Summary
The histone 3 lysine 79 (H3K79) methyltransferase Dot1l has been implicated in the development of leukemias bearing translocations of the Mixed Lineage Leukemia (MLL) gene. We identified the MLL-fusion targets in an MLL-AF9 leukemia model, and conducted epigenetic profiling for H3K79me2, H3K4me3, H3K27me3 and H3K36me3 in hematopoietic progenitor and leukemia stem cells (LSC). We found abnormal profiles only for H3K79me2 on MLL-AF9 fusion target loci in LSC. Inactivation of Dot1l lead to down-regulation of direct MLL-AF9 targets and an MLL-translocation associated gene expression signature, while global gene expression remained largely unaffected. Suppression of MLL-translocation associated gene expression corresponded with dependence of MLL-AF9 leukemia on Dot1l in vivo. These data point to DOT1L as a potential therapeutic target in MLL-rearranged leukemia.
Introduction
Chromatin modifying enzymes are central components of the epigenetic machinery that control developmental programs. It has become increasingly clear that these enzymes are also critical for cancer development (Albert and Helin, 2010; Chi et al., 2010). Genes encoding histone methyltransferase EZH2 and DNA methyltransferase DNMT3a are frequently mutated in hematopoietic malignancies (Ernst et al., 2010; Ley et al., 2010; Morin et al., 2010; Nikoloski et al., 2010), and recent trials have demonstrated clinical efficacy of epigenetic-directed therapies with histone deacetylase and DNA methyltransferase inhibitors (Griffiths and Gore, 2008; Issa, 2007). However, it remains unclear how to predict which patients will respond to epigenetic-directed therapies as clearly defined abnormalities in epigenetic pathways are often lacking. Identification of defined cancers that depend upon specific epigenetic abnormalities should lead to more informed use of therapies that target epigenetic programs.
Prominent examples of a cancer driven by mutations involving an epigenetic regulator are leukemias bearing translocations involving the Mixed Lineage Leukemia (MLL) gene. Wildtype MLL possesses a methyltransferase domain, which modifies histone H3 on lysine 4 (Milne et al., 2002; Nakamura et al., 2002), that is absent in the oncogenic fusion protein (Ayton and Cleary, 2001; Krivtsov and Armstrong, 2007). Biochemical studies have shown that several common MLL-fusion oncoproteins co-purify with protein complexes normally associated with transcriptional elongation (Mohan et al., 2010). At least three complexes, PAFc, DOT1L, and pTEFb (also designated AEP or SEC) bind MLL-fusion proteins. The polymerase associated factor complex (PAFc) regulates RNA polymerase II and associates with the NH3-terminal portion of wildtype MLL and MLL-fusion proteins (Muntean et al., 2010). The pTEFb complex (CDK9/cyclinT), which phosphorylates RNA polymerase II, interacts with MLL-fusion partners such as ENL, ELL (as well as ELL2, ELL3), AF4 and AF5 (Lin et al., 2010; Yokoyama et al., 2010) and co-purifies with several MLL-fusion proteins. The DOT1L complex consists of DOT1L, a histone methyltransferase that modifies Histone H3 on lysine 79 (H3K79), and multiple MLL-fusion partners such as AF9, ENL and AF10 (Bitoun et al., 2007; Mohan et al.; Mueller et al., 2007; Mueller et al., 2009; Okada et al., 2005; Zhang et al., 2006). Based on these biochemical data, it appears that MLL-fusions directly stimulate transcriptional elongation via recruitment of complexes that lead to deregulated transcription. However, how these complexes interact with each other and the extent to which each contributes to MLL-fusion mediated leukemia development remains unclear.
The interaction between MLL-fusion proteins and DOT1L has prompted assessment of H3K79 methylation in MLL-fusion leukemia models and patient samples. Enhanced H3K79 methylation has been associated with genes over expressed in MLL-fusion driven leukemias (Guenther et al., 2008; Krivtsov and Armstrong, 2007; Krivtsov et al., 2008). In particular, enhanced H3K79 methylation has been described on several known MLL-fusion target loci including the 5′ HoxA cluster genes (HoxA7-13) and Meis1 (Guenther et al., 2008; Krivtsov et al., 2008; Milne et al., 2005). Several groups have demonstrated that Dot1l is required for transformation of murine bone marrow cells with MLL-AF10 (Okada et al., 2005), MLL-AF9, (Chang et al., 2010; Jo et al., 2011; Nguyen et al., 2011) and possibly MLL-AFX (Chang et al., 2010; Okada et al., 2005).
As histone H3K79 dimethylation (H3K79me2) is a histone mark associated with actively transcribed genes, it has been hypothesized that DOT1L may play an active role in maintenance of MLL-fusion mediated gene expression (Krivtsov and Armstrong, 2007; Okada et al., 2005). Genome wide analysis of MLL-rearranged leukemia samples has revealed unique transcriptional and H3K79 methylation profiles that can distinguish MLL-rearranged leukemias from MLL-germline leukemias (Armstrong et al., 2002; Ross et al., 2004; Krivtsov et al., 2008). Thus, it is possible that the institution of abnormal histone methylation patterns at key loci is a crucial step in MLL-fusion mediated transformation. However, the near ubiquitous association of H3K79 methylation with transcription is an argument against a specific role for Dot1l in MLL-fusion mediated leukemogenesis (Steger et al., 2008). In addition, whether a single chromatin mark can specifically regulate a leukemogenic transcription program is unknown. In this study, we determine genome wide histone methylation profiles for multiple modifications in normal hematopoietic progenitor cells and MLL-AF9 driven leukemia stem cells, define the MLL-AF9 target genes with an associated aberrant H3K79me2 profile, and assess the dependence of MLL-fusion mediated gene expression and leukemia maintenance on DOT1L.
Results
MLL-AF9 target loci are associated with H3K79me2 in abnormal amount and distribution
Previous reports (Guenther et al., 2008) demonstrated the presence of aberrant levels of H3K79me2 and perhaps H3K4me3 at MLL-AF4 target genes in a human cell line harboring an MLL-AF4 translocation. We assessed for the presence of a similar epigenetic lesion in a mouse model of MLL-AF9 leukemia stem cells (LSC) defined previously as Il-7R−Lin−Sca-1−c-Kit+CD34+FcγRII/III+ leukemic granulocyte-macrophage progenitors (L-GMP) (Krivtsov et al., 2006). We identified the genome wide localization of H3K79me2 in L-GMP by chromatin immunoprecipitation (ChIP) followed by next generation sequencing (ChIP-Seq). In addition, we identified direct MLL-AF9 fusion protein target genes through use of metabolic labeling of MLL-AF9 with biotin followed by ChIP-Seq using streptavidin. Significant H3K79me2 was found associated with 5507 genes (Figure 1A, Table S1). MLL-AF9 was bound to the promoter region of 139 genes (Table S1), 120 of which also had significant H3K79me2 (Figure 1A, p=1.2e-14). The target genes include genes with known functional significance in MLL-leukemia biology such as HoxA cluster genes and Meis1, and less well defined targets such as Runx2 and Jmjd1c (Figure S1). Gene Set Enrichment Analysis (GSEA) showed expression of the 139 MLL-AF9 targets is highly enriched in L-GMP compared to the corresponding normal granulocyte-macrophage progenitors (GMP) (p=<0.001), suggesting that transduction with MLL-AF9 causes increased expression of its direct targets (Figure 1B). We analyzed the relationship between the amount of H3K79me2 and gene expression, and found a strong positive correlation (Figure 1C). Globally, H3K79me2 levels peaked sharply just downstream of the transcription start site (TSS), and the height of the peak correlated with the amount of mRNA transcribed from the respective locus (Figure 1C). We analyzed the H3K79me2 profile of MLL-AF9 target genes in L-GMP and observed a consistent perturbation of this pattern, with a higher peak, and wider spatial distribution of the H3K79me2 modification to both sides of the TSS (Figure 1D and 1E). To further assess the association of H3K79me2 with MLL-AF9 targets, we created a control set of another 139 genes that showed increased expression in L-GMP compared to GMP, but were not direct MLL-AF9 targets. The level of H3K79me2 in the control set is the same as expected for highly expressed genes (Figure 1D). Therefore an H3K79me2 epigenetic lesion is present at MLL-AF9 target loci in murine MLL-AF9 L-GMPs.
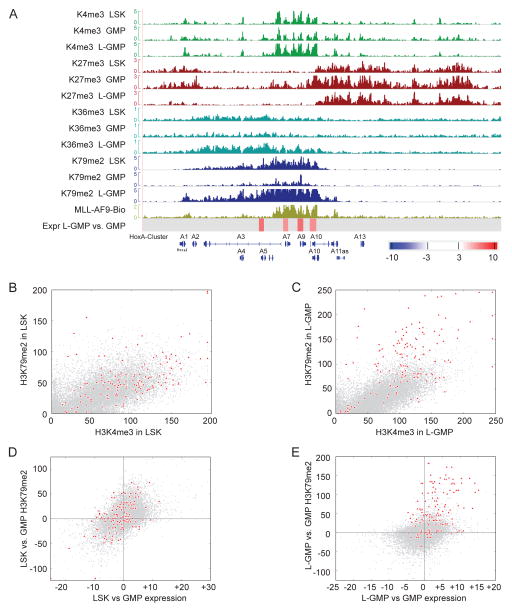
Figure 1. Presence of an H3K79me2 epigenetic lesion on direct MLL-AF9 fusion targets.
(A) ChIP-Sequencing for H3K79me2 and MLL-AF9 in MLL-AF9 transformed cells. The whole genome view denotes regions associated with H3K79me2 (5507 genes, p=0.02, light green tracks) and Bio-MLL-AF9 (MLL-AF9 direct targets, 139 genes, p=0.0005, dark green tracks). Venn Diagram demonstrating overlap of MLL-AF9 direct targets and genes associated with H3K79me2.
(B) GSEA of the 139 MLL-AF9 direct target genes demonstrating enrichment of gene expression for MLL-AF9 targets in leukemia stem cells (L-GMP) versus normal murine granulocyte macrophage progenitors (GMP) (p<0.001).
(C) H3K79me2 ChIP-Seq signal height and position are shown relative to transcription start site (TSS) for genes grouped according to their expression level in MLL-AF9 L-GMP (No = dark blue to high = red).
(D) Height and distribution of H3K79me2 profiles around the TSS of MLL-AF9 targets (brown) compared to non-targets with similar expression levels (green).
(E) H3K79me2 profiles of selected MLL-AF9 targets and non-targets.
(F) H3K79me2 signal height and position similar to (D) in human MLL-rearranged AML. H3K79me2 ChIP Sequencing was performed on a human MLL-AF9 rearranged primary AML sample. The 139 MLL-AF9 target loci determined in (A) converted to 120 MLL-AF9 target loci in the human genome, defining the MLL-AF9 direct targets (brown). Control gene sets with differential expression levels were created from a previously published expression array data set on human MLL-rearranged AML(Ross et al., 2004) (green).
(G) H3K79me2 profiles relative to the TSS for a list of “core” targets (turquoise) defined as the overlap of the direct MLL-AF9 targets with a previously published gene set of MLL-AF4 direct targets identified in a human MLL-AF4 rearranged cell line.
(H) H3K79me2 profiles relative to the TSS of MLL-fusion target genes expressed in a control AML patient sample with a normal karyotype (no MLL-rearrangement).
(I) H3K79me2 profiles relative to the TSS for “core targets” (defined as in G), in a control AML patient sample with a normal karyotype.
In order to test the relevance of this finding in human leukemias, we performed ChIP-Seq for H3K79me2 on a primary MLL–AF9 rearranged AML sample obtained at diagnosis. We created groups of genes with defined expression levels utilizing a published gene expression data set of MLL-rearranged human primary patient samples (Ross et al., 2004), and again found a strong correlation between global expression levels and the presence of H3K79me2 (Figure 1F, red and blue lines). Next, we analyzed H3K79me2 profiles at loci corresponding to the MLL-AF9 targets defined in the mouse model. We observed an increase in H3K79me2 peak signal height and distribution comparable to the murine H3K79me2 profiles (Figure 1F). For further characterization, we overlapped our murine MLL-AF9 target gene set with a recently published list of MLL-AF4 targets (Guenther et al., 2008), thus establishing a core set of common MLL-fusion targets (Table S1). When interrogating human leukemia H3K79me2 profiles at the core MLL-fusion target loci, the presence of an epigenetic lesion became even more pronounced (Figure 1G). By comparison, H3K79me2 patterns on MLL-AF9 targets in an AML control patient sample with a normal karyotype were normal and mirrored that of the non-target control gene set and other highly expressed genes (Figure 1H and 1I). These findings demonstrate an H3K79me2 epigenetic lesion at MLL-fusion target genes specifically in human primary leukemia cells bearing an MLL-translocation.
MLL-AF9 disrupts the relationship between H3K79me2 and other histone modifications
Previous studies have demonstrated a molecular connection between H3K4me3 and H3K79 methylation (Lee et al., 2007). Thus, we asked whether the abnormal H3K79me2 methylation patterns correlated with additional epigenetic abnormalities on MLL-AF9 target loci. There may be a concomitant increase of H3K79me2 with other transcription associated marks such as H3K4me3 and H3K36me3, which could reflect increased transcriptional activity at these loci. Alternatively, H3K79me2 hypermethylation could override a presumptive need for H3K4me3 and/or H3K36me3 for transcription initiation or elongation, resulting in lower association of MLL-fusion target loci with these modifications. We assessed H3K79me2, H3K4me3 H3K36me3, which are associated with active gene expression, and H3K27me3, which is associated with repression of gene expression, in MLL-AF9 L-GMP, as well as normal hematopoietic stem cell-enriched LSK cells (Lin−, Sca-1+, c-Kit+), and GMP (Figure 2A). In addition, we correlated histone modification profiles with previously published gene expression profiles for these same populations (Krivtsov et al., 2006). This afforded us an integrated view of epigenetic and gene expression changes as cells undergo transitions from LSK to GMP, and from GMP to L-GMP. Figure 2A shows the histone methylation patterns on the HoxA cluster for all assessed populations, and the region targeted by MLL-AF9. The 5′ HoxA cluster genes are expressed in LSK cells, and associated with H3K4me3, H3K36me3 and H3K79me2. As cells differentiate to GMPs, H3K4me3, H3K36me3 and H3K79me2 decreases as does gene expression. Furthermore, adjacent H3K27me3 spreads into the 5′ HoxA cluster. In L-GMP, the H3K27me3, H3K4me3 and H3K36me3 patterns are reversed to profiles comparable with LSK cells. In contrast, H3K79me2 showed an increased amount and abnormal pattern as defined above.
Figure 2. Relationship between H3K79me2 and other histone modifications on direct MLL-AF9 fusion targets.
(A) ChIP-Sequencing for H3K4me3, H3K27me3, H3K36me3, and H3K79me2, in sorted LSK, GMP and L-GMP populations. A screen shot of the HoxA cluster shows changes in these epigenetic marks during normal development (LSK and GMP), and in MLL-AF9 driven leukemogeneis (L-GMP). Also shown: MLL-AF9 fusion ChIP-Seq in MLL-AF9 tansformed cells (MLL-AF9-Bio); differential gene expression in L-GMP versus GMP as assessed by expression array (Expr L-GMP vs. GMP), color legend denotes fold change.
(B) Genome wide representation of the relation between H3K4me3 and H3K79me2 in LSK cells on fusion target genes (red) compared to non-fusion target genes (grey).
(C) Genome wide representation of the relation between H3K4me3 and H3K79me2 in L-GMP on fusion target genes (red) compared to non-fusion target genes (grey).
(D) Changes in expression between LSK and GMP (obtained by expression array, x-axis) in correlation to changes in H3K79me2 (Chip-seq, y-axis). This correlation is shown for MLL-AF9 targets in red, and non-targets in grey.
(E) Changes in expression between L-GMP and GMP in correlation with changes in H3K79me2. Shown is the amount of H3K79me2 signal difference for MLL-AF9 target genes (red) compared to genes that show similar differences in expression between L-GMP and GMP, but are not MLL-fusion targets (grey). See also Figure S2.
On a genome wide scale, there were no major differences in the number of loci associated with H3K4me3, H3K27me3, H3K36me3 or H3K79me2 in LSK, GMP and L-GMP populations (LSK: 4142 genes are associated with H3K79me2, GMP: 5002, L-GMP 5507). However, when looking at locus specific combinations, differences emerged (Figure 2B–C and S2A–L). In LSK and GMP, there is a positive correlation between H3K4me3 and H3K79me2 (Figure 2B and S2A), and H3K36me3 and H3K79me2 (Figure S2D–E). The presence of H3K27me3 and H3K79me2 was mutually exclusive (Figure S2G–H). There was also an inverse correlation between H3K27me3 and H3K36me3 (Figure S2J–K). In LSK and GMP, the relationship between H3K4me3 and H3K79me2 is preserved for MLL-fusion target loci (shown in red, Figure 2B and S2A). In contrast, MLL-fusion target loci in L-GMP acquire an abnormal histone methylation pattern characterized by increased H3K79me2, resulting in a disturbed ratio of H3K4me3 and H3K79me2 (Figure 2C). A similar pattern emerges for H3K79me2 and H3K36me3 (Figure S2F). The normal levels of H3K4me3 and H3K36me3 on MLL-AF9 target loci suggest that abnormal H3K79me2 does not substitute for H3K4me3 or H3K36me3. We also analyzed the distribution of H3K4me3, H3K27me3, and H3K36me3 with respect to the transcription start site in LSK, GMP and L-GMP populations (Figure S2M). In contrast to H3K79me2 (Figure 1D), there were no differences in H3K4me3, H3K27me3 and H3K36me3 profiles between MLL-fusion target genes and non-target genes in L-GMP, or between MLL-fusion target genes in L-GMP versus LSK or GMP populations.
Next, we assessed changes in H3K79me2 with respect to changes in gene expression either during normal development (LSK versus GMP), or during leukemia development (L-GMP versus GMP). As cells differentiate from LSK to GMP, changes in expression mirror changes in H3K79me2, and this correlation is maintained for MLL-AF9 targets (Figure 2D). Conversely, when comparing changes in expression and H3K79me2 in L-GMP versus GMP cells, MLL-AF9 targets stand out as acquiring H3K79me2 in excess of what would be expected based on changes in expression alone (Figure 2E). This is restricted to MLL-AF9 target genes in MLL-AF9 leukemias: non-target genes that are upregulated in L-GMP versus GMP, and MLL-AF9 target loci that are physiologically regulated during the LSK to GMP transition maintain normal H3K79me2 patterns. The H3K79me2 specific epigenetic lesion suggests that the expression of MLL-fusion target genes may depend on H3K79me2 to a far greater extent in MLL-rearranged cells than in other (normal or malignant) cells, and that inhibition of H3K79 methylation might specifically downregulate MLL-fusion driven transcriptional programs.
Loss of Dot1l leads to differentiation, cell cycle changes and apoptosis in MLL-fusion driven leukemia cells
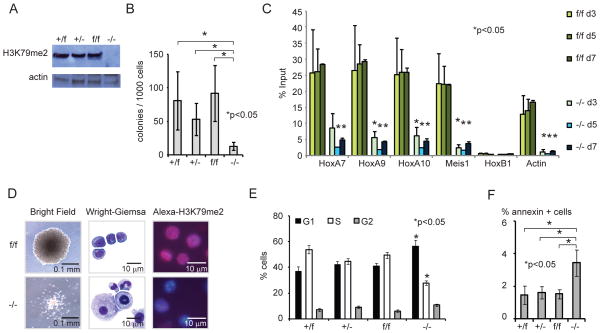
To establish that Dot1l and H3K79 methylation are indeed required for the maintenance of MLL-fusion driven transcriptional programs and leukemogenesis, we developed a conditional knockout mouse model for Dot1l. In this model, exon 5 of Dot1l, which encodes most of the methyltransferase active site (Min et al., 2003), is flanked by two loxP sites (Figure S3A–C). Homozygous Dot1lf/f mice are phenotypically normal and display normal blood counts (Figure S3D–F). We transformed lineage depleted (lin−) Dot1lf/f and Dot1l+/f bone marrow cells with retovirus expressing MLL-AF9 (Krivtsov et al., 2006). Transformed cells were subsequently transduced with retroviruses encoding either Cre-recombinase and YFP (MSCV-Cre-IRES-YFP, “Cre”) or YFP (MSCV-IRES-YFP, “MIY”) and sorted for GFP/YFP double positive cells. Introduction of Cre into transformed cells led to a near complete loss of H3K79me2 in Dot1l−/− cells (Figure 3A). Complete deletion of Dot1l profoundly decreased the number of blast-like colonies in methylcellulose that could be recovered after one week (Figure 3B). In addition, Dot1l−/− colonies were smaller compared to control colonies (data not shown).
Figure 3. Loss of Dot1l leads to decreased growth, differentiation and apoptosis of MLL-AF9 murine leukemia cells.
(A) Immunoblot analysis for H3K79me2 in MLL-AF9 transformed cells of the indicated geneotype 4 days after transduction with Cre or control retrovirus.
(B) Blast colony count of Dot1l-deleted MLL-AF9 transformed cells (−/−) in methylcellulose 10 days after transduction with Cre in comparison to controls. n= at least 3 independent experiments.
(C) H3K79me2 ChIP-qPCR from in vivo established MLL-AF9 leukemia cells on day 3, 5 and 7 after transduction with Cre. Loss of H3K79me2 is statistically significant on days 5 and 7 for HoxA7, HoxA9, HoxA11, Meis1 and Actin at p<0.05. n= 2–3 independent experiments for each time point.
(D) Morphologic changes (colony morphology in methylcellulose, Wright-Giemsa stain) and H3K79me2 immunofluorescence (Alexa594-H3K79me2 and DAPI nuclear stain) in established MLL-AF9 leukemia cells 10 days after transduction with Cre.
(E) Cell cycle changes (BrdU/7-AAD flow cytometry) in MLL-AF9 leukemia cells 7 days after transduction with Cre. n=3 independent experiments, *G0/1 increase significant at p<0.02 (+/f), p<0.04(+/−) and p<0.02 (f/f); decrease in S-phase significant at <0.0002 (+/f), p<0.0009(+/−) and p<0.0003 (f/f).
(F) Induction of apoptosis (annexin+/PI−) in MLL-AF9 leukemia cells 10 days after transduction with Cre. n=3 independent experiments.
Error bars: standard error (SEM)
See also Figure S3.
Next, we analyzed fully developed MLL-AF9 leukemias. Primary recipient mice were transplanted with MLL-AF9-GFP transformed lin− cells from either Dot1l+/f or Dot1lf/f mice. Established leukemias were isolated from moribund mice and transduced with Cre or control vector. Loss of H3K79me2 on several MLL-AF9 target genes (5′ HoxA cluster genes and Meis1) was assessed by ChIP followed by quantitative PCR. H3K79me2 at these loci was completely absent 5 days after retroviral transduction (Figure 3C). As observed for the in vitro transformed cells, colony formation of Dot1l−/− MLL-AF9 leukemia cells was greatly reduced, and colonies were smaller and less compact (Figure 3D). Control colonies were comprised of abundant cells with blast-like morphology, whereas loss of Dot1l induced profound morphologic changes consistent with terminal differentiation (Figure 3D). Loss of Dot1l also reduced the number of actively proliferating cells, with the majority of cells shifted towards G0/G1 (Figure 3E). In addition to morphologic and cell cycle changes, we observed a modest increase in the percentage of apoptotic cells after deletion of Dot1l (Figure 3F). We conducted additional studies in human MLL-rearranged leukemia cell using an shRNA approach, which yielded overall similar results with MLL-rearranged cells being more consistently affected by DOT1L suppression than MLL-germline cells (Figure S3G–M).
Loss of Dot1l has little effect on myeloid progenitors and HoxA9/Meis1a transformed cells
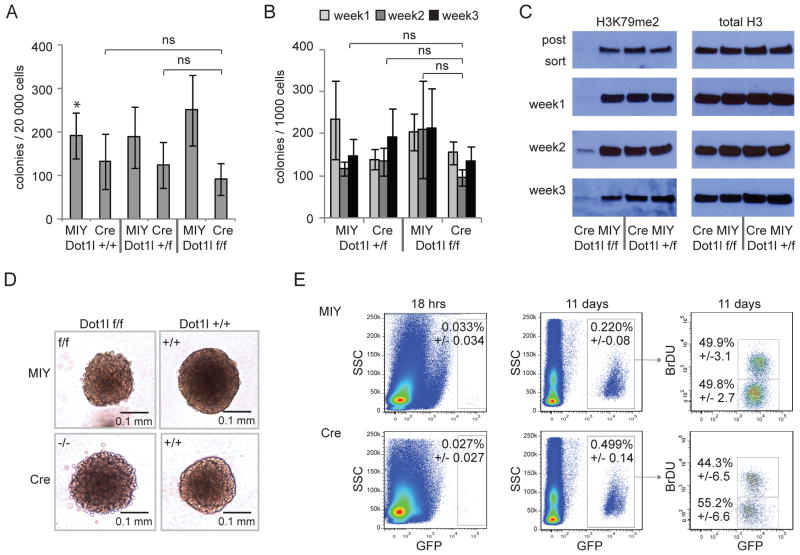
In order to determine if loss of Dot1l might non-specifically inhibit proliferation of all hematopoietic cells, we inactivated Dot1l in normal bone marrow and analyzed deleted cells in myeloid progenitor assays (Figure 4A). Dot1l+/+, Dot1l+/f and Dot1lf/f bone marrow transduced with Cre showed a moderate decrease in plating efficiency and colony size when compared to MIY transduced control populations. However, there was no significant difference between the three Cre-transduced populations (Figure 4A, S4A and S4B). Thus, in this short term assay we observed clear differential sensitivity between MLL-AF9 transformed cells (Figure 3B), and normal myeloid progenitors (Figure 4A).
Figure 4. Colony formation of normal hematopoietic progenitors and HoxA9/Meis1a transformed cells is unaffected by loss of Dot1l.
(A) Colony formation of normal hematopoietic progenitors 10 days after Cre-mediated excision of exon 5 of Dot1l (Dot1lf/f Cre) compared to controls. 3–5 independent experiments.
(B) Blast colony count of HoxA9/Meis1a transformed cells in methylcellulose after transduction with Cre in comparison to controls over 3 weeks of serial replating. 3–5 independent experiments.
(C) Serial assessment of H3K79me2 in HoxA9/Meis1a blast colonies over three weeks replating.
(D) Blast colony size and morphology in methylcellulose of Dot1l deleted and Dot1l wild type HoxA9/Meis1a transformed cells.
(E) Homing (18 hrs) and engraftment (11 days) of Dot1lf/f and Dot1l−/− cells after injection of 5×105 HoxA9/Meis1a transformed cells (3 days after transduction with Cre or MIY). In vivo BrdU labeling demonstrates actively dividing cells. All differences are not statistically significant.
Error bars: SEM, *p<0.05, ns = not significant.
See also Figure S4.
Given the limited replating potential of committed myeloid progenitors, we conducted additional experiments using a myeloid leukemia model where leukemic transformation is driven by introduction of HoxA9 and Meis1a (Wang et al., 2010) into LSK cells. In contrast to MLL-AF9 cells, complete deletion of H3K79me2 in HoxA9/Meis1a transformed cells had no impact on the number of blast-like colonies in methylcellulose during serial replating (Figure 4B). Tracking of H3K79me2 revealed outgrowth of a small proportion of cells that had escaped full deletion in later passages (Figure 4C). Morphologically, HoxA9/Meis1a transformed Dot1l−/− colonies were indistinguishable from control colonies (Figure 4D). When transplanted into mice, Dot1l−/− cells were able to efficiently home to the bone marrow (Figure 4E). 11 days after transplantation, both populations had dramatically expanded (~100-fold). BrdU incorporation revealed actively dividing cells in similar proportions in both groups. Western blot analysis of sorted cells from these animals revealed re-emergence of H3K79me2, albeit at a low level (Figure S4C). This suggests that while Dot1l−/− HoxA9/Meis1a driven leukemia cells are at a slight disadvantage to Dot1l heterozygous or wild type cells, they are still able to proliferate and expand substantially.
Loss of Dot1l specifically decreases expression of MLL-AF9 targets
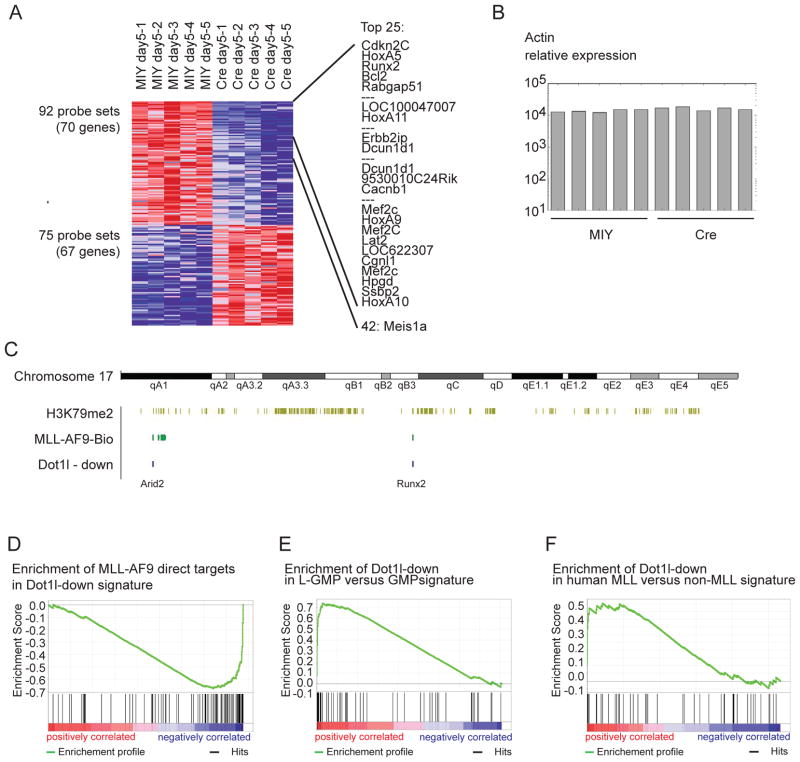
We next examined gene expression changes in MLL-AF9 leukemia cells after loss of Dot1l. H3K79 methylation has previously been reported to be ubiquitously coupled to transcription (Steger et al., 2008), raising the question whether loss of Dot1l would lead to widespread collapse of transcription. On the other hand, the aberrant amount and pattern of H3K79me2 could indicate that MLL-fusion targets would be more sensitive to Dot1l loss. Leukemic bone marrow from mice transplanted with Dot1lf/f MLL-AF9 cells was transduced with retroviral Cre or control vector. RNA was purified 3, 5 and 7 days after transduction, amplified and hybridized to murine Affymetrix 430 2.0 microarrays. We identified the genes differentially expressed at day 5 for further analysis as the major gene expression changes appeared firmly established by this time point (Figure S5). We found that despite the large number of loci associated with H3K79me2 (>5000, Figure 1A), only a small subset of genes showed a change in expression at p<0.01 (Figure 5A). In addition, we observed comparable numbers of genes with increased (75 probe sets (67 genes), Dot1l-up signature) and decreased (92 probe sets (70 genes), Dot1l-down signature) expression (p<0.01) (Figure 5A, Table S2). This indicates expression of the majority of genes associated with H3K79me2 did not significantly change after loss of this modification. As an example, the actin promoter is associated with H3K79me2, and although the methyl mark is lost after deletion of Dot1l (Figure 3C), actin expression remains largely unchanged (Figure 5B). In contrast, analysis of the set of genes with significantly decreased expression (Dot1l-down signature) revealed many genes known to be important in MLL-mediated leukemogenesis such as HoxA7-11 (Armstrong et al., 2002; Kawagoe et al., 1999; Zeisig et al., 2004), Mef2C (Krivtsov et al., 2006), and Meis1a (Zeisig et al., 2004) (Figure 5A), as well as newly identified MLL-AF9 direct targets such as Arid2 or Runx2 (Figure 5C). To investigate whether MLL-AF9 targets are enriched in the Dot1l-down set of genes, we performed GSEA and found significant enrichment of the MLL-AF9 targets in the Dot1l-down signature (p=0.01) (Figure 5D). We next used GSEA to assess enrichment of the Dot1l-down signature in several previously published gene expression data sets. Dot1l-down was highly enriched in a gene set distinguishing MLL-AF9 LSC (L-GMP) (Krivtsov et al., 2006) from their normal counterpart (GMP) (p<0.001) (Figure 5E). To assess the relevance of our signature to human disease, we performed GSEA on a dataset from human MLL and non-MLL rearranged AML (Ross et al., 2004). The Dot1l-down list was significantly enriched in the human MLL-rearranged AML samples (p=0.034) (Figure 5F). These results demonstrate that global expression patterns remain largely unaffected upon loss of Dot1l, while a small subset of genes whose expression is decreased are highly enriched for direct MLL-AF9 targets.
Figure 5. Loss of Dot1l specifically decreases expression of MLL-fusion driven transcriptional programs.
(A) Expression array of MLL-AF9 mouse leukemia cells 5 days after loss of Dot1l. Shown are all probe sets/genes with differential expression at p=0.01, as well as a list of the top 25 differentially expressed genes.
(B) Change in relative expression of Actin after loss of H3K79me2 associated with the actin promoter (Figure 3C).
(C) Example for overlay of ChIP-Seq results for H3K79me2 (light green) and MLL-AF9 direct targets (dark green) with expression array results for genes that are differentially downregulated (blue, Dot1l-down signature ) after loss of Dot1l (shown: chromosome 17).
(D) GSEA showing enrichment of MLL-AF9 direct targets in “Dot1l-down” signature, p=0.01.
(E) GSEA showing enrichment of “Dot1l-down” signature in leukemia stem cells (L-GMP) versus normal progenitors (GMP), p<0.01.
(F) GSEA showing enrichment of “Dot1l-down” signature in human MLL-rearranged versus non- MLL-rearranged primary patient samples, p=0.034.
Loss of Dot1l results in impaired, but not absent hematopoiesis
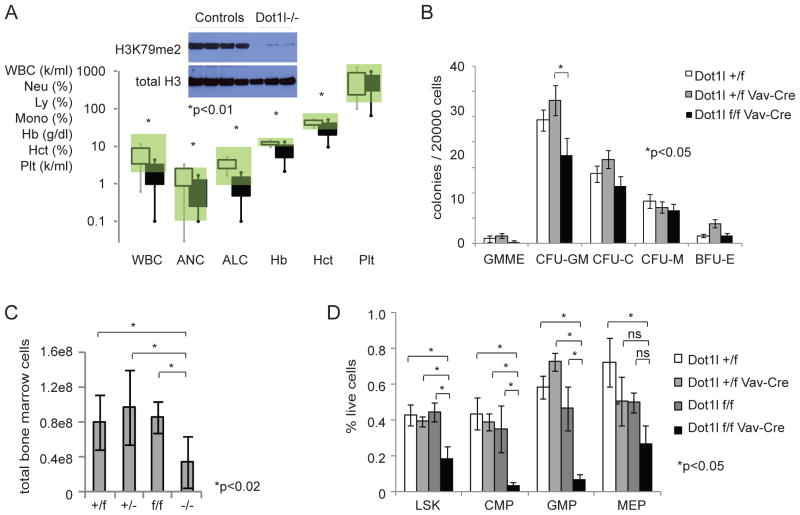
In order to investigate whether Dot1l is required for non-malignant hematopoietic development, we crossed Dot1lf/f mice with Vav-Cre mice (Georgiades et al., 2002) to delete Dot1l throughout the hematopoietic compartment starting during embryonal development (Figure 6). Dot1lf/f Vav-Cre mice were born in expected frequencies with essentially normal body and organ weights (Figure S6A–C). Young mice (3–6 weeks) displayed varying degrees of anemia (Figure 6A) and were mild to moderately neutropenic and lymphopenic. The colony forming activity of myeloid progenitors appeared relatively intact (Figure 6B), with a trend towards a more severe phenotype in earlier compartments. The total bone marrow cellularity in these mice was decreased (Figure 6C). However, loss of Dot1l did not lead to complete loss of myeloid or lymphoid development. Dot1l−/− peripheral blood leukocytes with confirmed absence of H3K79me2 developed and expanded up to 3–6 weeks postnatally and were readily isolated from these mice (Figure 6A). The LSK, GMP and CMP (common myeloid progenitors, Il-7R−Lin−Sca-1−c-Kit+CD34+FcγRII/IIIlo) compartments were moderately to severely decreased. Megakaryocyte/erythroid progenitors (MEP, Il-7R−Lin−Sca-1−c-Kit+CD34−FcγRII/III−) were less affected (Figure 6D). Analysis of older mice was challenging due to partial chimerism with non-deleted clones that over time contributed to varying degrees to the hematopoietic phenotype (data not shown). These results point to a role for Dot1l during hematopoietic development, but also demonstrate that hematopoiesis can develop to some extent in the absence of Dot1l.
Figure 6. Hematopoietic development in the absence of Dot1l.
(A) Peripheral blood counts of Dot1lf/f Vav-Cre (black squares, n=8) compared to littermate controls (open squares, n=12) at 3–6 weeks of age. Squares and error bars represent 1st and 2nd standard deviation. Normal range is shaded green. Insert: immunoblot for H3K79me2 in peripheral blood nucleated cells of Dot1lf/f Vav-Cre and control mice.
(B) Colony formation of normal hematopoietic progenitors from bone marrow of Dot1lf/f Vav-Cre mice. Colonies were scored 7–8 days after plating. Dot1lf/f Vav-Cre: n=5, Dot1l+/f: n=5, Dot1l+/f Vav-Cre: n=3, 3 independent experiments, error bars: SEM. All differences were not statistically significant except Dot1lf/f Vav-Cre versus Dot1l+/f Vav-Cre in CFU-GM.
(C) Bone marrow cellularity in Dot1lf/fVav-Cre mice shown as total cells recovered from front and hind legs + pelvis. Dot1lf/f Vav-Cre (−/−) (n=9), Dot1lf/f (f/f) (n=4), Dot1l+/f Vav-Cre (+/−) (n=3) and Dot1l+/f (+/f) (n=10) mice, error bars: SEM.
(D) Flow cytometric analysis of hematopoietic stem and progenitor cell compartments in Dot1lf/fVav-Cre mice. Dot1lf/f Vav-Cre (n=4), Dot1lf/f (n=3), Dot1l+/f Vav-Cre (n=4) and Dot1l+/f (n=3) mice from 3 separate litters, error bars: SEM.
See also Figure S6.
MLL-AF9 leukemia cells are dependent on Dot1l in vivo
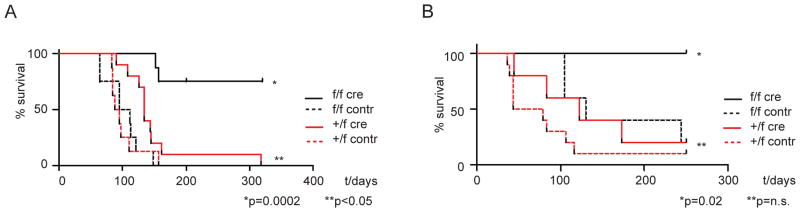
In order to assess the role of Dot1l in leukemia development in vivo, lin− Dot1lf/f and Dot1l+/f bone marrow cells were transformed using retroviral MLL-AF9. Subsequently, Dot1l was deleted in pre-leukemic transformed cells through retroviral delivery of Cre-recombinase. 2–4 days after transduction, before phenotypic changes became apparent, cells were sorted and injected into sublethally irradiated recipients. MLL-AF9 transformed Dot1lf/f and Dot1l+/f cells transduced with control retrovirus, and Dot1l+/f cells transduced with Cre rapidly and reliably caused clinically overt leukemia, with most animals having succumbed by 150 days (Figure 7A). Transduced Dot1l+/− cells exhibited a small but consistent increase in latency that reached statistical significance. In contrast, MLL-AF9 transformed Dot1l−/− cells failed to cause leukemia in the majority of recipients. Leukemia that did develop in one recipient of MLL-AF9 transformed Dot1l−/− cells was derived from cells that had failed to undergo full rearrangement of Dot1l alleles, and contained normal H3K79me2 levels (Figure S7C), a phenomenon we also observed in vitro (Figure S7A–B). These results demonstrate that Dot1l is necessary for MLL-AF9 mediated leukemia development in vivo.
Figure 7. Dot1l is required for transformation and maintenance of MLL-AF9 fusion driven leukemia in vivo.
(A) Survival curves for mice injected with 1×105 MLL-AF9 transformed lineage negative bone marrow cells 2–4 days after transduction with Cre or MIY-control retrovirus and resorting for GFP+/YFP+ cells.
(B) Survival curves for secondary recipient mice that received 1×105 MLL-AF9 leukemia cells isolated from leukemic mice that were subsequently transduced with Cre or MIY-control retrovirus 3 days prior to re-injection. Just prior to reinjection cells were resorted for GFP+/YFP+ cells.
See also Figure S7.
Next, we studied the consequence of loss of Dot1l in established MLL-AF9 leukemia. Dot1lf/f or Dot1l+/f AML was isolated from moribund mice and transduced with either Cre or control vector. Sorted cells were injected into sublethally irradiated secondary recipients 3 days after transduction. At this time point, neither the epigenetic changes (complete loss of H3K79me2, Figure 3C) nor the gene expression changes observed after loss of Dot1l (Figure S5) were fully established, and Dot1l−/− cell were indistinguishable from control cells with respect to growth pattern or the percentage of transduced cells. Transplantation of established MLL-AF9 AML into secondary recipients after deletion of Dot1l failed to cause disease (Figure 7B). A small difference was again observed between mice that received Dot1l+/− leukemia cells and those which received Dot1l+/f leukemias, but the difference did not reach statistical significance.
Discussion
The results presented here demonstrate that MLL-AF9 myeloid leukemia is driven by an aberrant epigenetic program involving H3K79 methylation. A potential role for Dot1l in MLL-rearranged leukemia is supported by multiple publications in recent years. However, the contribution of Dot1l and H3K79 methylation with respect to other hypotheses of MLL-fusion driven leukemogenesis, and the importance of H3K79me2 in the maintenance of gene expression remained unclear. H3K79 methylation has been reported to be near ubiquitously coupled to transcription (Steger et al., 2008), a fact that has raised serious concerns about the specificity of the dependence of MLL-rearranged leukemia cells on Dot1l. It was also unclear if a single and relatively abundant histone modification could exert locus specific effects on a highly selective gene set that controls the MLL-fusion associated leukemogenic transcription program.
We demonstrate an absolute dependence of MLL-AF9 leukemia cells on Dot1l in vitro and in vivo, and propose a molecular rationale for this finding by identifying a distinct epigenetic lesion specifically involving H3K79me2. We identified the direct targets of the MLL-AF9 fusion protein in a murine MLL-AF9 leukemia model, allowing us to assess chromatin states specifically at MLL-AF9 targets. We discovered highly abnormal H3K79me2 patterns at MLL-AF9 target loci, which were unique both with respect to other transcriptionally active loci within the same cell, and with respect to the same loci when expressed at the same level but under physiologic regulation in normal hematopoietic progenitors. These abnormalities could explain a specific and non-physiological dependence of the MLL-fusion driven leukemogenic transcription program on H3K79 methylation. Consistent with this hypothesis, only a small subgroup of genes, which was highly enriched for MLL-fusion targets and other genes with known functions in MLL-rearranged leukemias, showed any change in expression after loss of Dot1l and H3K79 methylation. This group of genes showed significant overlap with gene expression signatures that define murine MLL-AF9 leukemia stem cells and human MLL-rearranged clinical leukemia samples. On the other hand, transcription of the majority of genes associated with H3K79me2 did not depend on the presence of this chromatin mark. While our studies do not address the role of H3K79 methylation with respect to initiation or elongation, they do suggest that H3K79me2 is not uniformly involved in maintaining transcription. Rather, the exquisite and specific sensitivity of MLL-AF9 leukemia cells to loss of H3K79 methylation may be a consequence of the uniquely abnormal H3K79 methylation patterns at key loci. It is of tremendous interest to determine whether the dependence of MLL-fusion associated expression programs constitutes an over-reliance on an inappropriately targeted, but otherwise normal Dot1l, or whether Dot1l is aberrantly regulated in these leukemias.
Our data support a role for Dot1l in normal hematopoiesis. Conditional Dot1l knockout mice expressing Cre from a hematopoiesis specific promoter exhibit a moderate reduction in white blood cells, and a moderate to severe reduction in red blood cells as well as earlier progenitor compartments. This phenotype is similar to the hematopoietic phenotype recently reported in a conditional Dot1l CreER model (Jo et al., 2011). Analysis of embryonic blood development in Dot1l mutant mice revealed a severe defect in early erythroid, but not myeloid development (Feng et al., 2010). Earlier studies support a role for Dot1l in the transcriptional regulation of the alpha and beta globin loci (Fu et al., 2005; Sawado et al., 2008). Taken together, these studies provide evidence for a critical role of Dot1l in early hematopoietic and particularly erythroid development, therefore anemia or pancytopenia may be potential side effects of pharmacologic inhibition of Dot1l. However, complete deletion of c-kit results in severe anemia, bone marrow failure and embryonic lethality, while clinically effective pharmacologic inhibition of c-kit with imatinib is very well tolerated with only modest and manageable hematopoietic side effects (Broxmeyer et al., 1991). The fact that the isolation of near uniformly Dot1l−/− peripheral blood leukocytes from 3–6 week old mice is possible demonstrates that Dot1l is not absolutely required for all hematopoietic cells, and suggests an exploitable therapeutic window.
Dot1l is the only known methyltransferase for H3K79 and responsible for mono, -di and trimethylation of this residue. Mono- di and tri methylation may fulfill divergent biological roles. Genome occupancy of H3K79me2 and H3K79me3 modified histones in yeast is mutually exclusive, and associated with distinct biological processes (Schulze et al., 2009). Our study assessed only the genome wide methylation patterns for H3K79me2 due to lack of specific antibodies that recognize H3K79me3. Our study does not address whether loss of mono, di- or trimethylation is the most detrimental to MLL-rearranged leukemia cells. This question is not trivial, since the addition of methyl groups by Dot1l appears to be sequential and the conversion of H3K79me2 to H3K79me3 requires prior ubiquitination of histone H2B, hinting at a much more complex epigenetic network potentially involving additional histone modifications.
In summary, our findings demonstrate a strong rationale and imply a therapeutic window for pharmacologic inhibition of Dot1l as a strategy to target MLL-leukemias. Further studies will determine the exact molecular mechanisms of how abnormal H3K79 methylation patterns are established, and how mis-targeting of a single epigenetic modification may act as a master regulator of transcriptional programs that lead to leukemic transformation.
Experimental Procedures
For primer and sh-RNA sequences, antibodies and detailed experimental procedures please refer to the supplemental materials.
Human Samples
The samples from AML patients were provided by the German-Austrian AML Study Group (AMLSG) with patient informed consent for genetic analysis according to the Declaration of Helsinki, and institutional review board approval from all participating centers. Conventional chromosome banding and fluorescence-in-situ-hybridization (FISH) were performed as previously described (Schlenk et al., 2008). The samples included into the study contained at least 80% of leukemic cells following Ficoll-density gradient centrifugation based enrichment.
Generation of a Dot1l knockout mouse, breeding
Animals were maintained at the Animal Research Facility at Children’s Hospital Boston. Animal experiments were approved by the Internal Animal Care and Use Committee. A pFlexible© based targeting vector containing exon 5 of Dot1l flanked by loxP sites was used to target CJ9 ES cells (129 background). Conditional mice were maintained on a B6/129 (Taconic) background or crossed to Vav-Cre (kindly provided by Dr. Stuart Orkin).
Generation of transformed murine cells and leukemia
Ecotropic retroviral vectors containing murine MLL-AF9 IRES-GFP, HoxA9-IRES-GFP, Meis1a-pgk-Puro, Cre-IRES-YFP (Cre) and MSCV-IRES-YFP (MIY) were generated by cotransfection of 293 cells. Lin− or Lin−Sca-1+c-Kit+ cells were transduced with MLL-AF9-GFP or HoxA9 IRES-GFP and MEIS1a-pgk-Puro and maintained in methylcellulose with supplemental cytokines. After 2–6 days, GFP+ cells were sorted and transduced with Cre or MIY. 2–4 days after transduction, GFP+/YFP+ cells were sorted and transplanted into B6/129 syngeneic sublethally irradiated (600 rad) recipients at 1×105 or 5×105 cells/mouse. For secondary transplants, whole bone marrow from leukemic mice was isolated, GFP+ cells were sorted and blast colonies were allowed to grow out. Leukemic cells were transduced with Cre or MIY, sorted and transplanted as described above.
Biochemical Assays (cell growth, apoptosis, cell cycle analysis, western blotting, qPCR)
Cell growth and viability were followed by serial cell counts. Apoptosis and cell cycle analysis were performed using the Annexin-staining and BrdU-APC/7AAD kit from BD-Pharmingen (San Jose, CA). Antibodies used for immunoblot detection on whole cell lysates or histones are detailed in supplementary methods. For colony assays, sorted transduced leukemia cells were plated in methylcellulose M3234 containing IL3, IL6 and SCF at 1000 or 5000 cells per plate, and replated weekly at 1000 cells/plate. Normal, non-transformed bone marrow was plated in methylcellulose M3434 at 20000 cells per plate, and colonies were scored on day 8–9.
ChIP-qPCR and ChIP-Sequencing
Hematopoietic progenitor populations (LSK and GMP) and leukemia stem cells (L-GMP) were isolated by flowcytometry. For chromatin immunoprecipitation of MLL-AF9, 5-FU treated bone marrow was in vitro transformed with C-terminal HA-bio tagged MLL-AF9. Results using MLL-AF9-HAbio L-GMPs isolated from the bone marrow of transplanted mice yielded similar results, and for brevity are not included in this study. Chromatin immunoprecipitation was performed as previously described (Bracken et al., 2006; Krivtsov et al., 2008). Briefly, crosslinking was performed with 1% formalin, and the cells were lysed in SDS buffer. DNA was fragmented by sonication. Chromatin immunoprecipitation for H3K4me3, H3K27me3, H3K36me3, and H3K79me2 was performed using antibodies specific to the respective modifications. ChIP for MLL-AF9 was performed using streptavidin for precipitation of biotinylated MLL-AF9. Eluted DNA fragments were analyzed by quantitative PCR, or subjected to sequencing using next generation Solexa sequencing.
RNA amplification and gene expression array
RNA was isolated from 105 sorted GFP+/YFP+ cells using Trizol (invitrogen), amplified (Nugen Ovation Pico WTA, Nugen, San Carlos, CA), labeled (Nugen EncoreTM Biotin Module), and hybridized to Affymetrix 430 2.A murine microarrays.
Data analysis and statistical methods
Precipitated DNA from ChIP experiments was sequenced on Illumina Genome Aanalyzer IIx or HiSeq2000 platform. Sequence reads were aligned to mouse genome assembly mm8 or human genome assembly hg18 using Bowtie (Langmead et al., 2009). Reads that aligned to multiple loci in the genome were discarded. The chip-seq signal was quantified as total number of reads per million in the region 1 Kb upstream of transcription start site (TSS) to 2 Kb downstream of TSS for H3K79me2 and 1 Kb upstream of TSS to 1 KB downstream of TSS for MLL-AF9. An empirical background distribution model of reads was constructed to find the significance level of signal at a gene. Expression array data were analyzed with GenePattern (Reich et al., 2006) release 3.2.3 (http://www.broad.mit.edu/tools/software.html), and the probesets were mapped to genes using library files (ver 30) downloaded from Affymetrix website (affymetrix.com). Gene set enrichment analysis (GSEA) was performed using (www.broadinstitute.org/gsea). The integrative analysis of histone modification levels and gene expression was performed using iCanPlot (www.icanplot.org) (AU Sinha, in preparation). Microarray and ChIP-Sequencing data has been deposited at the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov.ezp-prod1.hul.harvard.edu/geo/). GSE25911 (Expression changes after loss of Dot1l in murine MLL-AF9 leukemia cells) and GSE29130 (Epigenetic profiling of hematopoietic stem cells and leukemia stem cells).
Supplementary Material
Significance.
MLL-rearranged leukemias carry a poor prognosis, and elucidating the biological processes underlying transformation by MLL-fusion proteins may facilitate the development of targeted therapies. Here we demonstrate an epigenetic lesion that specifically involves H3K79 methylation on MLL-fusion target genes. Loss of Dot1l selectively decreases expression of MLL-fusion driven transcriptional programs. The aberrant H3K79 methylation pattern, and the specific requirement of H3K79 methylation for the maintenance of the MLL-translocation associated oncogenic program demonstrates that MLL-rearranged leukemias are driven by an aberrant epigenetic program. This has profound therapeutic implications since Dot1l is one of few enzymes linked to MLL-fusion proteins, and as such may represent an important therapeutic target.
Highlights.
MLL-AF9 promotes enhanced H3K79me2 at fusion target genes.
H3K79me2 is specifically abnormal as compared to other histone modifications.
Loss of Dot1l selectively decreases leukemia-associated gene expression.
Dot1l is absolutely required for MLL-leukemia cell growth in vitro and in vivo.
Acknowledgments
We thank Yuko Fujiwara and Stuart Orkin for blastocyst injections and helpful suggestions, Konstanze Döhner and the German-Austrian AML Study Group for providing leukemia samples, and Ronald Mathieu for assistance with FACS. This work was supported by grants from the American Cancer Society, the Leukemia and Lymphoma Society, Gabrielle’s Angel Foundation and the National Cancer Institute (CA105423, 1RC2CA148222, CA140575, CA684841). KMB was supported by NHLBI Career Development Award K08 HL102264 and funding from the William Lawrence and Blanche Hughes Foundation. LB was supported in part by the Deutsche Forschungsgemeinschaft (Heisenberg-Stipendium BU 1339/3-1). VMR and RMP are employees and SAA is a consultant for Epizyme Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albert M, Helin K. Histone methyltransferases in cancer. Semin Cell Dev Biol. 2010;21:209–220. doi: 10.1016/j.semcdb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxmeyer HE, Maze R, Miyazawa K, Carow C, Hendrie PC, Cooper S, Hangoc G, Vadhan-Raj S, Lu L. The kit receptor and its ligand, steel factor, as regulators of hemopoiesis. Cancer Cells. 1991;3:480–487. [PubMed] [Google Scholar]
- Chang MJ, Wu H, Achille NJ, Reisenauer MR, Chou CW, Zeleznik-Le NJ, Hemenway CS, Zhang W. Histone H3 lysine 79 methyltransferase Dot1 is required for immortalization by MLL oncogenes. Cancer Res. 2010;70:10234–10242. doi: 10.1158/0008-5472.CAN-10-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- Feng Y, Yang Y, Ortega MM, Copeland JN, Zhang M, Jacob JB, Fields TA, Vivian JL, Fields PE. Early mammalian erythropoiesis requires the Dot1L methyltransferase. Blood. 2010;116:4483–4491. doi: 10.1182/blood-2010-03-276501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XH, Liu DP, Tang XB, Liu G, Lv X, Li YJ, Liang CC. A conserved, extended chromatin opening within alpha-globin locus during development. Exp Cell Res. 2005;309:174–184. doi: 10.1016/j.yexcr.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Georgiades P, Ogilvy S, Duval H, Licence DR, Charnock-Jones DS, Smith SK, Print CG. VavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis. 2002;34:251–256. doi: 10.1002/gene.10161. [DOI] [PubMed] [Google Scholar]
- Griffiths EA, Gore SD. DNA methyltransferase and histone deacetylase inhibitors in the treatment of myelodysplastic syndromes. Semin Hematol. 2008;45:23–30. doi: 10.1053/j.seminhematol.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lawton LN, Rozovskaia T, Frampton GM, Levine SS, Volkert TL, Croce CM, Nakamura T, Canaani E, Young RA. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 2008;22:3403–3408. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP. DNA methylation as a therapeutic target in cancer. Clin Cancer Res. 2007;13:1634–1637. doi: 10.1158/1078-0432.CCR-06-2076. [DOI] [PubMed] [Google Scholar]
- Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117:4759–4768. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagoe H, Humphries RK, Blair A, Sutherland HJ, Hogge DE. Expression of HOX genes, HOX cofactors, and MLL in phenotypically and functionally defined subpopulations of leukemic and normal human hematopoietic cells. Leukemia. 1999;13:687–698. doi: 10.1038/sj.leu.2401410. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005;65:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- Min J, Feng Q, Li Z, Zhang Y, Xu RM. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–723. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010;24:574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Lin C, Guest E, Shilatifard A. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer. 2010;10:721–728. doi: 10.1038/nrc2915. [DOI] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, Slany RK. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, Garcia-Cuellar MP, Bach C, Buhl S, Maethner E, Slany RK. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7:e1000249. doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, Connelly JA, Basrur V, Elenitoba-Johnson KS, Hess JL. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17:609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood. 2011 doi: 10.1182/blood-2011-02-334359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, van der Heijden A, Scheele TN, Vandenberghe P, de Witte T, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Ross ME, Mahfouz R, Onciu M, Liu HC, Zhou X, Song G, Shurtleff SA, Pounds S, Cheng C, Ma J, et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104:3679–3687. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- Sawado T, Halow J, Im H, Ragoczy T, Bresnick EH, Bender MA, Groudine M. H3 K79 dimethylation marks developmental activation of the beta-globin gene but is reduced upon LCR-mediated high-level transcription. Blood. 2008;112:406–414. doi: 10.1182/blood-2007-12-128983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenk RF, Dohner K, Krauter J, Frohling S, Corbacioglu A, Bullinger L, Habdank M, Spath D, Morgan M, Benner A, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, Haug J, Johnston M, Jaspersen SL, Kobor MS, Shilatifard A. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol Cell. 2009;35:626–641. doi: 10.1016/j.molcel.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, Zon LI, Armstrong SA. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisig BB, Milne T, Garcia-Cuellar MP, Schreiner S, Martin ME, Fuchs U, Borkhardt A, Chanda SK, Walker J, Soden R, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Xia X, Reisenauer MR, Hemenway CS, Kone BC. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. J Biol Chem. 2006;281:18059–18068. doi: 10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.