Abstract
The biological basis for regional and inter-species differences in cerebral cortical morphology is poorly understood. We focused on consanguineous Turkish families with a single affected member with complex bilateral occipital cortical gyration abnormalities. By using whole-exome sequencing, we initially identified a homozygous 2-bp deletion in LAMC3, the laminin γ3 gene, leading to an immediate premature termination codon. In two other affected individuals with nearly identical phenotypes, we identified a homozygous nonsense mutation and a compound heterozygous mutation. In human but not mouse fetal brain, LAMC3 is enriched in postmitotic cortical plate neurons, localizing primarily to the somatodendritic compartment. LAMC3 expression peaks between late gestation and late infancy, paralleling the expression of molecules that are important in dendritogenesis and synapse formation. The discovery of the molecular basis of this unusual occipital malformation furthers our understanding of the complex biology underlying the formation of cortical gyrations.
Extensive and stereotypic gyrations (convolutions) of the cerebral cortex are striking features of the human brain1,2. The cerebral cortex develops through a series of highly coordinated phases including proliferation of neural progenitors, migration of postmitotic cells from the germinal matrix to the newly forming cortex and organization of the mature cortical cytoarchitecture. Among these processes, the biological basis for the formation of cortical convolutions has been the subject of much debate3–7. The study of malformations of cortical development (MCD) that interfere with the proper formation of the typical cortical gyration pattern in humans provides a unique opportunity to understand this elegant process. We applied whole-exome capture and sequencing to the study of consanguineous, single-affected–member Turkish kindreds with malformations of occipital cortex gyration, notably characterized by the presence of both pachygyria and polymicrogyria8,9, entities traditionally thought to arise from different pathogenic mechanisms reflecting abnormalities in neuronal migration and cortical organization, respectively10.
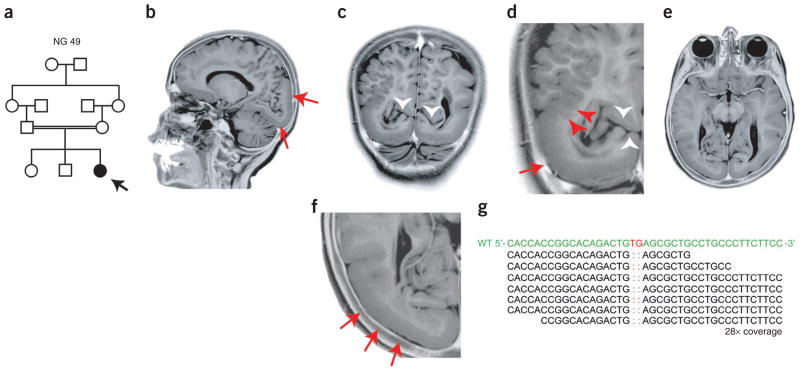
The first index case, designated NG 49-1 (Fig. 1a, Supplementary Fig. 1 and Supplementary Note), presented to medical attention with absence seizures and mild developmental delay during early childhood. Magnetic resonance imaging (MRI) revealed bilateral occipital pachygyria mainly localizing to the lateral surface, characterized by smoothening of the occipital cortices with loss of secondary and tertiary gyri, and polymicrogyric areas, characterized by the formation of numerous small gyri at the junction of the parieto-occipital lobes (Fig. 1b–f and Supplementary Videos 1 and 2). Following genome-wide genotyping, which confirmed consanguinity with an inbreeding coefficient of 5.77, and determination of homozygous genomic segments (>2.5 cM each) (Supplementary Table 1), we performed whole-exome capture and sequencing with high sensitivity and specificity using NimbleGen liquid-phase arrays and the Illumina Genome Analyzer-IIx instrument as described previously11 (Supplementary Tables 2 and 3). By focusing on the homozygous intervals (Supplementary Table 4), we identified a homozygous 2-bp deletion in LAMC3 (Laminin γ3), which is located on chromosome 9; this mutation is inferred to result in a frameshift at codon 301 (of the 1,575 total encoded amino acids) and lead to an immediate premature termination (p.Cys301X) (Fig. 1g and Supplementary Fig. 2). Both parents were heterozygous for the variant (Supplementary Fig. 2a). These results suggested that the homozygous LAMC3 variant was the disease-causing mutation in this family.
Figure 1.
LAMC3 mutation in NG 49 with occipital cortical malformation. (a) The pedigree structure of NG 49 (index case, NG 49-1, black arrow) is shown, which reveals a first cousin marriage. (b) A sagittal magnetic resonance image reveals occipital cortical pachygyria (red arrows). (c–f) Coronal (c,d) and axial (e,f) magnetic resonance images reveal thickening and smoothening of the occipital cortex (red arrows) along with polymicrogyria (red arrowheads). Loss of the clear distinction between the gray and white matter is visualized in c and d, in which the white arrowheads mark the preserved calcarine fissure. Panels d and f are enlarged views of c and e, respectively. Photographically inverted T2 images are shown. (g) Exome sequencing shows a 2-bp ‘TG’ deletion (marked in red in the wild-type (WT) sequence on top) in the LAMC3. The depth of coverage across the variant was 28×, and all but one of the reads showed the deletion.
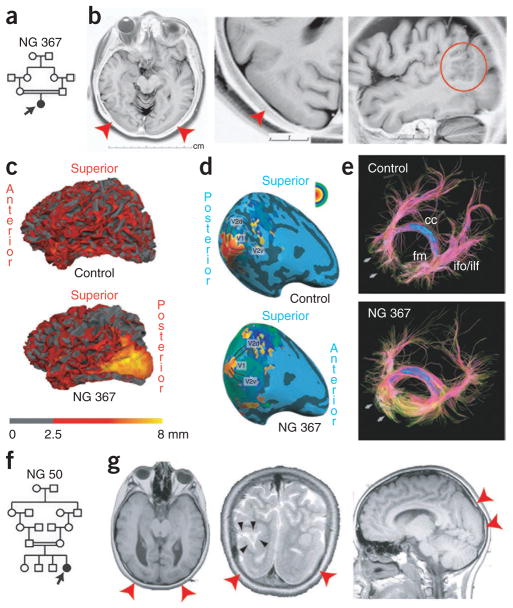
We then searched for additional mutations in a cohort of 12 individuals with MCD with homozygosity of at least 2.5 cM spanning LAMC3. Only one of these individuals (NG 367-1) had a diagnosis of occipital pachygyria, whereas the others were referred with various MCD phenotypes including polymicrogyria or lissencephaly, or were not otherwise specified. Sanger sequencing of the complete coding region of LAMC3 in these individuals revealed only one previously unidentified, homozygous mutation at codon 157 resulting in premature termination (p.Trp157X) in the subject with the diagnosis of occipital pachygyria (NG 367-1) (Fig. 2a,b and Supplementary Fig. 2b). Both parents of this individual were heterozygous for the variant (Supplementary Fig. 2b). Subject NG 367-1 had a phenotype almost identical to the first index case, NG 49-1 (Fig. 1), presenting to medical attention at age 10 years with seizures largely consisting of staring and blinking spells (Supplementary Note and Supplementary Video 3). On her current examination at age 33 years, this subject was found to be neurologically intact with normal visual acuity and reportedly average intelligence. MRI revealed prominent bilateral smoothening and thickening of the lateral occipital cortex, which is associated with polymicrogyria (Fig. 2b,c, Supplementary Fig. 3a and Supplementary Videos 4 and 5). However, despite these gross structural findings, retinotopic mapping using functional MRI indicated that the relative locations and functions of the primary visual areas were largely indistinguishable from control subjects (Fig. 2d, Supplementary Note and Supplementary Fig. 3b)12,13. Nonetheless, diffusion tensor imaging (DTI) tractography showed markedly lower fractional anisotropy and elevated radial diffusivity values in areas adjacent to the pachygyric region, suggesting microstructural changes of white matter, possibly caused by fiber disorganization (Fig. 2e and Supplementary Note).
Figure 2.
Additional LAMC3 mutations in individuals with occipital cortical malformation. (a) Simplified pedigree structure of NG 367 (arrow, index case) shows a first cousin consanguineous union. (b) Axial, enlarged axial and sagittal magnetic resonance images reveal cortical smoothening and thickening limited to the bilateral occipital cortices (red arrowheads). On the sagittal image, polymicrogyria (red circle) at the junction of the pachygyric occipital cortex with the parietal lobe is seen. (c) Three-dimensional reformatting of magnetic resonance images with cortical thickness color mapping shows the occipital cortex to be markedly thickened (yellow color, >8 mm) as compared to a control subject in which the cortex is less than 4 mm in thickness. (d) The retinotopic mapping reveals the anatomical locations of the primary visual areas and the spatial patterns of their activity in response to visual stimulation in NG 367-1 to be similar to a control subject. (e) Diffusion tensor imaging tractography reveals the disorganization of short association fibers in the posterior part of the occipital lobe in areas adjacent to the pachygyric regions (arrows). cc, corpus callosum; ifo, inferior fronto-occipital fasciculus; fm, forceps major; ilf, inferior longitudinal fasciculus. (f) Simplified pedigree structure of NG 50 (arrow, index case) shows a second cousin consanguineous union. (g) The axial, coronal and sagittal magnetic resonance imaging reveals occipital pachygyria (red arrowheads) in association with polymicrogyric areas. On the coronal image in the middle, a cerebrospinal fluid (CSF) cleft, which is typically associated with polymicrogyria, is visible (black arrowheads).
Finally, we investigated the remaining affected individuals in our neurogenetics cohort of over 600 people to assess whether any had similar MRI findings. We identified only one other individual, NG 50-1, who had an almost identical MRI picture. This subject was the product of a second cousin marriage (Fig. 2f) and presented to medical attention at the age of 11 with episodes of vision loss consistent with seizures; imaging studies again revealed bilateral occipital pachygyria associated with polymicrogyria (Fig. 2g, Supplementary Fig. 1b and Supplementary Note). We found NG 50-1 to be heterozygous for the interval surrounding LAMC3, suggesting potential genetic locus heterogeneity (Supplementary Table 5). However, exome sequencing revealed no homozygous nonsense or frameshift mutations, whereas all eight new homozygous mis-sense variants identified affected non-conserved domains of various proteins located within the autozygous intervals, suggesting that they were unlikely to be disease causing (Supplementary Table 6). Subsequent examination of the LAMC3 sequence available from the exome sequencing data followed by confirmation using PCR and Sanger sequencing identified a compound heterozygous mutation, with a premature termination mutation at codon 386 (resulting in p.Gln386X) and a missense mutation (resulting in p.Gly350Arg) at a position completely conserved among vertebrate orthologs (Supplementary Fig. 2c and Supplementary Fig. 4) and predicted to be damaging. One mutation was inherited from each parent (Supplementary Fig. 2c).
None of the four mutations newly discovered in LAMC3 had been previously reported in the dbSNP or 1000 Genomes databases and all were absent from our screening of 400 Turkish control subjects. They were also absent from our whole-exome sequence database of 1,023 subjects with non-neurological diseases in which we only identified four different heterozygous LAMC3 variants (p.Cys273Tyr, p.Gly275Glu, p.Gln928X and c.976+1G>A) with a collective allele frequency of less than 1 in 500, consistent with these deleterious alleles being under strong purifying selection. The rarity of such deleterious mutations and the finding of new homozygous and compound heterozygous mutations in LAMC3 in all three subjects with occipital pachygyria and polymicrogyria provide conclusive evidence that recessive LAMC3 mutations cause this syndrome.
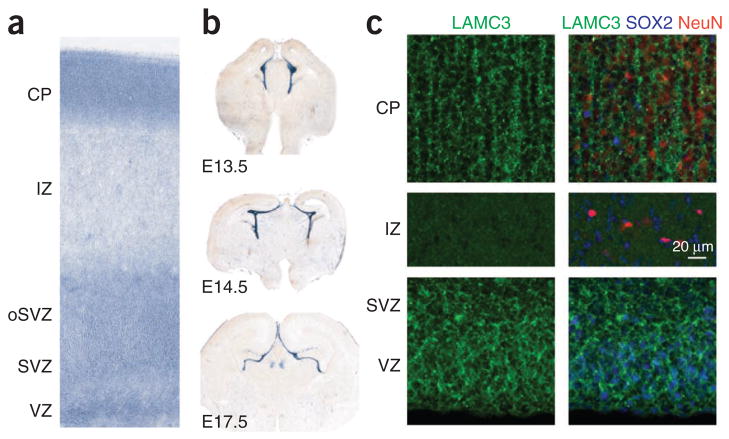
We next investigated the expression of LAMC3 in the developing human fetal brain at 20 post-conceptual weeks (PCW) and showed that it was enriched in the cortical plate, a dense layer of post-migratory pyramidal neurons, and was also present at lower levels in the ventricular and subventricular zones, the germinal layers of the cortex (Fig. 3 and Supplementary Fig. 5). In contrast, we observed a strikingly different expression pattern in the embryonic mouse brain, where Lamc3 expression was absent from the neuronal cells and was instead restricted to the embryonic cerebral vasculature and the meninges from embryonic day (E) 13.5 onwards (Fig. 3b and Supplementary Fig. 6). This finding, which is consistent with the previous observation that Lamc3 deletion in mice is silent with no apparent phenotype14, suggested that LAMC3 expression has diverged on the mammalian lineage, making the mouse a poor model for understanding LAMC3 function in human cortical development.
Figure 3.
Species differences in expression of LAMC3 in the cerebral cortex. (a) In situ hybridization of mid-fetal human brain (20 PCW) shows robust LAMC3 expression in the cortical plate (CP) and within the germinal zones of the neocortical wall, including the ventricular zone (VZ) and subventricular zone (SVZ), and the outer SVZ (oSVZ). IZ, intermediate zone. (b) In contrast, Lamc3 expression in the developing mouse brain (E13.5 to E17.5) is limited to the vasculature and meninges. (c) Human fetal neocortex (20 PCW) immunostained for LAMC3 (green), the post-mitotic neuronal marker NeuN (red) and the neural stem cell marker SOX2 (blue) shows LAMC3 to be strongly expressed in post-mitotic NeuN-positive neurons that have migrated and settled in the cortical plate (CP). Although LAMC3 is not expressed in the intermediate zone, which contains migrating neurons, expression is seen in SOX2-positive neural stem cells in the ventricular zone and subventricular zone. In addition, LAMC3 is expressed in the basal lamina of blood vessels. Composite images are shown on the right. See Supplementary Figure 5 for individual panels.
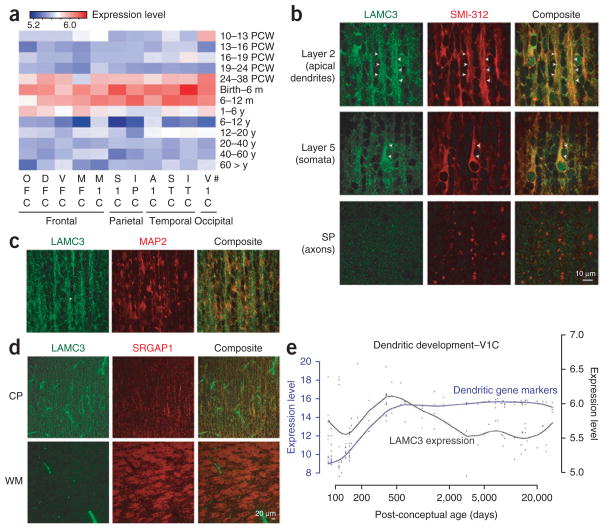
We then investigated the spatial and temporal changes in expression during human cortical development and first detected LAMC3 transcripts after mid-gestation, which peaked at an interval extending from late fetal development to late infancy (12 months) (Fig. 4a), coinciding with the period of cortical organization that takes place after neural stem cell proliferation and migration of postmitotic neurons to the cortical plate. Examination of LAMC3 expression at the subcellular level supported these observations, showing that LAMC3 primarily localizes to the soma and apical dendrite of pyramidal neurons (Fig. 4b,c and Supplementary Fig. 7) and is barely detected along or near the axons (Fig. 4d). A role for LAMC3 in extracellular matrix organization of the soma-todendritic compartment was further supported by an analysis of the Human Brain Transcriptome database15, showing that the developmental expression pattern of LAMC3 parallels that of other molecules important for dendritogenesis (Fig. 4e)16,17 and synapse formation (Supplementary Note, Supplementary Fig. 8 and Supplementary Table 7)18.
Figure 4.
Temporal and spatial LAMC3 expression pattern. (a) Heat map of LAMC3 expression in human brain. The strongest expression was seen between late gestation (24–38 PCW) and late infancy (6–12 post-natal months) and is more prominent within the temporo-occipital lobes as compared to frontal regions. PCW, post-conceptual weeks; m, month; y, year. #Brain regions sampled are listed under Methods. (b) Human fetal neocortex (20 PCW) immunostained for LAMC3 (green) and the neuronal marker SMI-312 (red) reveals high expression in pyramidal neurons in a punctate pattern primarily localizing to the somata and apical dendrites (arrowheads), both at the proximal and distal segments. LAMC3 expression is not detected on axons as they leave the cortical plate (CP) and enter the subplate (SP). (c) In cortical layer 2, LAMC3 (green) is localized to bundles of distal apical dendrites (arrowhead) immuno-positive for MAP2 (red), a marker for dendrites. (d) In the cortical plate, LAMC3 (green) is expressed in pyramidal neurons. In the white matter (WM), where bundles of SRGAP1-positive corticofugal axons (red) are abundantly present, LAMC3 is weak or absent on axons but highly expressed in the neighboring blood vessels. (e) LAMC3 expression (black line) parallels that of genes known to be expressed during the period of dendritogenesis (blue line) within the V1 visual cortex (V1C) during late fetal and early post-natal periods and declines after infancy. For a and e, log2-transformed expression values are shown (Online Methods).
Laminins are extracellular cell adhesion molecules mostly localizing to basement membranes19 and are essential for early embryonic development. They are cross-shaped heterotrimers consisting of α, β and γ chains. Mutations in various laminin molecules lead to diverse pathologies including Pierson syndrome (congenital nephrotic syndrome with or without ocular abnormalities caused by LAMB2 mutations), junctional epidermolysis bullosa (α3, β3 and γ2 chain mutations) and congenital merosin-deficient muscular dystrophy type 1A (LAMA2 mutations), which is sometimes associated with cobblestone lissencephaly and occipital pachygyria19–21. Mutations in LAMC3 have not previously been reported. LAMC3 is not thought to associate exclusively with the basement membrane—it can bind either to nidogen or to α6β1-integrin and is expressed in mouse retina and brain vessels22–25, with Lamc3 knockout mice lacking any overt phenotypes14,26.
Little is known about the precise molecular mechanisms underlying human cortical gyration. Here we present conclusive genetic evidence that recessive LAMC3 mutations cause human occipital cortical malformations characterized by complex gyration abnormalities and challenge the concept that cortical smoothening needs to be caused by disruption of early events in cortical neuronal migration exclusively. Further studies are needed to identify other molecules involved in the intricate process of cortical organization and to understand why the phenotype associated with LAMC3 mutations is restricted to the occipital lobes.
URLs
dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/; 1000 Genomes Project, http://www.1000genomes.org/; Human Brain Transcriptome database, http://www.humanbraintranscriptome.org/; RefSeq, http://www.ncbi.nlm.nih.gov/RefSeq/; DAVID Bioinformatics Resources, http://david.abcc.ncifcrf.gov/; Freesurfer, http://surfer.nmr.mgh.harvard.edu/; Java programming package, http://bilkent.edu.tr/~hboyaci/PsychWithJava; MedINRIA, http://www-sop.inria.fr/asclepios/software/MedINRIA/index.php.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturegenetics/.
Accession codes. LAMC3 data are deposited in RefSeq under accession number NM_006059.
Supplementary Material
Acknowledgments
We are indebted to the subjects and families who have contributed to this study. We would like to thank M. State and J. Noonan for critical comments regarding the study and C. Camputaro for her help with the imaging studies. We acknowledge the use of Yale University Biomedical High Performance Computing Center for data analysis and storage. This study was supported by the Yale Program on Neurogenetics, the Yale Center for Human Genetics and Genomics, and US National Institutes of Health grants RC2NS070477 (to M.G.), UL1RR024139NIH (Yale Clinical and Translational Science Award) and UO1MH081896 (to N.S.). SNP genotyping was supported in part by a US National Institutes of Health Neuroscience Microarray Consortium award U24 NS051869-02S1 (to S.M.).
Footnotes
AUTHOR CONTRIBUTIONS
M.G. designed the study, and T.B., K.Y.K., A.L., R.P.L., N.S., K.B. and M.G. designed the experiments. T.B., K.Y.K., A.L., K.B., S.Y., M.B., A.O.C., A.K.O. and S.M. performed the experiments. V.D., S.S., B.T., H.K. and C.Y. identified, consented and recruited the study subjects and provided clinical information. A.D. and R.A.B. performed and evaluated magnetic resonance imaging. T.O., H.B., K.D. and E.A. performed and evaluated three-dimensional cortical reconstruction and functional imaging studies. M.C. and R.P.L. developed the bioinformatics scripts for data analysis. W.J.B. provided critical reagents. T.B., T.O., K.Y., K.B., R.P.L. and M.G. analyzed the genetics data. K.Y.K., A.L., Y.Z., N.S. and M.G. analyzed the expression data. T.B., K.Y.K., A.L., R.P.L., N.S., K.B. and M.G. wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturegenetics/.
Published online at http://www.nature.com/naturegenetics/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
Note: Supplementary information is available on the Nature Genetics website.
References
- 1.Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 2.Hofman MA. Size and shape of the cerebral cortex in mammals. I The cortical surface. Brain Behav Evol. 1985;27:28–40. doi: 10.1159/000118718. [DOI] [PubMed] [Google Scholar]
- 3.Caviness VS., Jr Mechanical model of brain convolutional development. Science. 1975;189:18–21. doi: 10.1126/science.1135626. [DOI] [PubMed] [Google Scholar]
- 4.Van Essen DC. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature. 1997;385:313–318. doi: 10.1038/385313a0. [DOI] [PubMed] [Google Scholar]
- 5.Kriegstein A, Noctor S, Martinez-Cerdeno V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- 6.Piao X, et al. G protein-coupled receptor-dependent development of human frontal cortex. Science. 2004;303:2033–2036. doi: 10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- 7.Kostovic I, Rakic P. Developmental history of the transient subplate zone in the visual and somatosensory cortex of the macaque monkey and human brain. J Comp Neurol. 1990;297:441–470. doi: 10.1002/cne.902970309. [DOI] [PubMed] [Google Scholar]
- 8.Ferrie CD, Jackson GD, Giannakodimos S, Panayiotopoulos CP. Posterior agyria-pachygyria with polymicrogyria: evidence for an inherited neuronal migration disorder. Neurology. 1995;45:150–153. doi: 10.1212/wnl.45.1.150. [DOI] [PubMed] [Google Scholar]
- 9.Ben Cheikh BO, et al. A locus for bilateral occipital polymicrogyria maps to chromosome 6q16-q22. Neurogenetics. 2009;10:35–42. doi: 10.1007/s10048-008-0143-3. [DOI] [PubMed] [Google Scholar]
- 10.Barkovich AJ, Kuzniecky RI, Jackson GD, Guerrini R, Dobyns WB. A developmental and genetic classification for malformations of cortical development. Neurology. 2005;65:1873–1887. doi: 10.1212/01.wnl.0000183747.05269.2d. [DOI] [PubMed] [Google Scholar]
- 11.Bilgüvar K, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sereno MI, et al. Borders of multiple visual areas in humans revealed by functional magnetic resonance imaging. Science. 1995;268:889–893. doi: 10.1126/science.7754376. [DOI] [PubMed] [Google Scholar]
- 13.Wandell BA, Dumoulin SO, Brewer AA. Visual field maps in human cortex. Neuron. 2007;56:366–383. doi: 10.1016/j.neuron.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Dénes V, et al. Laminin deficits induce alterations in the development of dopaminergic neurons in the mouse retina. Vis Neurosci. 2007;24:549–562. doi: 10.1017/S0952523807070514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson MB, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mrzljak L, Uylings HB, Kostovic I, van Eden CG. Prenatal development of neurons in the human prefrontal cortex. II A quantitative Golgi study. J Comp Neurol. 1992;316:485–496. doi: 10.1002/cne.903160408. [DOI] [PubMed] [Google Scholar]
- 17.Petanjek Z, Judas M, Kostovic I, Uylings HB. Lifespan alterations of basal dendritic trees of pyramidal neurons in the human prefrontal cortex: a layer-specific pattern. Cereb Cortex. 2008;18:915–929. doi: 10.1093/cercor/bhm124. [DOI] [PubMed] [Google Scholar]
- 18.Huttenlocher PR, Dabholkar AS. Regional differences in synaptogenesis in human cerebral cortex. J Comp Neurol. 1997;387:167–178. doi: 10.1002/(sici)1096-9861(19971020)387:2<167::aid-cne1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 19.Durbeej M. Laminins. Cell Tissue Res. 2010;339:259–268. doi: 10.1007/s00441-009-0838-2. [DOI] [PubMed] [Google Scholar]
- 20.Helbling-Leclerc A, et al. Mutations in the laminin alpha 2-chain gene (LAMA2) cause merosin-deficient congenital muscular dystrophy. Nat Genet. 1995;11:216–218. doi: 10.1038/ng1095-216. [DOI] [PubMed] [Google Scholar]
- 21.Jones KJ, et al. The expanding phenotype of laminin alpha2 chain (merosin) abnormalities: case series and review. J Med Genet. 2001;38:649–657. doi: 10.1136/jmg.38.10.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gersdorff N, Kohfeldt E, Sasaki T, Timpl R, Miosge N. Laminin γ3 chain binds to nidogen and is located in murine basement membranes. J Biol Chem. 2005;280:22146–22153. doi: 10.1074/jbc.M501875200. [DOI] [PubMed] [Google Scholar]
- 23.Koch M, et al. Characterization and expression of the laminin γ3 chain: a novel, non-basement membrane-associated, laminin chain. J Cell Biol. 1999;145:605–618. doi: 10.1083/jcb.145.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Libby RT, et al. Laminin expression in adult and developing retinae: evidence of two novel CNS laminins. J Neurosci. 2000;20:6517–6528. doi: 10.1523/JNEUROSCI.20-17-06517.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan HH, Cheng CY. Laminin α3 forms a complex with β3 and γ3 chains that serves as the ligand for α 6β1-integrin at the apical ectoplasmic specialization in adult rat testes. J Biol Chem. 2006;281:17286–17303. doi: 10.1074/jbc.M513218200. [DOI] [PubMed] [Google Scholar]
- 26.Pinzón-Duarte G, Daly G, Li YN, Koch M, Brunken WJ. Defective formation of the inner limiting membrane in laminin α2- and γ3-null mice produces retinal dysplasia. Invest Ophthalmol Vis Sci. 2010;51:1773–1782. doi: 10.1167/iovs.09-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.