small round cell tumors, codes for an RNA binding protein with novel regulatory domains. Oncogene. 1994;9:3087–3097. [PubMed] [Google Scholar]
- 8.Prasad DD, Ouchida M, Lee L, Rao VN, Reddy ES. TLS/FUS fusion domain of TLS/FUS-erg chimeric protein resulting from the t(16;21) chromosomal translocation in human myeloid leukemia functions as a transcriptional activation domain. Oncogene. 1994;9:3717–3729. [PubMed] [Google Scholar]
- 9.Panagopoulos I, Aman P, Fioretos T, et al. Fusion of the FUS gene with ERG in acute myeloid leukemia with t(16;21)(p11;q22) Genes Chromosomes Cancer. 1994;11:256–262. doi: 10.1002/gcc.2870110408. [DOI] [PubMed] [Google Scholar]
- 10.Sorensen PH, Lessnick SL, Lopez-Terrada D, Liu XF, Triche TJ, Denny CT. A second Ewing’s sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nat Genet. 1994;6:146–151. doi: 10.1038/ng0294-146. [DOI] [PubMed] [Google Scholar]
- 11.Shing DC, McMullan DJ, Roberts P, et al. FUS/ERG gene fusions in Ewing’s tumors. Cancer Res. 2003;63:4568–4576. [PubMed] [Google Scholar]
- 12.Yi H, Fujimura Y, Ouchida M, Prasad DD, Rao VN, Reddy ES. Inhibition of apoptosis by normal and aberrant Fli-1 and erg proteins involved in human solid tumors and leukemias. Oncogene. 1997;14:1259–1268. doi: 10.1038/sj.onc.1201099. [DOI] [PubMed] [Google Scholar]
- 13.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrovics G, Liu A, Shaheduzzaman S, et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24:3847–3852. doi: 10.1038/sj.onc.1208518. [DOI] [PubMed] [Google Scholar]
- 15.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 16.Vanaja DK, Cheville JC, Iturria SJ, Young CY. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res. 2003;63:3877–3882. [PubMed] [Google Scholar]
- 17.Mosquera JM, Mehra R, Regan MM, et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res. 2009;15:4706–4711. doi: 10.1158/1078-0432.CCR-08-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomlins SA, Mehra R, Rhodes DR, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 19.Helgeson BE, Tomlins SA, Shah N, et al. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68:73–80. doi: 10.1158/0008-5472.CAN-07-5352. [DOI] [PubMed] [Google Scholar]
- 20.Dhordain P, Dewitte F, Desbiens X, Stehelin D, Duterque-Coquillaud M. Mesodermal expression of the chicken erg gene associated with precartilaginous condensation and cartilage differentiation. Mech Dev. 1995;50:17–28. doi: 10.1016/0925-4773(94)00322-e. [DOI] [PubMed] [Google Scholar]
- 21.Birdsey GM, Dryden NH, Amsellem V, et al. Transcription factor Erg regulates angiogenesis and endothelial apoptosis through VE-cadherin. Blood. 2008;111:3498–3506. doi: 10.1182/blood-2007-08-105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwamoto M, Tamamura Y, Koyama E, et al. Transcription factor ERG and joint and articular cartilage formation during mouse limb and spine skeletogenesis. Dev Biol. 2007;305:40–51. doi: 10.1016/j.ydbio.2007.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rainis L, Toki T, Pimanda JE, et al. The proto-oncogene ERG in megakaryoblastic leukemias. Cancer Res. 2005;65:7596–7602. doi: 10.1158/0008-5472.CAN-05-0147. [DOI] [PubMed] [Google Scholar]
- 24.Loughran SJ, Kruse EA, Hacking DF, et al. The transcription factorErgisessentialfor the function of adult hematopoietic stem cells. Nat Immunol. 2008;9:810–819. doi: 10.1038/ni.1617. [DOI] [PubMed] [Google Scholar]
- 25.Hart AH, Corrick CM, Tymms MJ, Hertzog PJ, Kola I. Human ERG is a proto-oncogene with mitogenic and transforming activity. Oncogene. 1995;10:1423–1430. [PubMed] [Google Scholar]
- 26.Codrington R, Pannell R, Forster A, et al. The Ews-ERG fusion protein can initiate neoplasia from lineage-committed haematopoietic cells. PLoS Biol. 2005;3:E242. doi: 10.1371/journal.pbio.0030242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buttice G, Duterque-Coquillaud M, Basuyaux JP, Carrere S, Kurkinen M, Stehelin D. Erg, an Ets-family member, differentially regulates human collagenase1 (MMP1) and stromelysin1 (MMP3) gene expression by physically interacting with the Fos/Jun complex. Oncogene. 1996;13:2297–2306. [PubMed] [Google Scholar]
- 28.Klezovitch O, Risk M, Coleman I, et al. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci USA. 2008;105:2105–2110. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Cai Y, Ren C, Ittmann M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66:8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 30.Soller MJ, Isaksson M, Elfving P, Soller W, Lundgren R, Panagopoulos I. Confirmation of the high frequency of the TMPRSS2/ERG fusion gene in prostate cancer. Genes Chromosomes Cancer. 2006;45:717–719. doi: 10.1002/gcc.20329. [DOI] [PubMed] [Google Scholar]
- 31.Tomlins SA, Laxman B, Varambally S, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King JC, Xu J, Wongvipat J, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carver BS, Tran J, Gopalan A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zong Y, Xin L, Goldstein AS, Lawson DA, Teitell MA, Witte ON. ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proc Natl Acad Sci USA. 2009;106:12465–12470. doi: 10.1073/pnas.0905931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 37.Miller RW, Rubinstein JH. Tumors in Rubinstein-Taybi syndrome. Am J Med Genet. 1995;56:112–115. doi: 10.1002/ajmg.1320560125. [DOI] [PubMed] [Google Scholar]
- 38.Borrow J, Stanton VP, Jr, Andresen JM, et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 39.Rowley JD, Reshmi S, Sobulo O, et al. All patients with the T(11;16)(q23;p13.3) that involves MLL and CBP have treatment-related hematologic disorders. Blood. 1997;90:535–541. [PubMed] [Google Scholar]
- 40.Panagopoulos I, Fioretos T, Isaksson M, et al. Fusion of the MORF and CBP genes in acute myeloid leukemia with the t(10;16)(q22;p13) Hum Mol Genet. 2001;10:395–404. doi: 10.1093/hmg/10.4.395. [DOI] [PubMed] [Google Scholar]
- 41.Avantaggiati ML, Carbone M, Graessmann A, Nakatani Y, Howard B, Levine AS. The SV40 large T antigen and adenovirus E1a oncoproteins interact with distinct isoforms of the transcriptional co-activator, p300. EMBO J. 1996;15:2236–2248. [PMC free article] [PubMed] [Google Scholar]
- 42.Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki T, Uchida-Toita M, Yoshida M. Tax protein of HTLV-1 inhibits CBP/p300-mediated transcription by interfering with recruitment of CBP/p300 onto DNA element of E-box or p53 binding site. Oncogene. 1999;18:4137–4143. doi: 10.1038/sj.onc.1202766. [DOI] [PubMed] [Google Scholar]
- 44.Patel D, Huang SM, Baglia LA, McCance DJ. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 1999;18:5061–5072. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Kalkum M, Yamamura S, Chait BT, Roeder RG. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science. 2004;305:1286–1289. doi: 10.1126/science.1097937. [DOI] [PubMed] [Google Scholar]
- 46.Bayly R, Murase T, Hyndman BD, et al. Critical role for a single leucine residue in leukemia induction by E2A-PBX1. Mol Cell Biol. 2006;26:6442–6452. doi: 10.1128/MCB.02025-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujimura Y, Siddique H, Lee L, Rao VN, Reddy ES. EWS-ATF-1 chimeric protein in soft tissue clear cell sarcoma associates with CREB-binding protein and interferes with p53-mediated trans-activation function. Oncogene. 2001;20:6653–6659. doi: 10.1038/sj.onc.1204684. [DOI] [PubMed] [Google Scholar]
- 48.Nakatani F, Tanaka K, Sakimura R, et al. Identification of p21WAF1/CIP1 as a direct target of EWS-Fli1 oncogenic fusion protein. J Biol Chem. 2003;278:15105–15115. doi: 10.1074/jbc.M211470200. [DOI] [PubMed] [Google Scholar]
- 49.Ramakrishnan R, Fujimura Y, Zou JP, et al. Role of protein-protein interactions in the antiapoptotic function of EWS-Fli-1. Oncogene. 2004;23:7087–7094. doi: 10.1038/sj.onc.1207927. [DOI] [PubMed] [Google Scholar]
- 50.Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci USA. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Saunthararajah Y, Redner RL, Liu JM. Inhibitors of histone deacetylase relieve ETO-mediated repression and induce differentiation of AML1-ETO leukemia cells. Cancer Res. 1999;59:2766–2769. [PubMed] [Google Scholar]
- 52.Altucci L, Gronemeyer H. The promise of retinoids to fight against cancer. Nat Rev Cancer. 2001;1:181–193. doi: 10.1038/35106036. [DOI] [PubMed] [Google Scholar]
- 53.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 54.Halkidou K, Gaughan L, Cook S, Leung HY, Neal DE, Robson CN. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate. 2004;59:177–189. doi: 10.1002/pros.20022. [DOI] [PubMed] [Google Scholar]
- 55.Weichert W, Roske A, Gekeler V, et al. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer. 2008;98:604–610. doi: 10.1038/sj.bjc.6604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Halkidou K, Cook S, Leung HY, Neal DE, Robson CN. Nuclear accumulation of histone deacetylase 4 (HDAC4) coincides with the loss of androgen sensitivity in hormone refractory cancer of the prostate. Eur Urol. 2004;45:382–389. doi: 10.1016/j.eururo.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 57.Peterson GM, Naunton M. Valproate: a simple chemical with so much to offer. J Clin Pharm Ther. 2005;30:417–421. doi: 10.1111/j.1365-2710.2005.00671.x. [DOI] [PubMed] [Google Scholar]
- 58.Gurvich N, Tsygankova OM, Meinkoth JL, Klein PS. Histone deacetylase is a target of valproic acid-mediated cellular differentiation. Cancer Res. 2004;64:1079–1086. doi: 10.1158/0008-5472.can-03-0799. [DOI] [PubMed] [Google Scholar]
- 59.Gottlicher M, Minucci S, Zhu P, et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Korenchuk S, Lehr JE, MClean L, et al. VCaP, a cell-based model system of human prostate cancer. In Vivo. 2001;15:163–168. [PubMed] [Google Scholar]
- 61.Xia Q, Sung J, Chowdhury W, et al. Chronic administration of valproic acid inhibits prostate cancer cell growth in vitro and in vivo. Cancer Res. 2006;66:7237–7244. doi: 10.1158/0008-5472.CAN-05-0487. [DOI] [PubMed] [Google Scholar]
- 62.Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci USA. 2004;101:1241–1246. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lagger G, Doetzlhofer A, Schuettengruber B, et al. The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol Cell Biol. 2003;23:2669–2679. doi: 10.1128/MCB.23.8.2669-2679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qian X, Ara G, Mills E, LaRochelle WJ, Lichenstein HS, Jeffers M. Activity of the histone deacetylase inhibitor belinostat (PXD101) in preclinical models of prostate cancer. Int J Cancer. 2008;122:1400–1410. doi: 10.1002/ijc.23243. [DOI] [PubMed] [Google Scholar]
- 65.Bjorkman M, Iljin K, Halonen P, et al. Defining the molecular action of HDAC inhibitors and synergism with androgen deprivation in ERG-positive prostate cancer. Int J Cancer. 2008;123:2774–2781. doi: 10.1002/ijc.23885. [DOI] [PubMed] [Google Scholar]
- 66.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 67.Wang J, Cai Y, Yu W, Ren C, Spencer DM, Ittmann M. Pleiotropic biological activities of alternatively spliced TMPRSS2/ERG fusion gene transcripts. Cancer Res. 2008;68:8516–8524. doi: 10.1158/0008-5472.CAN-08-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carducci MA, Nelson JB, Chan-Tack KM, et al. Phenylbutyrate induces apoptosis in human prostate cancer and is more potent than phenylacetate. Clin Cancer Res. 1996;2:379–387. [PubMed] [Google Scholar]
- 69.Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65:3980–3985. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- 70.Matsumoto Y, Tanaka K, Nakatani F, Matsunobu T, Matsuda S, Iwamoto Y. Downregulation and forced expression of EWS-Fli1 fusion gene results in changes in the expression of G(1) regulatory genes. Br J Cancer. 2001;84:768–775. doi: 10.1054/bjoc.2000.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsui Y, Chansky HA, Barahmand-Pour F, et al. COL11A2 collagen gene transcription is differentially regulated by EWS/ERG sarcoma fusion protein and wild-type ERG. J Biol Chem. 2003;278:11369–11375. doi: 10.1074/jbc.M300164200. [DOI] [PubMed] [Google Scholar]
- 72.Yang L, Mei Q, Zielinska-Kwiatkowska A, et al. An ERG (ets-related gene)-associated histone methyltransferase interacts with histone deacetylases 1/2 and transcription co-repressors mSin3A/B. Biochem J. 2003;369:651–657. doi: 10.1042/BJ20020854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 74.Iljin K, Wolf M, Edgren H, et al. TMPRSS2 fusions with oncogenic ETS factors in prostate cancer involve unbalanced genomic rearrangements and are associated with HDAC1 and epigenetic reprogramming. Cancer Res. 2006;66:10242–10246. doi: 10.1158/0008-5472.CAN-06-1986. [DOI] [PubMed] [Google Scholar]