Abstract
We have analyzed the possible relevance of HPV infection for breast cancer risk among Iranian women from north part of Iran. Among women with breast cancer, 25.9% had positive test results for HPV DNA in breast tumor samples in contrast to 2.4% of women with noncancer status (P = 0.002). The infection of HPV has increased the risk of breast cancer (OR 14.247; 95% CI 1.558–130.284, P = 0.019). The high-risk HPV genotypes (types 16 and 18) in samples of breast cancer patients were the predominant types (53.34%). Other genotypes detected in breast cancer were HPV-6, HPV-11, HPV-15, HPV-23, and HPV-124, and one isolate could not be genotyped compared to HPV reference sequences. While the sole detected HPV in control specimens was HPV-124. Our study reveals that HPV infection and age are the risk factors in breast cancer development in the north part of Iran.
1. Introduction
Breast cancer is the most common cancer in women worldwide, and there were representing 22.9% of all new cancers in 2008 (an estimated 1.378 million new cases) and ranking second overall when both sexes are considered together [1]. Despite of good prognosis of breast cancer, it was also the most common cause of death from cancer, with 13.7% of deaths the world total in woman [1, 2]. Mazandaran province is located in north of Iran and beneath the Caspian Sea. According to the annual report of Health Deputy of Ministry of Health and Medical Education in Iran, breast cancer in women was representing 23.38% of all new cancers in Mazandaran province in 2006-2007 and was the most frequent cancer of women [3].
The incidence of breast cancer is increasing almost everywhere, and this is due in part to increases in risk factors including decreased childbearing and breast feeding, increased exogenous hormone exposure, and harmful dietary and lifestyle changes [2, 4]. On the other hand, breast cancer development might be a consequence of different environmental exposures, including viral infection [4, 5]. Worldwide an estimated 12.1% of all human cancers (about 1.3 million cancer cases) were etiologically related to viral agents in 2002 [5]. Hence, the correlation of viral agents with breast cancer cannot be excluded.
There are controversial reports on the aetiology of HPV in breast cancer around the world. In the last two decades, considerable evidence has been found for a role for HPV in human breast cancer [6–13], but some studies suggested negative relationships [14–16]. In vitro studies have shown that the main oncoproteins E6 and E7 from HPV16 are able to immortalize primary mammary epithelial cells and provided additional evidence for a possible role of this virus in breast carcinogenesis [17–19].
To our knowledge, this is the first study on the presence of HPVs and their relation with in breast cancer in Iranian women. Our previous studies revealed that HPV infection could be considered as a risk factor for the development of lung cancer in Mazandaran population [20, 21]. In the present study, we conducted a case-control study of breast cancer patients and controls and tried to find a viral aetiology by the detection of HPV genome, and to evaluate the possible relevance of this factor for breast cancer risk among north female Iranian population from Mazandaran province.
2. Materials and Methods
2.1. Tissue Samples
The study protocol was approved by the scientific and ethics committee of the National Research Institute of Tuberculosis and Lung disease, Tehran. A total of 130 blocks of paraffin-embedded tissue including 79 samples diagnosed as breast carcinomas, and 51 noncancer samples as control were retrieved from archive of Imam Khomeini Hospital, Medicine Faculty of Sari city, Mazandaran University of Medical Sciences, Iran between 2002 and 2009. It is worth mentioning that the consents were acquired from the patients during the healthcare and clinical services. Breast fibroadenomas served as control subjects in this study. Furthermore to adjust the environmental confounders, we tried to match the subject's residence place in Mazandaran province between both groups.
2.2. DNA Extraction and HPV DNA Detection
Genomic DNAs from tissue sections were prepared according to the methods that previously described [22]. To avoid contamination of the DNA, great care was taken during extraction and PCR (sectioning the blocks to several small groups at different times, using new surgical blade for each sample and filter tips during extraction and PCR).
The adequacy of the DNA in each specimen for PCR amplification was determined by the detection of a 268-base pair (bp) fragment of the β-globin gene using the GH20/PC04 primer set [23]. In case of negative results, the PC03/PC04 primer set was used to amplify the 110 bp fragment. For the detection of HPV genome, 3 different primer sets including the GP5+/GP6+ primers [24], the CP primers [25], and the FAP primers [26] were applied in a way that described before [10].
2.3. Sequencing and Phylogenetic Analysis
For genotyping of HPV, the positive PCR products were analyzed by sequencing. The DNA sequence was determined with the Big-Dye terminator cycle sequence kit and an ABI 377A sequencer (Applied Biosystems Inc.).
The HPV partial sequences were edited with the BioEdit software program version 7.0.5.2, and then phylogenetic and molecular evolutionary analyses were conducted using MEGA software version 4.0.2 [27]. The UPGMA method was used for phylogenetic reconstructions that were implemented in the MEGA 4.0.2 program. For the UPGMA method, the nucleotide substitution model employed was the Maximum Composite Likelihood. Statistical confidence for the evolutionary trees was assessed by bootstrap (500 replicates). The phylogenetic trees were drawn using the MEGA 4.0.2 program.
2.4. Nucleotide Sequence Accession Number
The nucleotide sequences of HPV isolates that determined in this study have been deposited in GenBank data base [accession numbers HM748606–HM748622]. The GenBank accession numbers for the reference HPV nucleotide sequences are as follows:
HPV-1 [V01116], HPV-2 [X55964], HPV-3 [X74462], HPV-4 [X70827], HPV-5 [M17463], HPV-6 [X00203], HPV-7 [X74463], HPV-8 [M12737], HPV-9 [X74464], HPV-10 [X74465], HPV-11 [M14119], HPV-12 [X74466], HPV-13 [X62843], HPV-14 [X74467], HPV-15 [X74468], HPV-16 [K02718], HPV-17 [X74469], HPV-18 [X05015], HPV-19 [X74470], HPV-20 [U31778], HPV-21 [U31779], HPV-22 [U31780], HPV-23 [U31781], HPV-24 [U31782], HPV-25 [X74471], HPV-26 [X74472], HPV-27 [AB211993], HPV-28 [U31783], HPV-29 [U31784], HPV-30 [X74474], HPV-31 [J04353], HPV-32 [X74475], HPV-33 [M12732], HPV-34 [X74476], HPV-36 [U31785], HPV-37 [U31786], HPV-38 [U31787], HPV-39 [M62849], HPV-40 [X74478], HPV-41 [X56147], HPV-42 [M73236], HPV-43 [AJ620205], HPV-44 [U31788], HPV-45 [X74479], HPV-47 [M32305], HPV-48 [U31789], HPV-49 [X74480], HPV-50 [U31790], HPV-51 [M62877], HPV-52 [X74481], HPV-53 [X74482], HPV-54 [U37488], HPV-55 [U31791], HPV-56 [X74483], HPV-57 [X55965], HPV-58 [D90400], HPV-59 [X77858], HPV-60 [U31792], HPV-61 [U31793], HPV-62 [AY395706], HPV-63 [X70828], HPV-64 [U12495], HPV-65 [X70829], HPV-66 [U31794], HPV-67 [D21208], HPV-68 [X67161], HPV-69 [AB027020], HPV-70 [U21941], HPV-71 [AB040456], HPV-72 [X94164], HPV-73 [X94165], HPV-74 [U40822], HPV-75 [Y15173], HPV-76 [Y15174], HPV-80 [Y15176], HPV-81 [AJ620209], HPV-82 [AB027021], HPV-83 [AF151983], HPV-84 [AF293960], HPV-85 [AF131950], HPV-86 [AF349909], HPV-87 [AJ400628], HPV-88 [EF467176], HPV-89 [AF436128], HPV-90 [AY057438], HPV-91 [AF419318], HPV-92 [AF531420], HPV-93 [AY382778], HPV-94 [AJ620211], HPV-95 [AJ620210], HPV-96 [AY382779], HPV-97 [DQ080080], HPV-98 [NC_012744], HPV-99 [NC_012745], HPV-100 [NC_012746], HPV-101 [NC_008189], HPV-102 [DQ080083], HPV-103 [NC_012750], HPV-104 [NC_012750], HPV-105 [NC_012747], HPV-106 [DQ080082], HPV-107 [EF422221], HPV-108 [NC_012213], HPV-109 [NC_012485], HPV-110 [EU410348], HPV-111 [EU410349], HPV-112 [NC_012486], HPV-113 [NC_012748], HPV-114 [NC_013931], HPV-115 [NC_013591], HPV-116 [NC_013035], HPV-117 [GQ246950], HPV-119 [GQ845441], HPV-120 [GQ845442], HPV-121 [NC_014185], HPV-122 [GQ845444], HPV-123 [GQ845445], HPV-124 [GQ845446], PcPV [X62844], RhPV1 [M60184].
2.5. Statistical Data Processing
Data were processed by SPSS statistical software program version 16.0. The correlations were subjected to χ 2 (Pearson chi-square) and Fisher's exact test. Odds ratios and logistic regression were also calculated. Statistical significance was set as a P-value less than 0.05.
3. Results
The characteristics of study subjects including age, HPV DNA, HPV genotype, and breast cancer histopathologic types are shown in Table 1. A total of 130 individuals, including 79 breast cancer patients and 51 noncancer controls, were recruited into this study (Table 1).
Table 1.
The characteristics of study subjects and prevalence of HPV DNA status in breast cancer patients and noncancer controls.
| Parameter | Casesa (N = 79) | Controlsa (N = 51) | P-value |
|---|---|---|---|
| Age (year ± SD) | 47.77 ± 12.552 | 34.20 ± 9.704 | ≤0.0001 (t-test) |
|
| |||
| HPV | 0.002 | ||
| Positive | 15 (25.9)b | 1 (2.4) | O R 13.953 |
| Negative | 43 (74.1) | 40 (97.6) | (95% CI 1.762–110.526) |
|
| |||
| HPV genotype (% within type) | |||
| HPV-124 | 1 (6.25) | 1 (100) | |
| HPV-23 | 2 (12.5) | ||
| HPV-18 | 4 (25) | ||
| HPV-16 | 4 (25) | ||
| HPV-15 | 1 (6.25) | ||
| HPV-11 | 1 (6.25) | ||
| HPV-6 | 2 (12.5) | ||
| Unknown | 1 (6.25) | ||
|
| |||
| HPV genotype (% within subjects) | |||
| High-risk type | 8 (14) | 0 | 0.005 |
| Low-risk type | 6 (10.5) | 1 (2.4) | |
| Negative | 43 (75.5) | 40 (97.6) | |
|
| |||
| HPV genotype (% within subjects) | |||
| Mucosal type | 11 (19) | 0 | 0.002 |
| EV-cutaneous type | 4 (6.9) | 1 (2.4) | |
| Negative | 43 (74.1) | 40 (97.6) | |
|
| |||
| Tumor type (% within tumor type) | |||
| IDC | 67 (84.8) | ||
| ILC | 8 (10.1) | ||
| IDC-ILC mix | 1 (1.3) | ||
| MC | 2 (2.5) | ||
| DC | 1 (1.3) | ||
aSome of the subjects have been considered as missing value after quality examination of nucleic acid extraction.
bNumbers in parentheses are percentages.
The mean ages were 47.77 ± 12.552 (S.D.) and 34.20 ± 9.704 (S.D.) years in breast cancer and control groups, respectively (P ≤ 0.0001, Table 1). The most abundant type of breast cancer determined histologically was IDC (84.8%), followed by ILC (10.1%), MC (2.5%), DC, and IDL-ILC (1.3%) (Table 1).
Statistical difference was observed in the HPV DNA status (25.9% versus 2.4 0%, P = 0.002) and HPV genotype (caser risk type, P = 0.005; tissue tropism type, P = 0.002) between these two groups (Table 1). HPV infection has increased the risk of breast cancer and had an OR of 13.953 (95% CI 1.762–110.526; P = 0.002, Table 1).
The high-risk HPV types (HPV 16 and 18) were more prevalent than other HPV types in the cases (14.0% versus 10.5%, Table 1). The HPV genotypes in samples of breast cancer patients were 26.67% for HPV-16 (4 isolates) and HPV-18 (4 isolates), 13.3% for HPV-23 (2 isolates) and HPV-6 (2 isolates), 6.67% for HPV-11 (1 isolate), HPV-15 (1 isolate), and HPV-124 (1 isolate), and one isolate could not be genotyped compared to HPV reference sequences (Table 1) while the sole detected HPV in control specimens was HPV-124 (Table 1). The constructed phylogenetic trees of the HPV isolates are shown in Figure 1 in 3 separate parts. The co-infection with different HPV types (HPV-16 and HPV-23) was observed only in one sample, B32, which was detected by GP and CP primer sets (Figures 1(a) and 1(b)).
Figure 1.
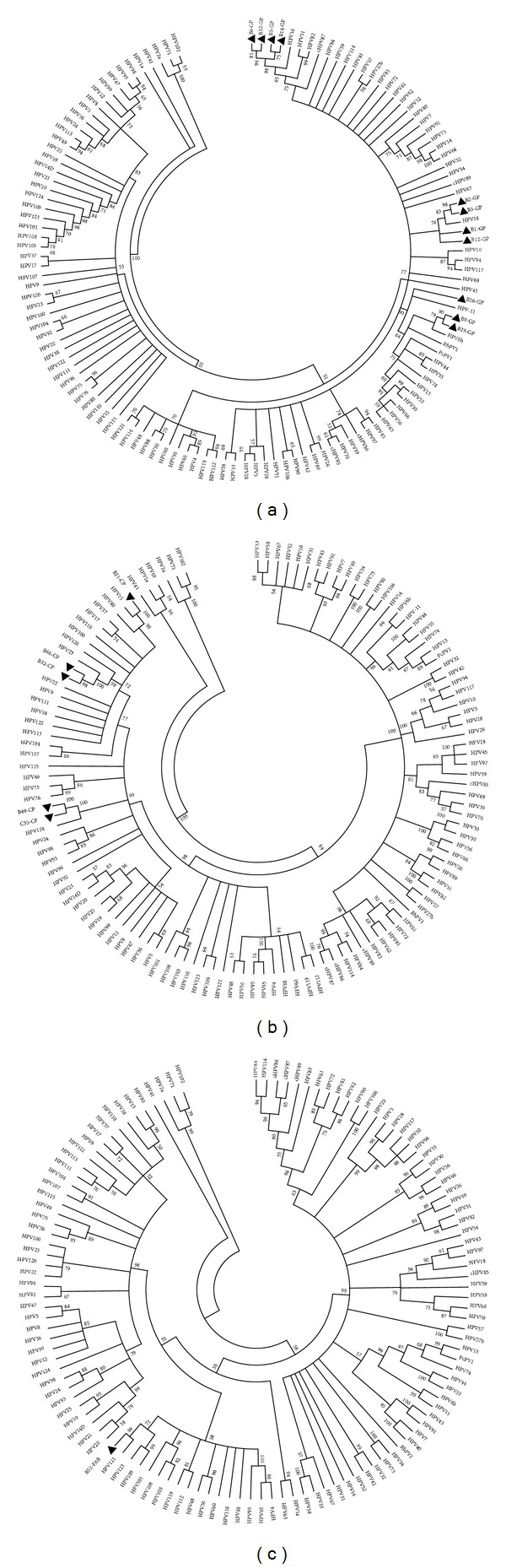
Phylogenetic analysis of L1 gene partial sequences amplified by (a) GP5+/GP6+, (b) CP primer set, and (c) FAB primer set in HPV isolates. The trees were constructed by the UPGMA method using MEGA 4.0.2. The HPV isolates are marked with black triangles.
In Table 2, HPV DNA detection rate is stratified by age and breast cancer histopathologic types. No significant difference was observed among breast tumor types considering HPV status (Table 2). Due to incomparability of the age means of case and control and to avoid the effect of the age parameter, a logistic regression model was run. Data shown that age (OR 0.873, 95% CI 0.822–0.927; P ≤ 0.0001) and HPV infection (OR 14.247, 95% CI 1.558–130.284; P = 0.019) are the significant risk factors in breast carcinogenesis in the studied women in north part of Iran, Mazandaran province (Table 3).
Table 2.
HPV status according to age groups and breast cancer histopathologic types.
| Variable | Cases | Controls | P-value | ||
|---|---|---|---|---|---|
| HPV positive (%) | Number of subjects | HPV positive (%) | Number of subjects | ||
| Age (year) | |||||
| <35 | 3 (49.9) | 7 | 0 | 22 | 0.01 |
| ≥35 | 12 (23.5) | 51 | 1 (7.7) | 19 | 0.097 |
|
| |||||
| P-value | 0.360 | 0.463 | |||
|
| |||||
| Tumor type | |||||
| IDC | 11 (22.9) | 48 | |||
| ILC | 4 (66.7) | 6 | |||
| IDC-ILC | 0 | 1 | |||
| MC | 0 | 2 | |||
| DC | 0 | 1 | |||
|
| |||||
| P-value | 0.144 | ||||
Table 3.
The Breast cancer risk estimation using the logistic regression model according to HPV status and Age in Iran, Mazandaran province.
| Variable | Number | OR | CI 95% | P-value |
|---|---|---|---|---|
| Age | 99 | 0.873 | 0.822–0.927 | 0.000 |
| HPV | 130 | 14.247 | 1.558–130.284 | 0.019 |
4. Discussion
HPVs belong to Papillomaviridae family, and epidemiological studies have shown that a persistent HPV infection is the most important risk factor for cervical cancer [28, 29]. HPVs are also considered to be one of the risk factors for human breast carcinogenesis. The concept of the relationship between HPV and breast cancer is based on the identification of HPV genome sequence in breast cancer tissues and immortalization of primary mammary epithelial cells by high-risk HPV [17–19]. However, involvement of HPV in breast cancer is controversial. Since 1992, a growing number of studies had identified HPVs in breast tumors by PCR around the world, with a positivity variation from 4% to 86% for suggesting negative [14–16, 30, 31] or positive relationships [6–13, 32–36]. These results reflect the controversy in the role of HPV in the pathogenesis of breast cancer. The controversy is influenced by the technical limitations and the epidemiology of HPV in different geographical area [36–38].
In our previous studies [20, 21], we showed that HPV infection may be associated with the development of lung cancer in Mazandaran, north part of Iran. It was worth mentioning that the HPV infection was the risk factor only in male gender, not in females [20, 21]. In this study, we have evaluated the association between the breast cancer and HPV infection in the north part of Iran. For the detection of HPV DNA, FFPE tissues were analyzed by PCR using 3 different primer sets. As shown in Table 1, the detection frequency of HPV DNA in breast cancer patients was significantly higher than that of control patients (25.9% versus 2.40%, P = 0.002). We suggest that HPV infection is significantly related to breast cancer in women live in Mazandaran province (OR 14.247, 95% CI 1.558–130.284; P = 0.019, Table 3). Regarding to HPV prevalence in breast cancer, a meta-analysis study that explores the correlation between HPV infection and risk of breast carcinoma revealed that HPV prevalence was lowest in Europe (12.91%) and highest in Oceania (42.11%) followed by Asia (32.42%) [38].
High-risk HPV types, such as HPV-16 and HPV-18, were detected in our study, and they comprised the majority of isolates (Table 1). Although there are great variability in the HPV detection rate worldwide, the majority of HPV types that are detected are the oncogenic types, HPV-16 and -18 [6–8, 11, 33, 38]. Our results are in agreement with these previous reports (Table 1). In addition, we detected cutaneous HPV types (HPV-15, -23, and -124). The presence of cutaneous HPV types was reported in the previous studies among women with breast cancer or at increased risk for breast cancer [10, 39]. The only isolate that screened by the FAB primer set has no significant similarity to any known HPV types. Pairwise distance calculation revealed that the HPV type 121 is the most similar type to the isolate (Figure 1(c)) and the distance estimation is 0.235.
5. Conclusions
The pathogenesis of breast cancer is complex. This study demonstrates the presence of HPV genome in tumor tissues in women with breast cancer in north part of Iran. HPV infection is associated with the development of breast cancer in women live in Mazandaran province. Further studies are needed to clarify the role and the risk assessment of HPV in human breast cancer. Confirming an etiologic role for HPV in breast cancer in Iranian females may help develop vaccine strategies for combating this increasingly common cancer.
Conflict of Interests
The authors declare that they have no competing interests.
Acknowledgments
The authors would like to thank the staff of pathology laboratory of Imam Khomeini Hospital in City of Sari and the staff of Virology Laboratory of Virology research Center at Masih Daneshvari Hospital for their kind cooperation. This study was financially supported by the Virology Research Centre, Academic Pivot for Education and Research, National Research Institute for Tuberculosis and Lung Disease.
Abbreviations
- HPV:
Human papillomavirus
- PCR:
Polymerase chain reaction
- FFPE:
Formalin-fixed paraffin-embedded
- bp:
Base pair
- SD:
Standard deviation
- OR:
Odds ratio
- CI:
Confidence interval
- IDC:
Invasive ductal carcinoma
- ILC:
Invasive lubular carcinoma
- MC:
Medullary carcinoma
- DC:
Ductal carcinoma in situ.
References
- 1. GLOBOCAN 2008, Cancer Incidence and Mortality Worldwide: IARC http://globocan.iarc.fr/factsheets/populations/factsheet.asp?uno=900.
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA-A Cancer Journal for Clinicians. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Center for Disease Control & Prevention. Iran: Ministry of Health and Medical Education Office of Deputy Minister for Health Center for disease control; 2010. Cancer office: Iranian annual of national cancer registration report 2006-2007. Tech. Rep. [Google Scholar]
- 4.Parkin DM, Fernández LM. Use of statistics to assess the global burden of breast cancer. Breast Journal. 2006;12(supplement 1):S70–S80. doi: 10.1111/j.1075-122X.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- 5.Parkin DM. The global health burden of infection-associated cancers in the year 2002. International Journal of Cancer. 2006;118(12):3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 6.di Lonardo A, Venuti A, Marcante ML. Human papillomavirus in breast cancer. Breast Cancer Research and Treatment. 1992;21(2):95–100. doi: 10.1007/BF01836955. [DOI] [PubMed] [Google Scholar]
- 7.Hennig EM, Suo Z, Thoresen S, Holm R, Kvinnsland S, Nesland JM. Human papillomavirus 16 in breast cancer of women treated for high grade cervical intraepithelial neoplasia (CIN III) Breast Cancer Research and Treatment. 1999;53(2):121–135. doi: 10.1023/a:1006162609420. [DOI] [PubMed] [Google Scholar]
- 8.Damin AP, Karam R, Zettler CG, Caleffi M, Alexandre CO. Evidence for an association of human papillomavirus and breast carcinomas. Breast Cancer Research and Treatment. 2004;84(2):131–137. doi: 10.1023/B:BREA.0000018411.89667.0d. [DOI] [PubMed] [Google Scholar]
- 9.Widschwendter A, Brunhuber T, Wiedemair A, Mueller-Holzner E, Marth C. Detection of human papillomavirus DNA in breast cancer of patients with cervical cancer history. Journal of Clinical Virology. 2004;31(4):292–297. doi: 10.1016/j.jcv.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 10.de Villiers EM, Sandstrom RE, zur Hausen H, Buck CE. Presence of papillomavirus sequences in condylomatous lesions of the mamillae and in invasive carcinoma of the breast. Breast Cancer Research. 2005;7(1):R1–R11. doi: 10.1186/bcr940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kan CY, Iacopetta BJ, Lawson JS, Whitaker NJ. Identification of human papillomavirus DNA gene sequences in human breast cancer. British Journal of Cancer. 2005;93(8):946–948. doi: 10.1038/sj.bjc.6602778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heng B, Glenn WK, Ye Y, et al. Human papilloma virus is associated with breast cancer. British Journal of Cancer. 2009;101(8):1345–1350. doi: 10.1038/sj.bjc.6605282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai JH, Hsu CS, Tsai CH, et al. Relationship between viral factors, axillary lymph node status and survival in breast cancer. Journal of Cancer Research and Clinical Oncology. 2007;133(1):13–21. doi: 10.1007/s00432-006-0141-5. [DOI] [PubMed] [Google Scholar]
- 14.Gopalkrishna V, Singh UR, Sodhani P, et al. Absence of human papillomavirus DNA in breast cancer as revealed by polymerase chain reaction. Breast Cancer Research and Treatment. 1996;39(2):197–202. doi: 10.1007/BF01806186. [DOI] [PubMed] [Google Scholar]
- 15.Lindel K, Forster A, Altermatt HJ, Greiner R, Gruber G. Breast cancer and human papillomavirus (HPV) infection: no evidence of a viral etiology in a group of Swiss women. Breast. 2007;16(2):172–177. doi: 10.1016/j.breast.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 16.de Cremoux P, Thioux M, Lebigot I, Sigal-Zafrani B, Salmon R, Sastre-Garau X. No evidence of Human papillomavirus DNA sequences in invasive breast carcinoma. Breast Cancer Research and Treatment. 2008;109(1):55–58. doi: 10.1007/s10549-007-9626-4. [DOI] [PubMed] [Google Scholar]
- 17.Band V, Zajchowski D, Kulesa V, Sager R. Human papilloma virus DNAs immortalize normal human mammary epithelial cells and reduce their growth factor requirements. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(1):463–467. doi: 10.1073/pnas.87.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Chen JJ, Gao Q, et al. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. Journal of Virology. 1999;73(9):7297–7307. doi: 10.1128/jvi.73.9.7297-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimri G, Band H, Band V. Mammary epithelial cell transformation: insights from cell culture and mouse models. Breast Cancer Research. 2005;7(4):171–179. doi: 10.1186/bcr1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadji SA, Mokhtari-Azad T, Mahmoodi M, et al. Relationship between lung cancer and human papillomavirus in north of Iran, Mazandaran province. Cancer Letters. 2007;248(1):41–46. doi: 10.1016/j.canlet.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 21.Nadji SA, Mahmoodi M, Ziaee AA, et al. An increased lung cancer risk associated with codon 72 polymorphism in the TP53 gene and human papillomavirus infection in Mazandaran province, Iran. Lung Cancer. 2007;56(2):145–151. doi: 10.1016/j.lungcan.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Impraim CC, Saiki RK, Erlich HA, Teplitz RL. Analysis of DNA extracted from formalin-fixed, paraffin-embedded tissues by enzymatic amplification and hybridization with sequence-specific oligonucleotides. Biochemical and Biophysical Research Communications. 1987;142(3):710–716. doi: 10.1016/0006-291x(87)91472-0. [DOI] [PubMed] [Google Scholar]
- 23.Saiki RK, Bugawan TL, Horn GT, Mullis KB, Erlich HA. Analysis of enzymatically amplified β-globin and HLA-DQ α DNA with allele-specific oligonucleotide probes. Nature. 1986;324(6093):163–166. doi: 10.1038/324163a0. [DOI] [PubMed] [Google Scholar]
- 24.Husman AMDR, Walboomers JMM, van den Brule AJ, Meijer CJ, Snijders PJ. The use of general primers GP5 and GP6 elongated at their 3’ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. Journal of General Virology. 1995;76(4):1057–1062. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- 25.de Villiers EM, Laverone D, McLaren K, Benton EC. Prevailing papillomavirus types in non-melanoma carcinomas of the skin in renal allograft recipients. International Journal of Cancer. 1997;73(3):356–361. doi: 10.1002/(sici)1097-0215(19971104)73:3<356::aid-ijc9>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 26.Forslund O, Antonsson A, Nordin P, Stenquist B, Hansson BG. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. Journal of General Virology. 1999;80(9):2437–2443. doi: 10.1099/0022-1317-80-9-2437. [DOI] [PubMed] [Google Scholar]
- 27.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 28.Baseman JG, Koutsky LA. The epidemiology of human papillomavirus infections. Journal of Clinical Virology. 2005;32:S16–S24. doi: 10.1016/j.jcv.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 29.Smith JS, Lindsay L, Hoots B, et al. Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: a meta-analysis update. International Journal of Cancer. 2007;121(3):621–632. doi: 10.1002/ijc.22527. [DOI] [PubMed] [Google Scholar]
- 30.Bratthauer GL, Tavassoli FA, O’Leary TJ. Etiology of breast carcinoma: no apparent role for papillomavirus types 6/11/16/18. Pathology Research and Practice. 1992;188(3):384–386. doi: 10.1016/S0344-0338(11)81229-X. [DOI] [PubMed] [Google Scholar]
- 31.Wrede D, Luqmani YA, Coombes RC, Vousden KH. Absence of HPV 16 and 18 DNA in breast cancer. British Journal of Cancer. 1992;65(6):891–894. doi: 10.1038/bjc.1992.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu Y, Morimoto T, Sasa M, et al. HPV33 DNA in premalignant and malignant breast lesions in Chinese and Japanese populations. Anticancer Research. 1999;19(6):5057–5061. [PubMed] [Google Scholar]
- 33.Liu Y, Klimberg VS, Andrews NR, et al. Human papillomavirus DNA is present in a subset of unselected breast cancers. Journal of Human Virology. 2001;4(6):329–334. [PubMed] [Google Scholar]
- 34.Gumus M, Yumuk PF, Salepci T, et al. HPV DNA frequency and subset analysis in human breast cancer patients’ normal and tumoral tissue samples. Journal of Experimental and Clinical Cancer Research. 2006;25(4):515–521. [PubMed] [Google Scholar]
- 35.Kroupis C, Markou A, Vourlidis N, Dionyssiou-Asteriou A, Lianidou ES. Presence of high-risk human papillomavirus sequences in breast cancer tissues and association with histopathological characteristics. Clinical Biochemistry. 2006;39(7):727–731. doi: 10.1016/j.clinbiochem.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 36.Akil N, Yasmeen A, Kassab A, Ghabreau L, Darnel AD, Al Moustafa AE. High-risk human papillomavirus infections in breast cancer in Syrian women and their association with Id-1 expression: a tissue microarray study. British Journal of Cancer. 2008;99(3):404–407. doi: 10.1038/sj.bjc.6604503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Damin AP, Damin DC, Alexandre COP. HPV infection and breast cancer. The Breast. 2007;16(3):p. 222. doi: 10.1016/j.breast.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Li N, Bi X, Zhang Y, Zhao P, Zheng T, Dai M. Human papillomavirus infection and sporadic breast carcinoma risk: a meta-analysis. Breast Cancer Research and Treatment. 2011;126(2):515–520. doi: 10.1007/s10549-010-1128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cazzaniga M, Gheit T, Casadio C, et al. Analysis of the presence of cutaneous and mucosal papillomavirus types in ductal lavage fluid, milk and colostrum to evaluate its role in breast carcinogenesis. Breast Cancer Research and Treatment. 2009;114(3):599–605. doi: 10.1007/s10549-008-0040-3. [DOI] [PubMed] [Google Scholar]


