Abstract
Ribosome assembly involves rRNA transcription, modification, folding and cleavage from precursor transcripts, and association of ribosomal proteins (Rps). In bacteria, this complex process requires only a handful of proteins in addition to those needed for rRNA transcription, modification and cleavage, while in eukaryotes a large machinery comprising ~ 200 proteins in the yeast S. cerevisiae has been identified. Furthermore, while the bacterial assembly factors generally produce only cold-sensitive phenotypes upon deletion, most of the eukaryotic assembly factors are essential, comprising ~ 20% of essential yeast proteins. This review explores recent rapid progress in the structural and functional dissection of the 40S assembly machinery.
Over the last 10 years, the ribosome assembly field has undergone a dramatic transformation. Once essentially considered a solved question (after all, ribosomes could be reconstituted in vitro) the field was pushed wide open by ground-breaking proteomic studies revealing the large complexity of the eukaryotic ribosome assembly machinery [1-3]. Once the dizzying number of ribosome assembly factors became evident, with almost no functional data available, making sense of ribosome assembly seemed almost impossible. Excitingly, the dust is now settling and we can see the possibility of understanding the vast ribosome assembly machinery. This review focuses on structural insights into small subunit assembly that have been recently garnered. Importantly, we believe that some general features deduced here are relevant for large subunit assembly as well, and some references to that are made. Interested readers are also referred to other recent reviews with more functional focus [3-5].
snoRNAs bind co-transcriptionally and delay tertiary folding of rRNA
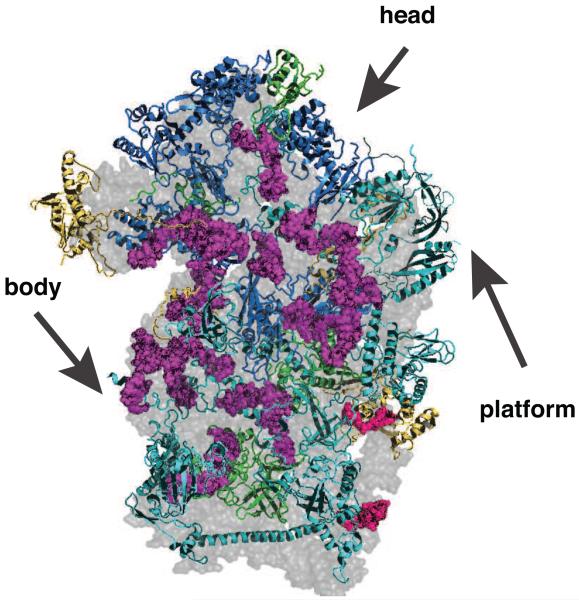
Box C/D snoRNAs bind co-transcriptionally to promote pre-rRNA methylation at certain 2′-OH residues [6*], as suggested by their binding to components of the PolI transcription machinery [3]. Deletion of individual snoRNAs has, with the exception of U3, U14 and snR30 [7-9], no effect on yeast growth or ribosome assembly. However the concurrent deletion of several snoRNAs often produces synthetic phenotypes [10,11]. Mapping of the box C/D snoRNA binding sites within ribosomes indicates that their binding prevents the formation of secondary and sometimes tertiary structure elements (Figure 1), perhaps to keep the rRNA structure more fluid and facilitate the insertion of Rps, many of which have unstructured extensions that insert into the rRNA core [12, *13, 14]. snR30 prevents the premature formation of a large eukaryote-specific structure, which could lock in the tertiary structure of the body [15*]. Furthermore, several rRNA duplexes whose formation is inhibited by snoRNA binding interact with later-binding Rps (Figure 1), indicating that snoRNA binding might delay the incorporation of these proteins.1
Figure 1.
Binding of box C/D snoRNAs prevents the formation of secondary and tertiary structure in ribosomes and could affect binding of ribosomal proteins (Rps). The subunit interface of the 40S ribosome is shown, and head, body and platform are indicated. Nucleotides that base pair with box C/D snoRNAs are highlighted in magenta, those that bind snR30 in red. Rps required for co-transcriptional processing steps are shown in cyan, Rps required for post-transcriptional steps are shown in blue, and proteins not required for assembly are shown in yellow. Proteins for which no information is available are shown in green.
Upon methylation snoRNPs are removed from pre-rRNAs by Prp43 [17-20]. It is not known if all snoRNAs dissociate prior to early pre-rRNA cleavage steps, or if some remain bound longer. Quantitative studies in vertebrates have shown that the snoRNA-dependent 2′-methylations at G1448 in 18S and A394 in 28S are not complete during transcription, while essentially all 20S (pre-18S) and 32S (pre-28S) rRNA molecules are methylated at these sites [21]. Furthermore, Prp43 genetically interacts with proteins required for late cytoplasmic 40S assembly steps [22], indicating that perhaps some snoRNAs are not removed until later in assembly. The finding that all pre-18S and pre-28S rRNAs are methylated at the two investigated residues, if confirmed for additional methylation sites, also indicates an impressive efficiency of the snoRNP system. Perhaps the transcription machinery includes factors that facilitate the annealing of snoRNAs. Alternatively, it is possible that transcripts not bound to snoRNAs could be efficiently degraded, consistent with recent observations [*6].
Assembly of Rps in the body is cotranscriptional while Rps in the head bind late
Early work with bacterial ribosomes uncovered the order in which Rps bind, and how binding of a first set of proteins (primary binders) promotes rRNA structural changes that facilitate binding of a secondary and then tertiary set of proteins [23,24]. More recently, mechanistic studies have shown that Rps binding to rRNA is not a simple two-state process, in which the individual interactions of a single protein are made all at once; instead, even the interactions of a single Rps occur in multiple steps [**25].
More globally, a recent EM study of in vitro reconstituted assembly intermediates has shown that binding of proteins to the body precedes binding to the platform and head, with the formation of stable assembly intermediates that lack the head and sometimes even the platform, or are missing individual protein components from these regions [**26].
Excitingly, these in vitro results with bacterial 30S subunits are highly similar to results from yeast in vivo studies. Most Rps in the body are required for early co-transcriptional processing steps, while only a subset of platform binding proteins is required (Figure 1). Head-binding proteins are not required until the later export and cytoplasmic cleavage steps [13,*27]. Even more strikingly, intermediates in the bacterial assembly reaction lack the homologs of the head binders Rps3 and Rps20, as well as the platform binders Rps14 and Rps26, but are otherwise completely assembled [**26]. Similarly, we have recently shown that in a late yeast assembly intermediate Rps26 is absent, and Rps14 is not at its final position. Additionally, Rps3 is present but not correctly positioned, Rps20 and Rps29 are only bound to a fraction of molecules and Rps10, which also binds to the head but has no homolog in bacteria, is absent. The absence of Rps10 and Rps26 is explained by the overlap of their binding sites with assembly factors suggesting that their incorporation is delayed until assembly factors dissociate, perhaps even after rRNA cleavage is complete [**28].
The similarity in the assembly of Rps in vitro and in vivo and from different kingdoms of life, suggests that the main factors governing protein binding are conserved and must be intrinsic to rRNA and proteins. Furthermore, assembly factors are not producing these patterns although they might modulate them. For example, by delaying Rps incorporation until snoRNAs are dissociated or 18S maturation is complete, as described above.
Interestingly, deletions of a single copy of most Rps are synthetically lethal or slow growing with deletion of the cytoplasmic Dom34, required for degradation of stalled 80S ribosomes, including those containing 18S precursors [*29,**30]. This finding suggests that ribosome assembly can continue in the absence of an early binding Rps. However, because these ribosomes cannot function, they stall at the 80S stage, and are then degraded.
Insights on the structures of U3-containing early assembly intermediates
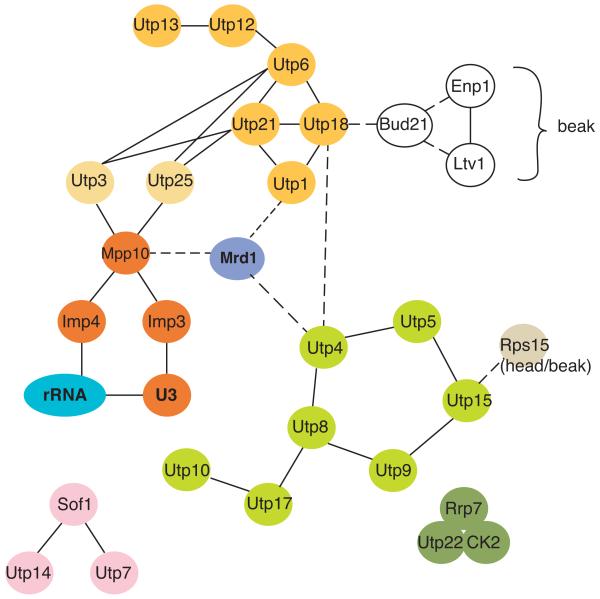
In addition to Rps from the body and platform, a large set of assembly factors is required for the early, co-transcriptional rRNA cleavage steps. These proteins, as well as the essential snoRNA U3, are collectively referred to as the small subunit (SSU) processosome, and comprise several subcomplexes: UtpA, UtpB, UtpC and Mpp10/U3 [31-35]. Data in the literature indicate that there is at least one additional complex, consisting of Utp7, Utp14 and Sof1. Interactions within the UtpA and UtpB subcomplexes, as well as among complexes have been mapped by yeast-2-hybrid screens and the conceptually related protein complementation assay (PCA) [36,37], and are summarized in Figure 2 (see also: [34]). This and other work suggests the following assembly steps for the SSU processosome: The UtpA subcomplex binds first to nascent pre-rRNA transcripts [*38], somehow changing its structure to facilitate subsequent processing steps. This function is required under normal conditions but becomes dispensible when quality control mechanisms that degrade slowly-assembling particles are inactivated [39]. Base pairing between U3 and two regions in the pre-rRNA 5′ to the 18S rRNA (5′-ETS, Figure 3A) is required for incorporation of the U3/Mpp10 complex, which is bound to UtpB [40,41]. At this later stage, the base pairs between U3 and 18S rRNA sequences are formed [*42], stabilized by Imp4 [*43]. These base pairs are mutually exclusive with the formation of the central pseudoknot. Incorporation of the UtpC complex is independent of these events but requires Rrp5 [38], which binds near the 3′-end of 18S rRNA and within the ITS1 spacer sequence between 18S and 5.8S rRNA [44].
Figure 2.
Protein-protein interactions within the SSU processosome. Members of the UtpA, B and C complexes are shown in light green, dark green, and yellow, respectively. Utp3 and Utp25 are shown in light yellow, as they have not formally been described as members of UtpB; however, they have intimate connections to this complex. Members of the Imp3/Imp4/Mpp10 complex are shown in orange. Interactions between complexes are shown as dotted lines. Data are from [36,37,40,41,62].
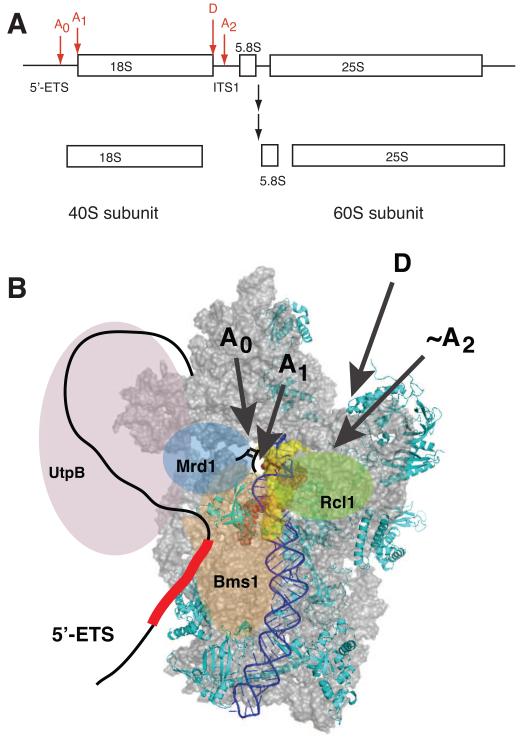
Figure 3.
(A) pre-rRNA precursor and cleavage sites for 18S rRNA production. (B) Structural model of early pre-40S ribosomes, viewed from the subunit interface. Only Rps required for early steps are depicted [*27]. H44 is highlighted in blue. Nucleotides that form base pairs with U3 snoRNA and ITS1 are shown in red or yellow spacefill, respectively. In early pre-ribosomes the top of H44, which otherwise covers the U3-interacting regions, is disrupted and might be dislocated to allow access to U3 snoRNA [*45]. The nucleotides that form base pairs with the yellow region in H44 are directly downstream of cleavage site A2 [*45], which allows the rough location of that site. The 5′- and 3′-ends of 18S rRNA (sites A1 and D) point toward the back, as indicated with arrows. Bms1 and Rcl1 are positioned where Tsr1 binds, and near the A2 site, respectively. U3 snoRNA binds the red regions in 18S rRNA, as well as regions in the 5′-ETS, schematically depicted as a red box, and for clarity is not explicitly shown. Mrd1 is positioned to bind near the U3 binding site, as well as near UtpB, in keeping with interactions with both ([58], see Figure 2). The UtpB complex, shown in lavender, forms interactions with the U3/Mpp10 complex, and Enp1 and Ltv1 [36,37,40,41,62], located at the back of the beak [**28,52,*53].
These interactions, as well as existing structural and biochemical data, provide insight into the organization of early pre-40S ribosomes despite the lack of any direct structural data (Figure 3): Early SSU-dependent cleavages occur in a body-like particle, with a partially assembled platform and head, but with formation of the central pseudoknot inhibited by U3 snoRNA binding. The 5′-end of 18S rRNA is located on the solvent side, suggesting that the hairpin with cleavage sites A0 and A1 extends in that direction. On the subunit interface of the mature particle, the central pseudoknot is rendered inaccessible by the top of the decoding site helix, H44 (Figure 3B). However, in early pre-40S particles, the top of H44 is not yet formed, leaving the central pseudoknot accessible to U3 binding [*45]. Furthermore, in the early pre-40S structure cleavage site A2 in ITS1 is located near the top of H44. Cleavage site A2 is recognized by the nuclease Rcl1 (D.M. Horn, S.L. Mason & K.K., in revision), which binds the GTPase Bms1. Bms1 interacts with U3 snoRNA [46] and H44 (K.K., unpublished), and based on the substantial sequence similarity with Tsr1 [47], is likely positioned to the left of H44, where Tsr1 is bound in later pre-40S particles [**28]. Together these data suggest that the U3 snoRNP is located on the subunit interface, where it interacts with the Bms1/Rcl1 complex. As a result, the 5′-ETS sequence between the U3 binding sites and the A0/A1 cleavage sites must wrap around from the solvent side to the subunit interface. As ITS1 binds near the platform [**28], it is likely that the 5′-ETS binds near the assembling beak, which could explain the interactions between the UtpB component Utp18, and the Ltv1/Enp1 subcomplex located at the beak. It will be exiting to obtain structures of such particles in the next several years.
Early co-transcriptional and late cytoplasmic events are separated by a conformational switch and involve a different set of proteins
While the early assembly events described above occur co-transcriptionally in the nucleolus, a post-transcriptional stage in 40S ribosome assembly takes place in the cytoplasm. These two stages are not only separated physically, their hierarchy is imposed by a structural switch in pre-40S ribosomes, and the almost complete exchange of the bound assembly factors: Early in 40S assembly, base pairing between the 3′-strand of H44 and ITS1 prevents premature cleavage at the 3′-end, formation of the decoding site, while allowing access of U3 snoRNA to the central pseudoknot [*45]. Additionally, while early assembly events require a large number of SSU components [35], the assembly intermediate that accumulates in the cytoplasm contains only seven assembly factors [*48, **28]; of these, only Dim1 and Pno1 bind early assembly intermediates. This suggests that at some point between cleavage at site A1 (to generate the mature 5′-end) and before nuclear export, the SSU components must dissociate to allow for binding of the late 40S assembly factors. What triggers this remodeling (perhaps cleavage at sites A1 or A2), or whether proteins catalyze SSU processome removal, akin to the AAA+-ATPases, which perform similar roles during 60S assembly [49, **50, 51], is unknown. Similarly, the inhibitory H44-ITS1 duplexes must be unwound to allow for the conformational switch in H44, required for cleavage at the 3′-end of 18S rRNA [*45].
Late Cytoplasmic Events in Assembly
Cytoplasmic 40S assembly requires seven stably bound assembly factors: the methylase Dim1, the nuclease Nob1 and its regulator Pno1, the export adaptor Ltv1 and its binding partner Enp1, the kinase Rio2 and the GTPase-like protein Tsr1. In addition, the kinase Rio1 and the ATPase Fap7 also transiently interact with this particle to produce mature 40S subunits. While the binding sites for these factors have been recently mapped [52,*53], only a few have known functions. Dim1 methylates two conserved adenosines near the 3′-end in a universally conserved reaction [54]. Pno1 modulates Nob1′s RNA binding activity and is required for efficient Nob1-dependent formation of the 3′-end of 18S rRNA [55].
Cytoplasmic pre-40S subunits are inundated with large concentrations of mRNAs, translation initiation factors and 60S subunits, but premature formation of 80S subunits leads to efficient degradation of the pre-18S rRNA [**30]. To chaperone the pre-40S particle all seven cytoplasmic assembly factors cooperate to inhibit each step in the translation initiation pathway [**28]. Assembly factors bound to the subunit interface (Tsr1, Rio2 and Dim1) and the platform (Pno1 and Nob1) prevent binding of translation initiation factors eIF1, eIF1A, and eIF3, while Enp1 and Ltv1 prevent opening of the mRNA channel. Tsr1 blocks joining of 60S subunits and, finally, the decoding site is deformed. Interestingly, with the exception of Dim1, there are no bacterial homologs of these proteins. Nevertheless there are non-homologous assembly factors, which could have analogous functions. E.g., mRNA recruitment in bacteria requires the Shine-Dalgarno sequence, which is bound and possibly blocked by the GTPase Era [56]. Similarly, the bacterial KH-domain containing protein RbfA deforms the decoding site [57].
If Tsr1 blocks premature joining of 60S subunits with assembling 40S subunits, then how can pre-18S rRNA-containing 40S subunits enter polysomes as described [**30]? This apparent paradox indicates that there is more than one cytoplasmic 40S assembly intermediate: one or more that contain Tsr1 and at least one later particle, which enters polysomes and does not contain Tsr1. Consistent with this hypothesis, it has long been known that cytoplasmic 40S ribosome maturation involves two chemical changes to the 18S rRNA. These include cleavage at the 3′-end to produce the mature 18S rRNA [22,*45] and the universally conserved methylation of two neighboring adenosines in H45, the terminal stemloop [54]. In addition, Rps10, Rps26, and Rps3 must be incorporated, the latter from a salt-labile conformation [*48, **28]. Furthermore, two conformational rearrangements, the formation of the decoding site [**28] as well as the formation of contacts between the very C-terminus of Rps14 and 18S rRNA [58] take place. Finally, the seven stably bound assembly factors dissociate. This extensive (and likely incomplete) list of distinct steps in cytoplasmic 40S assembly suggests that this stage is substantially more complex than previously thought. What is the order of these steps and how are they regulated? The answer to these questions awaits further study but promises to be as intricate as the recently unraveled 60S maturation pathway [*59].
Future Directions
Towards a better understanding of 40S assembly, we must now provide richer functional as well as structural data. Drawing on the identification and mapping of subcomplexes, this will likely require generating of point mutations that more subtly interfere with protein function, perhaps by disrupting a protein-protein or protein-RNA interface [36,46,50,55]. This will be useful because depletion of one assembly factor often leads to co-depletion of many others, thereby obscuring the role of any single protein. Furthermore, novel intermediates need to be identified, as assembly is clearly much more complex than Northern blots reveal. This will require the development of new techniques such as CRAC crosslinking [*53, 60] or the revisiting of old ones, such as RNA structure probing [*42,*45]. High-resolution structures of individual assembly factors, or smaller complexes, as well as medium-resolution electron microscopy-derived structures of pre-ribosomal complexes will also have a tremendous impact on better understanding the assembly process.
As the ribosome assembly field matures its interface with other fields will also be explored: How do ribosomes transition from assembly to translation? Does the first round of translation initiation utilize the same mechanism and translation initiation factors as any subsequent one? If there are specialized ribosomes, as suggested [61], are there specialized assembly factors to generate certain subpopulations? And finally, how exactly is the ribosome assembly machinery integrated with the cell cycle?
Highlights.
snoRNAs prevent formation of secondary structure, may modulate insertion of proteins
co-transcriptional binding of proteins to the body and platform
conserved pathway of protein assembly
co-transcriptional and cytoplasmic maturation steps separated by conformational switch
cytoplasmic assembly comprises many novel steps
Acknowledgements
I thank the members of my laboratory for many helpful discussions and comments on the manuscript. Work on 40S ribosome assembly in my lab is funded by the NIH (1R01GM086451), NSF (MCB0845156), and a Beginning Investigator grant from the AHA.
Footnotes
The same conclusion can be drawn for box H/ACA snoRNAs, which introduce pseudouridylation into rRNA [16], but which are not discussed for space reasons. Interestingly, snoRNA binding sites can overlap, indicating either that they function one after the other, or that they occur alternatively, thereby combinatorially increasing the diversity in the ribosome pool.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 2.Hage AE, Tollervey D. A surfeit of factors: why is ribosome assembly so much more complicated in eukaryotes than bacteria? RNA Biol. 2004;1:10–15. [PubMed] [Google Scholar]
- 3.Strunk BS, Karbstein K. Powering through ribosome assembly. RNA. 2009;15:2083–2104. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochimica et biophysica acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends in biochemical sciences. 2010;35:260–266. doi: 10.1016/j.tibs.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *6.Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. In this work, the authors describe a modified version of “pulse-chase” labeling that provides improved time-resolution for rRNA processing. By combining this technical innovation with complex mathematical modeling, they show that, akin to cleavage of the 35S pre-rRNA, the 2′-O-methyl modifications are acquired co-transcriptionally.
- 7.Hughes JM, Ares M., Jr. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. The EMBO journal. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li HD, Zagorski J, Fournier MJ. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morrissey JP, Tollervey D. Yeast Snr30 Is a Small Nucleolar RNA Required for 18S Ribosomal-RNA Synthesis. Mol. Cell. Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Molecular cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA. 2009;15:1716–1728. doi: 10.1261/rna.1724409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- *13.Rabl J, Leibundgut M, Ataide S, Haag A, Ban N. Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. This work is the culmination of the efforts from many labs, and provides atomic-resolution structures of 40S ribosomes, which show the placement of all 32 Rps.
- 14.Wimberly BT, Brodersen DE, Clemons WM, Jr., Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- *15.Fayet-Lebaron E, Atzorn V, Henry Y, Kiss T. 18S rRNA processing requires base pairings of snR30 H/ACA snoRNA to eukaryote-specific 18S sequences. EMBO J. 2009;28:1260–1270. doi: 10.1038/emboj.2009.79. In this work, the authors map the binding site of the essential snoRNA snR30 to the eukaryotic extension segment ES6. Interestingly, the data show that the snR30 binding site overlaps the site of a tertiary interaction with ES3, indicating that snR30 binding is required to prevent the premature formation of this tertiary structure element, which, according to the recent 40S structures, could serve to stabilize the body.
- 16.Kiss T, Fayet-Lebaron E, Jady BE. Box H/ACA small ribonucleoproteins. Mol. Cell. 2010;37:597–606. doi: 10.1016/j.molcel.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 17.Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Combs DJ, Nagel RJ, Ares M, Jr., Stevens SW. Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol Cell Biol. 2006;26:523–534. doi: 10.1128/MCB.26.2.523-534.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lebaron S, Froment C, Fromont-Racine M, Rain JC, Monsarrat B, Caizergues-Ferrer M, Henry Y. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol Cell Biol. 2005;25:9269–9282. doi: 10.1128/MCB.25.21.9269-9282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leeds NB, Small EC, Hiley SL, Hughes TR, Staley JP. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol Cell Biol. 2006;26:513–522. doi: 10.1128/MCB.26.2.513-522.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu YT, Shu MD, Steitz JA. A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA. 1997;3:324–331. [PMC free article] [PubMed] [Google Scholar]
- 22.Pertschy B, Schneider C, Gnadig M, Schafer T, Tollervey D, Hurt E. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem. 2009;284:35079–35091. doi: 10.1074/jbc.M109.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Held WA, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J. Biol. Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- 24.Mizushima S, Nomura M. Assembly mapping of 30S ribosomal proteins from E. coli. Nature. 1970;226:1214–1218. doi: 10.1038/2261214a0. [DOI] [PubMed] [Google Scholar]
- **25.Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008;455:1268–1272. doi: 10.1038/nature07298. In this article, the authors use time-resolved hydroxylradical footpinting to determine the kinetics of protein-mediated folding of bacterial 16S rRNA. Surprisingly, the data show that not only do contacts attributed to different proteins occur on different timescales, but even individual contacts attributed by a single protein are made over different timescales. This indicates that an initial “encounter” complex is remodeled so that additional contacts are successively acquired.
- **26.Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science. 2010;330:673–677. doi: 10.1126/science.1193220. In this ground-breaking work, the authors use electron microscopy to determine the 3D structures of 30S ribosome assembly intermediates prepared in vitro. These structures are correlated with kinetic measurements to show that binding of Rps to the body results in the stable and rapid accumulation of an assembly intermediate, followed by formation of the platform and head. Even largely assembled subunits often lack the homologs for Rps3, Rps14, Rps20 and Rps29.
- *27.Ferreira-Cerca S, Poll G, Gleizes PE, Tschochner H, Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 2005;20:263–275. doi: 10.1016/j.molcel.2005.09.005. This is the first of two beautiful papers in which the effects of deletions of individual Rps on pre-rRNA processing and export are investigated. This work, together with the recent 40S structures, shows that assembly of eukaryotic Rps in vivo and prokaryotic Rps in vitro follow very similar pathways.
- **28.Strunk BS, Loucks CR, Su M, Vashtish H, Cheng S, Brooks CL, 3rd, Karbstein K, Skiniotis G. Ribosome Assembly Factors Prevent Premature Translation Initiation by 40S Assembly Intermediates. Science. 2011 doi: 10.1126/science.1208245. In this work we describe the cryo-EM structure of a late 40S assembly intermediate, containing seven-late binding assembly factors. By solving the structures of several intermediates lacking individual assembly factors their position is assigned. The data suggest how all seven assembly factors cooperate to prevent premature translation initiation on this cytoplasmic pre-40S assembly intermediate. Furthermore, it is shown that Rps from the platform and head are still assembling.
- *29.Bhattacharya A, McIntosh KB, Willis IM, Warner JR. Why Dom34 stimulates growth of cells with defects of 40S ribosomal subunit biosynthesis. Molecular and cellular biology. 2010;30:5562–5571. doi: 10.1128/MCB.00618-10. In this work, the authors use a genetic screen to identify the dom34 gene as synthetically lethal with the deletions of individual copies of most Rps genes. Subsequent biochemical analysis demonstrates that this effect is due to accumulation of inactive 80S ribosomes, which could titrate essential components. Dom34 is known to break apart such 80S complexes. This work indicates that assembly intermediates lacking an essential Rps proceed through the assembly line until they remain inactive in the cytoplasm.
- **30.Soudet J, Gelugne JP, Belhabich-Baumas K, Caizergues-Ferrer M, Mougin A. Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. EMBO J. 2010;29:80–92. doi: 10.1038/emboj.2009.307. In this paradigm-shifting work, the authors show that premature 40S ribosomes, containing 20S rRNA, Nob1, and possibly other factors, can enter the polysomes. These precursors were accumulated in the absence of the Rio1 kinase.
- 31.Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 33.Rudra D, Mallick J, Zhao Y, Warner JR. Potential interface between ribosomal protein production and pre-rRNA processing. Mol Cell Biol. 2007;27:4815–4824. doi: 10.1128/MCB.02062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim YH, Charette JM, Baserga SJ. Assembling a Protein-Protein Interaction Map of the SSU Processome from Existing Datasets. PloS one. 2011;6:e17701. doi: 10.1371/journal.pone.0017701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phipps KR, Charette JM, Baserga SJ. The SSU Processome in Ribosome Biogenesis-Progress and Prospects. WIREs RNA. 2011;2:1–21. doi: 10.1002/wrna.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Champion EA, Lane BH, Jackrel ME, Regan L, Baserga SJ. A direct interaction between the Utp6 half-a-tetratricopeptide repeat domain and a specific peptide in Utp21 is essential for efficient pre-rRNA processing. Mol Cell Biol. 2008;28:6547–6556. doi: 10.1128/MCB.00906-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freed EF, Baserga SJ. The C-terminus of Utp4, mutated in childhood cirrhosis, is essential for ribosome biogenesis. Nucleic Acids Res. 2010;38:4798–4806. doi: 10.1093/nar/gkq185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *38.Perez-Fernandez J, Roman A, De Las Rivas J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol Cell Biol. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. In this tour-de force manuscript, the authors dissect the assembly pathway of SSU components, providing evidence for both parallel assembly pathways as well as hierarchy in the assembly of the SSU processosome.
- 39.Wery M, Ruidant S, Schillewaert S, Lepore N, Lafontaine DL. The nuclear poly(A) polymerase and Exosome cofactor Trf5 is recruited cotranscriptionally to nucleolar surveillance. RNA. 2009;15:406–419. doi: 10.1261/rna.1402709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charette JM, Baserga SJ. The DEAD-box RNA helicase-like Utp25 is an SSU processome component. RNA. 2010;16:2156–2169. doi: 10.1261/rna.2359810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldfeder MB, Oliveira CC. Utp25p, a nucleolar Saccharomyces cerevisiae protein, interacts with U3 snoRNP subunits and affects processing of the 35S pre-rRNA. The FEBS journal. 2010;277:2838–2852. doi: 10.1111/j.1742-4658.2010.07701.x. [DOI] [PubMed] [Google Scholar]
- *42.Dutca LM, Gallagher JE, Baserga SJ. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res. 2011;39:5164–5180. doi: 10.1093/nar/gkr044. In this article, the authors use creative in vivo DMS modification experiments to identify novel base pairing interactions between the essential snoRNA U3 and pre-rRNA. Excitingly, the authors also dissect the pathway of U3-pre-rRNA binding to show that individual base pairing interactions are made independently, with the ones formed with the 5′-most elements forming first and producing distinct intermediates.
- *43.Gerczei T, Shah BN, Manzo AJ, Walter NG, Correll CC. RNA chaperones stimulate formation and yield of the U3 snoRNA-Pre-rRNA duplexes needed for eukaryotic ribosome biogenesis. Journal Mol. Biol. 2009;390:991–1006. doi: 10.1016/j.jmb.2009.05.072. In this work, the authors use quantitative kinetic and thermodynamic measurements to determine the roles of Imp3 and Imp4 in promoting the formation of the U3/pre-18S interaction. They achieve this by destabilizing an internal structure in U3 and stabilizing the U3/rRNA interaction.
- 44.Young CL, Karbstein K. The roles of S1 RNA-binding domains in Rrp5’s interactions with pre-rRNA. RNA. 2011;17:512–521. doi: 10.1261/rna.2458811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *45.Lamanna AC, Karbstein K. A Conformational Change Regulates Pre-18S Cleavage. J. Mol. Biol. 2011;405:3–17. doi: 10.1016/j.jmb.2010.09.064. In this work, we use quantitative RNA binding experiments, in vivo and in vitro DMS probing, RNA cleavage experiments, and in vivo mutagenesis to show that prior to cleavage at site A2, sequence elements in the non-coding ITS1 spacer between 18S and 5.8S rRNAs form base pairs with the 3′-end of H44. These base pairs prevent premature formation of the decoding site as well as premature 3′-end maturation, thereby imposing a hierarchy on pre-rRNA cleavage events.
- 46.Karbstein K, Jonas S, Doudna JA. An essential GTPase promotes assembly of preribosomal RNA processing complexes. Mol. Cell. 2005;20:633–643. doi: 10.1016/j.molcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 47.Gelperin D, Horton L, Beckman J, Hensold J, Lemmon SK. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA. 2001;7:1268–1283. doi: 10.1017/s1355838201013073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *48.Schafer T, Maco B, Petfalski E, Tollervey D, Bottcher B, Aebi U, Hurt E. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006;441:651–655. doi: 10.1038/nature04840. In this article, the authors show that the assembly factors Enp1 and Ltv1, and the ribosomal protein Rps3 form a salt-labile complex on late cytoplasmic 40S ribosomes. Furthermore, the authors also demonstrate that the casein kinase Hrr25 can phosphorylate members of this complex. It is suggested that this could regulate 40S export, akin to the regulation of 60S export by Hrr25.
- 49.Gadal O, Strauss D, Braspenning J, Hoepfner D, Petfalski E, Philippsen P, Tollervey D, Hurt E. A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. Embo J. 2001;20:3695–3704. doi: 10.1093/emboj/20.14.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **50.Ulbrich C, Diepholz M, Bassler J, Kressler D, Pertschy B, Galani K, Bottcher B, Hurt E. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell. 2009;138:911–922. doi: 10.1016/j.cell.2009.06.045. In this cutting-edge manuscript, the authors combine beautiful biochemistry and elegant electronmicroscopy to show that the Rea1 ATPase and Rsa4 interact via a MIDAS-motif. This interaction is required for Rea1 to remove Rsa4 and a whole group of additional proteins from nascent 60S particles.
- 51.Pertschy B, Saveanu C, Zisser G, Lebreton A, Tengg M, Jacquier A, Liebminger E, Nobis B, Kappel L, van der Klei I, et al. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol Cell Biol. 2007;27:6581–6592. doi: 10.1128/MCB.00668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campbell MG, Karbstein K. Protein-Protein Interactions within Late Pre-40S Ribosomes. PLoS One. 2011;6:e16194. doi: 10.1371/journal.pone.0016194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *53.Granneman S, Petfalski E, Swiatkowska A, Tollervey D. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein crosslinking. EMBO J. 2010;29:2026–2036. doi: 10.1038/emboj.2010.86. In this manuscript, the authors use the recently developed CRAC crosslinking technique to map late pre-40S assembly factors onto the pre-rRNA structure. The results illustrate both the tremendous potential of this technique, as the binding sites for most ribosome assembly factors remain unknown, but also potential pitfalls, as multiple sites of crosslinking are sometimes found. These may be hard to reconcile with single structures or specific crosslinks. See also: references 28&52
- 54.Lafontaine D, Delcour J, Glasser AL, Desgres J, Vandenhaute J. The DIM1 gene responsible for the conserved m6(2)Am6(2)A dimethylation in the 3′-terminal loop of 18 S rRNA is essential in yeast. J. Mol. Biol. 1994;241:492–497. doi: 10.1006/jmbi.1994.1525. [DOI] [PubMed] [Google Scholar]
- 55.Woolls HA, Lamanna AC, Karbstein K. The Roles of Dim2 in Ribosome Assembly. J. Biol. Chem. 2011;286:2578–2586. doi: 10.1074/jbc.M110.191494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tu C, Zhou X, Tropea JE, Austin BP, Waugh DS, Court DL, Ji X. Structure of ERA in complex with the 3′ end of 16S rRNA: implications for ribosome biogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14843–14848. doi: 10.1073/pnas.0904032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Datta PP, Wilson DN, Kawazoe M, Swami NK, Kaminishi T, Sharma MR, Booth TM, Takemoto C, Fucini P, Yokoyama S, et al. Structural aspects of RbfA action during small ribosomal subunit assembly. Mol Cell. 2007;28:434–445. doi: 10.1016/j.molcel.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jakovljevic J, de Mayolo PA, Miles TD, Nguyen TM, Leger-Silvestre I, Gas N, Woolford JL., Jr. The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol. Cell. 2004;14:331–342. doi: 10.1016/s1097-2765(04)00215-1. [DOI] [PubMed] [Google Scholar]
- *59.Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Molecular cell. 2010;39:196–208. doi: 10.1016/j.molcel.2010.06.018. In this inspiring piece of work, the authors use yeast genetics to order the known events in cytoplasmic 60S assembly. The data provide evidence for both parallel as well as hierarchical maturation events.
- 60.Granneman S, Kudla G, Petfalski E, Tollervey D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci U S A. 2009;106:9613–9618. doi: 10.1073/pnas.0901997106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gilbert WV. Functional specialization of ribosomes? Trends in biochemical sciences. 2011;36:127–132. doi: 10.1016/j.tibs.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]