Abstract
The largest biological fractionations of stable carbon isotopes observed in nature occur during production of methane by methanogenic archaea. These fractionations result in substantial (as much as ≈70‰) shifts in δ13C relative to the initial substrate. We now report that a stable carbon isotopic fractionation of comparable magnitude (up to 70‰) occurs during oxidation of methyl halides by methylotrophic bacteria. We have demonstrated biological fractionation with whole cells of three methylotrophs (strain IMB-1, strain CC495, and strain MB2) and, to a lesser extent, with the purified cobalamin-dependent methyltransferase enzyme obtained from strain CC495. Thus, the genetic similarities recently reported between methylotrophs, and methanogens with respect to their pathways for C1-unit metabolism are also reflected in the carbon isotopic fractionations achieved by these organisms. We found that only part of the observed fractionation of carbon isotopes could be accounted for by the activity of the corrinoid methyltransferase enzyme, suggesting fractionation by enzymes further along the degradation pathway. These observations are of potential biogeochemical significance in the application of stable carbon isotope ratios to constrain the tropospheric budgets for the ozone-depleting halocarbons, methyl bromide and methyl chloride.
Methyl bromide (MeBr) and methyl chloride (MeCl) are, respectively, the most abundant volatile bromo- and chlorocarbons in the troposphere and are major contributors to stratospheric ozone destruction (1). Both compounds have natural and human-influenced sources and a predominant sink by reaction with OH in the troposphere (2–4). MeBr also has a bacterial soil sink (5) that represents about 20% of the estimated total removal from the troposphere, and it is likely that a soil sink of similar magnitude exists for MeCl (6). Hence, if an isotopic fractionation is associated with the soil sink, it will influence the isotopic compositions of MeBr and MeCl in the lower atmosphere (7). The δ 13C value of industrially produced MeBr ranges between −43.5‰ and −66.4‰ (7), but δ 13C values of tropospheric MeBr and natural sources are not yet known. The δ 13C of atmospheric MeCl has been measured from −22‰ to −45‰ (8, 9). If carbon isotope ratios are to be used to constrain the budgets of these methyl halides, it is essential to determine the extent of carbon isotope fractionation that occurs during biological degradation of these compounds.
Methylotrophic bacteria use C1 compounds, which are simple organic molecules that contain no carbon–carbon bonds. Strains IMB-1, CC495, and MB2 are as-yet-unnamed facultative methylotrophs isolated from agricultural soil, woodland leaf litter, and coastal seawater, respectively (10–13), environments where methyl halides are produced. They are members of the α subgroup of the Proteobacteria. On the basis of 16S rRNA gene sequences, strains IMB-1 and CC495 show some phylogenetic alignment with the genus Rhizobium (10, 11) and are very closely related to the new genus Pseudoaminobacter (I. McDonald, personal communication). Strain MB2 aligns within the Ruegeria clade [J. K. Schaefer, K. D. Goodwin, I. R. McDonald, J. C. Murrell and R.S.O., unpublished work]. All of these aerobic bacteria are methylotrophs in that they can grow by using MeBr or MeCl as their sole carbon source, but they do not metabolize methane. They oxidize MeBr, MeCl, and methyl iodide (MeI) to CO2.
Soil bacteria are known to consume MeBr at the ambient tropospheric mixing ratio of around 10 parts per trillion by volume (5). Preliminary experiments with strain IMB-1 indicate that it can oxidize MeBr at these mixing ratios** and is therefore likely to be characteristic of bacteria associated with MeBr uptake by soils. We examined δ 13C of MeCl, MeBr, and MeI during oxidation by whole-cell suspensions of IMB-1 and CC495 and also the change in δ 13C values of the three methyl halides during oxidation by the marine strain MB2. In addition, we measured the fractionation of carbon isotopes during formation of methane thiol (MeSH) from MeCl by the purified cobalamin-dependent enzyme, halomethane:bisulphide/halide ion methyltransferase (11) from CC495, to determine whether this initial step in MeCl degradation could account for the observed fractionation by whole cells. Finally, we determined the fractionation associated with the degradation of MeBr during field studies with agricultural soil by monitoring MeBr concentration and δ 13C of MeBr in the headspace of flux chambers under fumigation conditions.
Methods
GC/Isotope Ratio Mass Spectrometry.
We manipulated the cell concentrations in our experiments to achieve slow (low-cell-density) or fast (high-cell-density) degradation rates for methyl halides. Experiments with low cell densities (<5 × 107 cells ml−1) of MeBr-grown strains IMB-1 and MB2, as well as agricultural field studies, were conducted at the U.S. Geological Survey and the University of California, Berkeley. Values of δ 13C of compounds under investigation were determined by isotope-ratio-monitoring GC-combustion-mass spectrometry (irmGCMS) at the Lawrence Berkeley National Laboratory Center for Isotope Geochemistry (7). Methyl halides were quantified by flame ionization detection or electron capture detection GC (14). Investigations with high cell densities (>1 × 1011 cells ml−1) of MeCl-grown strains IMB-1, CC495, and MB2, and experiments with the enzyme halomethane:bisulphide/halide ion methyltransferase, were conducted at The Queen's University of Belfast. Values of δ 13C for the compounds under investigation were determined on a continuous-flow irmGCMS at the Environmental Engineering Research Centre, and the compounds, including methyl halides, CO2, MeSH, and dimethyl sulfide, were concurrently identified and quantified by quadrupole mass spectrometry (15).
Calculations.
The isotope fractionation associated with each bacterial degradation was determined from the slope (b) of the regression of δ 13C values of the reactant methyl halide on the logarithm of the fraction of reactant remaining (ln f) as follows:
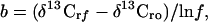 |
1 |
where rf and ro refer to the reactant at f and to the initial reactant. The fractionation factor (α = k12/k13) is then:
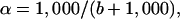 |
2 |
which, for clarity is reported as ɛ, the deviation from unity, where:
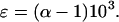 |
3 |
The number of measurements (n = 8) made during low-cell-density experiments with the three methyl halides and high-cell-density experiments by using IMB-1 and MeI was small, hence data from replicate experiments were pooled to obtain b and the standard error (SE) of b for the three methyl halides studied. The number (13 ≤ n ≤ 42) of measurements made during each of the other high-cell-density experiments was higher, which allowed the determination of a unique slope and its SE for each experiment. The mean slope and SE of b for replicate experiments were used to calculate ɛ and its error.
Corrections for Abiotic Reactions During Biological Oxidation.
Chemical reactions [e.g., hydrolysis and transhalogenation (16, 17)] competed with biological oxidation for substrate during incubations with strains IMB-1, CC495, and MB2 with MeBr and MeI. Experiments where buffer was incubated without cells were conducted to allow correction for the effects of chemical reactions. A correction was also made for a slight loss of MeBr without concomitant isotope fractionation during incubations with high-density cell suspensions. No correction was made for chemical reaction during incubations of whole cells with MeCl or during enzymatic conversion of MeCl to MeSH, as chemical reaction was insignificant in the short time period of these experiments. Incubations with MeBr and MeI using low cell densities were of sufficient duration to require correction of both concentration and isotope values. The maximum corrections applied were <0.2, or about 10%, for ln f and <15‰ for δ 13C.
Low-Cell-Density Suspensions.
Strains IMB-1 and MB2 were grown with MeBr as sole carbon source and harvested during late logarithm-phase growth (10, 12). Cells were washed twice with 16 mM KH2PO4, pH 7.2, medium for IMB-1 (18, 19) or with marine mineral salts, pH 7.2, medium containing 0.27 M NaCl for MB2, and resuspended in these buffer solutions. Incubations were conducted in 159-ml serum bottles containing 51 ml of either buffer solution or cells suspended in buffer solution [0.9 and 1.4 mg wet weight cells per bottle for IMB-1 and MB2, respectively (10)]. After sealing with rubber stoppers, the bottles and solutions were flushed with CO2-free air for 5 min. Subsequently, MeBr (6.8 mg) or MeCl (3.6 mg) was added directly as a pure gas. MeI (3.4 mg) was added as pure liquid. Incubations were conducted in the dark at 26°C with shaking. Headspace was sampled daily for determination of methyl halide concentrations (0.1 ml) and δ 13C values (0.25 ml).
High-Cell-Density Suspensions.
Strains IMB-1, CC495, and MB2 were grown with MeCl as sole carbon source and harvested in late logarithm-phase growth (10, 12, 18). Cells were washed twice with 16 mM phosphate buffer, pH 7.2, for IMB-1; 50 mM phosphate buffer, pH 7.2, for CC495; and 16 mM phosphate buffer, pH 7.2, containing 0.27 M NaCl, for MB2. Cells of each bacterium were then resuspended in these buffers. Replicate screw-capped vials (20 ml), each fitted with a Mininert sampling port (Alltech, Deerfield, IL) and containing 2.5-ml cell suspensions (25- to 100-mg wet weight cells), were flushed with CO2-free air for 5 min. Cell densities were chosen to achieve greater than 90% substrate utilization within 4 h. MeCl (30 μg), MeBr (200 μg), or MeI (200 μg) was added in aqueous solution (20–30 μl) to the sealed vials, which were incubated at 26°C with shaking. Headspace was sampled every 30 min for determination of methyl halide and CO2 concentrations and δ 13C values (0.25 ml). Concurrent measurements were made on methyl halide abiotic controls (no cells added), live cell controls (no methyl halide added), and heat-killed controls (cell suspensions heated at 100°C for 5 min before addition of methyl halide). Incubations continued for between 2 and 4 h and, on termination, phosphoric acid (0.5 ml) was added to an aliquot (0.5 ml) of each cell suspension in a helium-flushed sealed tube. After 1 h, the δ 13C value of the CO2 released was determined by using a Europa (Cheshire, U.K.) 20–20 isotope ratio mass spectrometer with a trace gas concentrator.
Enzyme.
Halomethane:bisulphide/halide ion methyltransferase was extracted from strain CC 495 (grown with MeCl as sole carbon source), and the purified enzyme was activated in the presence of 2.5 mM DTT and 0.5 mM MeCl, as previously described (11). MeCl (30 μg) in aqueous solution (20 μl) was added to duplicate screw-capped vials (20 ml), each fitted with a Miniinert sampling port and containing 1 ml of a solution of purified enzyme (0.1 mg of protein) in 50 mM phosphate buffer, pH 7.2, in the presence of 0.5 mM Na2S. Incubations were conducted at 26°C, and the headspace was sampled every 30 min for determination of MeCl concentration and δ 13C. Production of MeSH and dimethyl sulfide was monitored by GC-quadrupole MS.
Soil Flux.
To determine whether naturally occurring populations of soil bacteria can fractionate elevated (fumigation) levels of methyl bromide, we deployed flux-chamber collars over experimental soil plots (University of California Bay Area Research and Extension Center, Santa Clara, CA), into which we injected MeBr (34 g m−2 at 12 cm depth). The chamber collars were polyvinylchloride (PVC) cylinders (diameter = 30 cm, height = 14 cm) with removable lids constructed of PVC rings fitted with 0.0025-cm-thick polyethylene covers stretched as a drum skin (20). The chamber lids were placed over the collars and, after an accumulation period of 4 h, samples of the gas phase were collected 4 times over the next 24 h.
Results
Cell Suspensions.
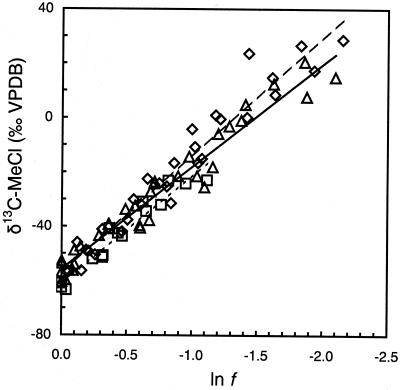
The δ 13C of MeCl increased from an initial value of −60‰ to +30‰ when 90% of the compound was degraded by high-cell-density suspensions of all three methylotrophs (Fig. 1). Over the same period, no significant loss of MeCl was noted for the abiotic controls (<1%) or for heat-killed controls (<1%), hence no correction for chemical reaction was necessary. The cultures produced CO2 with exceptionally depleted δ 13C values, ranging from −76‰ for CC495 to −94‰ for IMB-1 at the point where half the MeCl had been used (f = 0.5; data not shown).
Figure 1.
δ13C of MeCl during incubations of high-cell-density suspensions of strains IMB-1 (◊), CC495 (▵), and MB2 (□) at 26°C in relation to the fraction of reactant remaining (f). Linear regressions shown in this composite plot are for visual reference only; IMB-1 (dashed line), CC495 (solid line), and MB2 (dotted line). Isotope effects were calculated from regressions of individual incubations.
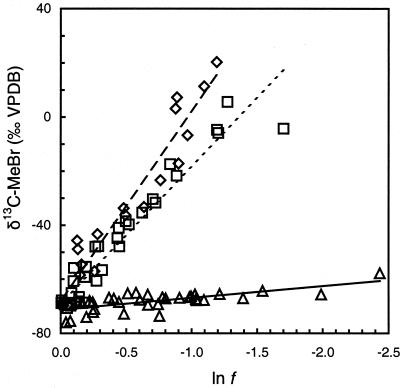
The δ 13C of MeBr increased from about −70‰ to approximately +15‰ when 70% of the compound was degraded by high-cell-density suspensions of IMB-1 and MB2 (Fig. 2). However, little isotopic enrichment was observed during degradation of MeBr by strain CC495. Some loss of MeBr was noted in abiotic controls (<10%) and for heat-killed controls (<10%). The calculated ɛ for each experiment was therefore corrected for the changes because of hydrolysis (16) and other losses recorded in the controls. These corrections were similarly applied to the determination of ɛ for the oxidation of MeI by cultures. Very similar ɛ values were obtained for the degradation of methyl halides in both laboratories (Table 1).
Figure 2.
δ13C of MeBr during incubations of high-density cell suspensions of strains IMB-1 (◊), CC495 (▵), and MB2 (□) at 26°C in relation to the fraction of reactant remaining (f), corrected for loss because of abiotic processes. Linear regressions shown in this composite plot are for visual reference only; IMB-1 (dashed line), CC495 (solid line), and MB2 (dotted line). Isotope effects were calculated from regressions of individual incubations.
Table 1.
Fractionation factors [ɛ = (α-1)103] for bacterial and enzymatic processes at 26°C
| Reaction | ɛ MeCl | ɛ MeBr | ɛ MeI |
|---|---|---|---|
| IMB-1 oxidation (low cell density) | 50 ± 2* | 66 ± 6* | 40 ± 3* |
| IMB-1 oxidation (high cell density) | 47 ± 4† | 72 ± 3† | 29 ± 3* |
| MB2 oxidation (low cell density) | 63 ± 5* | ||
| MB2 oxidation (high cell density) | 44 ± 4† | 57 ± 5† | 30 ± 4† |
| CC495 oxidation (high cell density) | 42 ± 2† | 4 ± 2† | 9 ± 1† |
| Enzyme/CC495 0.5 mM HS− | 21 ± 1‡ |
Data were pooled from replicate experiments to obtain a single composite slope; ɛ and SE of the slope are reported.
Discrete slopes were determined for each experiment; mean ɛ and SE of the slopes are reported.
Single experiment; ɛ and SE of the slope are reported.
Enzyme Reactions.
The δ 13C of MeCl increased from about −60‰ to −25‰ during the enzyme-mediated reaction of the compound with HS−. This reaction was complete within several hours. During this time, enzyme-free abiotic controls showed negligible reaction of MeCl with HS−. The ɛ value calculated from the loss of MeCl and the increase in observed δ 13C of residual MeCl is reported in Table 1.
Soil Flux.
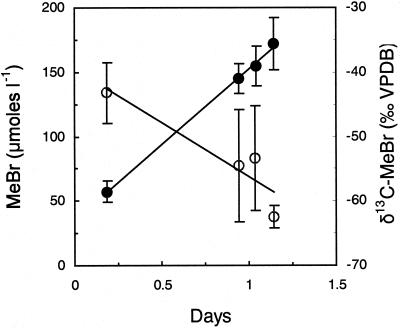
MeBr concentrations in the flux chambers reached maximum values within several hours of injection. We report only on the subsequent decrease in concentration (Fig. 3). During this time, the δ 13C values of the residual MeBr in the flux chambers increased. Although not quantified, much of the loss of MeBr occurred by advection, which does not result in stable isotope fractionation. Differential diffusion of 12C- and 13C-MeBr should result in only slight enrichment in δ 13C of residual MeBr [<4‰ (21)]. Ignoring this effect allows us to calculate an isotope fractionation for the removal of MeBr (ɛ = 17‰). This is a minimum value, because it is based on the total MeBr loss including that caused by mass transport and redistribution as well as that caused by bacterial consumption. Sizable isotopic fractionation is expected to be associated with only the latter process. The soil studied had resident microflora with the ability to oxidize MeBr, as we detected biological oxidation of 14C-MeBr to 14CO2 (data not shown) by using methods previously described (14).
Figure 3.
Changes in chamber headspace MeBr concentration (○) and δ13C-MeBr (●) after injection of fumigation levels of MeBr (34 g m−2) below the soil surface. Error bars indicate ± 1 SD of the mean of 3 chambers.
Discussion
Oxidation of methyl halides by methylotophic bacteria is accompanied by large carbon isotope fractionations of up to 70‰. These fractionations are comparable to those in two important methanogenic pathways: CO2 reduction with H2 (22, 23) and metabolism of methylated C1 compounds (24) and exceed those reported for cultures of aceticlastic methanogens (25). Methylotrophs and methanogens have recently been shown to have significant genetic similarities, including common expression of cofactors that shuttle C1 units of various oxidation states between enzymes. These cofactors include corrinoids (26), methanopterins and folates (27–29), and methylcoenzyme M (30). Two or more of these cofactors found in methylotrophs are associated with every substrate pathway in methanogenesis (31).
Although understanding of the biochemistry of methylotrophs and methanogens has advanced substantially over the past two decades, no measurements of carbon isotope fractionation in experiments with purified enzymes have been reported. Strains IMB-1, CC495, and MB2 oxidize methyl halides to CO2 through a series of enzyme-mediated reactions. For irreversible processes, the isotope effect associated with consumption is calculated by regression of the δ 13C values of the residual substrate on ln f, where f is the fraction of substrate remaining. Additional information regarding specific steps in the oxidation pathway can be obtained by isolating the kinetic isotope effects attributed solely to the initial enzyme reaction.
The initial step in the biological degradation of MeCl or MeBr by all three strains is probably catalyzed by a corrinoid enzyme (6, 11, 26, 32). One such enzyme from strain CC495, halomethane:bisulphide/halide ion methyltransferase, has recently been purified and, in the presence of HS−, mediates the conversion of MeCl, MeBr, or MeI to MeSH. It was not possible to accurately determine ɛ for the enzymatic transformation of MeBr or MeI to MeSH, because these methyl halides rapidly react chemically with DTT, which is required for in vitro activation of the enzyme. However, MeCl showed no detectable reaction with DTT, and we were able to determine ɛ for its enzymatic conversion to MeSH. The ɛ value for this reaction, 21‰, was considerably lower than those observed for whole cells (all strains tested >40‰; Table 1). The discrepancy between fractionations measured for the enzyme and for whole cells may be because of differences in behavior of the enzyme in vitro compared with in vivo. Alternatively, the discrepancy may indicate that the enzyme reaction is not entirely irreversible and hence may be responsible for only part of the total fractionation observed in the complete bacterial oxidation of methyl halides. Additional fractionation would then be associated with subsequent enzyme reactions in the catabolism of MeSH to CO2.
Isotope effects associated with the consumption of MeCl by cell suspensions were similar for all three bacterial species (Table 1). Strains IMB-1 and MB2 also showed similar ɛ values for consumption of MeBr and MeI. However, strain CC495 exhibited little fractionation of MeI and almost no fractionation of MeBr. There are at least two possible explanations for this effect, both of which are speculative. It is conceivable that dehalogenation by each strain is catalyzed by different enzymes, each capable of producing a different isotopic effect for a given methyl halide. However, the enzyme halomethane:bisulphide/halide ion methyltransferase isolated from strain CC495 has broadly similar affinities for the three methyl halides (11). An alternative possibility is that MeBr and MeI, which readily methylate enzyme thiol groups (33), are inhibiting one or more downstream enzymatic processes in strain CC495 that may be responsible for the fractionation observed during the complete metabolism of MeCl.
The results of our flux-chamber experiments indicate that uptake of fumigation levels of MeBr by field soil is accompanied by significant fractionation (ɛ > 17‰). This net fractionation is not as extensive as that displayed by cell suspensions (up to 70‰). It is not clear what proportion of MeBr degradation in this soil is because of biological compared with abiotic processes. Regardless of the degradation mechanism, the MeBr remaining in the soil during fumigation and available for transport to the atmosphere is enriched in 13C relative to the initial MeBr (7).
What is yet unproven is whether a significant carbon isotope fractionation is associated with the microbial degradation of MeBr and other methyl halides at their respective atmospheric mixing ratios, as opposed to the higher concentrations used in these experiments. Because we observed substantial isotopic fractionation of MeBr by soils during fumigation, we suspect there might also be fractionation during the bacterial oxidation of ambient (10 parts per trillion by volume) tropospheric levels of MeBr in soils. This suspicion can be confirmed only after we identify the bacteria responsible for MeBr consumption in soils at ambient concentration and determine the fractionation associated with this process.
Soils represent 20% of the total sink for tropospheric MeBr (5). If fractionation during oxidation of MeBr by soils is substantial (for example, 70‰, as we have determined for low-affinity methylotrophs), the microbial soil sink for MeBr should significantly increase the stable isotopic composition of MeBr in the troposphere. As recent reports indicate that the microbial soil sink for atmospheric MeCl is also substantial (6, 34), it is possible that it exerts a similar effect on δ 13C of atmospheric MeCl. Thus, the microbiological oxidation of methyl halides by terrestrial soils could significantly influence the stable carbon isotopic composition of these compounds in the troposphere.
Acknowledgments
We thank J. Schaefer, A. Downey, T. Kennedy, N. Ogle, S. Silva, R. Huddleston, N. Wall, Z. Mousli, and S. Davis for assistance, and S. D. Lennox for advice on statistical analysis. The manuscript benefited from comments by J. Hayes, R. Dias, K. Goodwin, and D. Des Marais. We are grateful to the reviewers for their contributions. Financial support was from U. S. Geological Survey Projects to Develop New Technologies, the National Aeronautics and Space Administration Upper Atmospheric Research Program (5188-AU-0080), the National Science Foundation Atmospheric Chemistry Program (ATM-9729110), the U. K. Engineering and Physical Sciences Research Council (GR/L85183 and GR/M26374), and the Department of Agriculture for Northern Ireland.
Abbreviations
- MeBr
methyl bromide
- MeCl
methyl chloride
- MeI
methyl iodide
- MeSH
methane thiol
Footnotes
Goodwin, K. D., Varner, R. A., Crill, P. M. & Oremland, R. S. (1999) Eos Trans. Am. Geophys. Union, Spring Meeting Abstract 80, S64.
References
- 1.Butler J H. Nature (London) 2000;403:260–261. doi: 10.1038/35002232. [DOI] [PubMed] [Google Scholar]
- 2.Mellouki A, Talukdar R K, Schmoltner A-M, Gierczak T, Mills M J, Soloman S, Ravishankara A R. Geophys Res Lett. 1992;19:2059–2062. [Google Scholar]
- 3.Khalil M A K, Rasmussen R A, Gunawardena R. J Geophys Res. 1993;98:2887–2896. [Google Scholar]
- 4.Khalil M A K, Rasmussen R A. Atmos Environ. 1999;33:1305–1321. [Google Scholar]
- 5.Shorter J H, Kolb C E, Crill P M, Kerwin R A, Talbot R W, Hines M E, Harriss R C. Nature (London) 1995;377:717–719. [Google Scholar]
- 6.Harper D B. Nat Prod Rep. 2000;17:337–348. doi: 10.1039/a809400d. [DOI] [PubMed] [Google Scholar]
- 7.McCauley S E, Goldstein A H, DePaolo D J. Proc Natl Acad Sci USA. 1999;96:10006–10009. doi: 10.1073/pnas.96.18.10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rudolph J, Lowe D C, Martin R J, Clarkson T S. Geophys. Res. Lett. 1997. 24659–24662. [Google Scholar]
- 9.Tsunogai U, Yoshida N, Gamo T. J Geophys Res. 1999;104:16033–16039. [Google Scholar]
- 10.Connell Hancock T L, Costello A M, Lidstrom M E, Oremland R S. Appl Environ Microbiol. 1998;64:2899–2905. doi: 10.1128/aem.64.8.2899-2905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coulter C, Hamilton J T G, McRoberts W C, Kulakov L, Larkin M J, Harper D B. Appl Environ Microbiol. 1999;65:4301–4312. doi: 10.1128/aem.65.10.4301-4312.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodwin K D, Schaefer J K, Oremland R S. Appl Environ Microbiol. 1998;64:4629–4636. doi: 10.1128/aem.64.12.4629-4636.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harper D B, Kalin R M, Larkin M J, Hamilton J T G, Coulter C. Environ Sci Technol. 2000;34:2525–2527. [Google Scholar]
- 14.Miller L G, Connell T L, Guidetti J R, Oremland R S. Appl Environ Microbiol. 1997;63:4346–4354. doi: 10.1128/aem.63.11.4346-4354.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalin R M, Hamilton J T G, Harper D B, Miller L G, Lamb C, Kennedy J T, Downey A, McCauley S E, Goldstein A H. Rapid Commun Mass Spectrom. 2001;15:357–363. doi: 10.1002/rcm.219. [DOI] [PubMed] [Google Scholar]
- 16.Elliott S, Rowland F S. J Atmos Chem. 1995;20:229–236. [Google Scholar]
- 17.Elliott S, Rowland F S. Geophys Res Lett. 1993;20:1043–1046. [Google Scholar]
- 18.Schaefer J K, Oremland R S. Appl Environ Microbiol. 1999;65:5035–5041. doi: 10.1128/aem.65.11.5035-5041.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doronina N V, Sokolov A P, Trotsenko Y A. FEMS Microbiol Lett. 1996;142:179–183. [Google Scholar]
- 20.Yagi K, Williams J, Wang N-Y, Cicerone R J. Proc Natl Acad Sci USA. 1993;90:8420–8423. doi: 10.1073/pnas.90.18.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerling T H, Solomon D K, Quade J, Bowman J R. Geochim Cosmochim Acta. 1991;55:3403–3405. [Google Scholar]
- 22.Games L M, Hayes J M, Gunsalus R P. Geochim Cosmochim Acta. 1978;42:1295–1297. [Google Scholar]
- 23.Fuchs G, Thauer R, Ziegler H, Stichler W. Arch Microbiol. 1979;120:135–139. [Google Scholar]
- 24.Summons R E, Franzmann P D, Nichols P D. Org Geochem. 1978;28:465–475. [Google Scholar]
- 25.Gelwicks J T, Risatti J B, Hayes J M. Appl Environ Microbiol. 1994;60:467–472. doi: 10.1128/aem.60.2.467-472.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vannelli T, Messmer M, Studer A, Vuilleumier S, Leisinger T. Proc Natl Acad Sci USA. 1999;96:4615–4620. doi: 10.1073/pnas.96.8.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christoserdova L, Vorholt J A, Thauer R K, Lidstrom M E. Science. 1998;281:99–102. doi: 10.1126/science.281.5373.99. [DOI] [PubMed] [Google Scholar]
- 28.Pomper B K, Vorholt J A, Christoserdova L, Lidstrom M E, Thauer R K. Eur J Biochem. 1999;261:475–480. doi: 10.1046/j.1432-1327.1999.00291.x. [DOI] [PubMed] [Google Scholar]
- 29.Vorholt J A, Christoserdova L, Stolyar S M, Thauer R K, Lidstrom M E. J Bacteriol. 1999;181:5750–5757. doi: 10.1128/jb.181.18.5750-5757.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krum J G, Ensign S A. J Bacteriol. 2000;182:2629–2634. doi: 10.1128/jb.182.9.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferry J G. FEMS Microbiol Rev. 1999;23:13–38. doi: 10.1111/j.1574-6976.1999.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 32.Woodall C A, Warner K L, Oremland R S, Murrell J C, McDonald J R. Appl Environ Microbiol. 2001;67:1959–1963. doi: 10.1128/AEM.67.4.1959-1963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacDonald O C, Reichmuth C. In: The Methyl Bromide Issue. Bell C H, Price N, Chakrabarti B, editors. Chichester, U.K.: Wiley; 1996. pp. 149–189. [Google Scholar]
- 34.Khalil M A K, Rasmussen R A. Environ. Sci. Pollut. Res. 2000. 719–782. [Google Scholar]