Summary
We present a strategy to examine the chromatin conformation of individual loci in specific cell types during Drosophila embryogenesis. Regulatory DNA is tagged with binding sites (lacO) for LacI, which is used to immunopreciptiate the tagged chromatin from specific cell types. We applied this approach to Distalless (Dll), a gene required for limb development in Drosophila. We show that the local chromatin conformation at Dll depends on the cell type: in cells that express Dll, the 5’ regulatory region is in close proximity to the Dll promoter. In Dll nonexpressing cells this DNA is in a more extended configuration. In addition, transcriptional activators and repressors are bound to Dll regulatory DNA in a cell type specific manner. The pattern of binding by GAGA factor and the variant histone H2Av suggest that they play a role in the regulation of Dll chromatin conformation in expressing and non-expressing cell types, respectively.
Introduction
The regulation of transcription in higher eukaryotes depends on cis-regulatory modules (CRMs), DNA sequences that integrate temporal and spatial information by binding groups of transcription factors (Istrail and Davidson, 2005). CRMs can be very far – even tens or hundreds of kilobases – from a gene’s promoter, where transcription initiates (Bartkuhn and Renkawitz, 2008). Moreover, in some cases, CRMs have been shown to regulate the transcription of genes located on other chromosomes (Apostolou and Thanos, 2008; Cavalli, 2007; Dekker, 2008; Ling et al., 2006; Lomvardas et al., 2006; Simonis et al., 2006). In many cases, communication between distant CRMs and promoters has been observed as a physical interaction between these elements, with intervening DNA looped out (Gothard et al., 1996; Heintzman and Ren, 2009; Liu and Garrard, 2005; Nolis et al., 2009; Petrascheck et al., 2005; Schneider and Grosschedl, 2007). Several transcription factors, such as GAGA factor (GAF) and CTCF, have been implicated in mediating such long-range interactions, which are thought to underlie much of gene regulation in eukaryotes (Ling et al., 2006; Mahmoudi et al., 2002; Ohtsuki and Levine, 1998).
Although chromatin structure can have a profound influence on gene expression, most approaches for analyzing chromatin during animal embryogenesis do not have cell type specific resolution and thus cannot reveal biologically relevant differences if they exist. Capturing chromosome conformation (3C), for example, is capable of detecting interactions between DNA elements but, when applied to a whole embryo, cannot reveal in which cells these interactions occur (Dekker et al., 2002). Similarly, chromatin immunoprecipitation (ChIP) can also identify interactions between DNA elements, but unless some method is used to purify cell types (for example, by cell sorting), it also cannot determine if such interactions are cell-type specific (Kadauke and Blobel, 2009). ChIP assays also suffer from the problem that it is difficult to determine if a DNA element is immunoprecipitated because of an interaction with another element or because both elements have a binding site for the immunoprecipitated transcription factor. In one study, a solution to this problem was made possible by knocking in binding sites for the yeast transcription factor Gal4 into the imprinted Igf-H19 locus (Murrell et al., 2004; Reik et al., 2004). Using antibodies against Gal4 to specifically ChIP this DNA, it was discovered that the pattern of long-range interactions differed depending on whether the locus was paternally or maternally inherited. Tissue-dependent differences in chromatin conformations have also been observed in Drosophila at the Abd-B locus (Cleard et al., 2006), as well as at Sonic hedgehog (Shh) (Amano et al., 2009), β-globin (Palstra et al., 2003), and vertebrate Hox gene complexes (Montavon et al., 2011; Noordermeer et al., 2011). However, these studies generally have limited resolution and compared tissues that have very distinct developmental origins. Moreover, most of the approaches used to identify long-range interactions in these studies cannot be used in a second step to identify the factors that mediate these interactions. Thus it remains an open question whether changes in CRM-promoter interactions are used to regulate gene expression on a finer scale and, if so, which factors may be involved.
Distalless (Dll) is required for appendage development in Drosophila (Cohen et al., 1989; Cohen and Jurgens, 1989), and depends on multiple CRMs for its correct expression during embryogenesis and larval development (Estella et al., 2012; Galindo et al., 2011; McKay et al., 2009; Vachon et al., 1992). Two of these CRMs, Dll304 and LT, are located next to each other and ~12 kb 5’ to the start of Dll transcription, suggesting that there is long-range communication between these CRMs and the Dll promoter (Estella et al., 2008) (Figure 1A). Dll304 is the first Dll CRM to be active at ~stage 10 (~5 hours) of embryogenesis in a group of ~30 cells/thoracic hemisegment. Dll304 is activated by Wingless (Wg) signaling, but is repressed in abdominal segments by the abdominal Hox factors, Ultrabithorax (Ubx) and Abdominal-A (Abd-A) (Gebelein et al., 2002; Vachon et al., 1992) (Figure 1A). Ubx and Abd-A directly and cooperatively bind to Dll304 with two Hox cofactors, Extradenticle (Exd) and Homothorax (Hth) (Gebelein et al., 2004). LT, which is activated later in embryogenesis (stage 13), requires direct input from both Wg and Decapentaplegic (Dpp) signaling, as well as input from the Zn-finger transcription factors, Buttonhead (Btd) and Sp1 (Estella et al., 2003; McKay et al., 2009). In addition, LT requires Dll input, derived from the earlier acting Dll304 CRM. As a consequence, direct Hox-mediated repression of Dll304 is a key reason that LT is not activated and Dll is not expressed in the abdomen. Once LT is activated Dll expression is maintained via a positive autoregulatory loop that requires direct binding of Dll to a maintenance (M) element, which encompasses the Dll promoter (Estella et al., 2003; McKay et al., 2009) (Figure 1A). In the experiments described here, we confirm that Dll CRMs interact with the Dll promoter. More interestingly, we show that this interaction depends on the cell type. Our results suggest that Hox proteins regulate Dll transcription in part by locally modifying chromatin structure at the Dll locus.
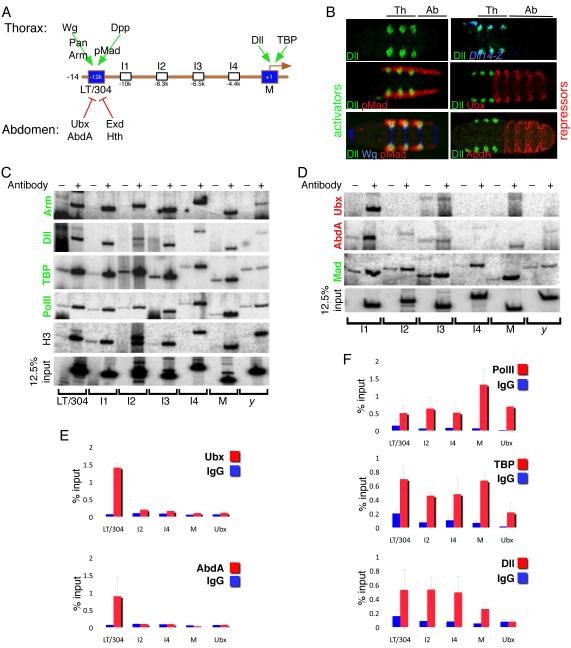
Figure 1. Whole embryo ChIPs show unique distributions of activators and repressors bound to Dll regulatory regions.
(A) Schematic of the 14 kb of DNA 5’ to the start of Dll transcription, showing the positions of the LT/304 CRMs and M element. Positive inputs in the thorax (above the DNA) and negative inputs in the abdomen (below the DNA) are indicated. I1 to I4 are intermediate regions that were monitored by PCR in ChIP experiments.
(B) Expression patterns of known Dll activators (Wg, blue; pMad, red) and repressors (Ubx and AbdA, red) relative to Dll expression in the thorax (green).
(C) Whole embryo ChIPs using unprogrammed IgG (−) or antibodies (+) to known activators (Arm, Dll, TBP, PolII), Histone3 (H3). IPd chromatin was used as a template for 32P PCRs with the amplicons indicated below the gels. y refers to an amplicon in the yellow gene and serves as a negative control. 12.5 % input shows the signal obtained with input chromatin.
(D) Whole embryo ChIPs using unprogrammed IgG (−) or antibodies (+) to a known activator (Mad) or two known repressors (Ubx and AbdA). IPd chromatin was used as a template for 32P PCRs with the amplicons shown below the gels.
(E) qPCRs of whole embryo ChIPs comparing the signals obtained with IgG and either anti-Ubx or anti-AbdA for a subset of Dll amplicons. For these repressors, a strong signal was only obtained for LT/304. An amplicon close to the Ubx promoter serves as a negative control. In these and all subsequent qPCRs the error bars represent the standard error of the mean.
(F) qPCRs of whole embryo ChIPs comparing the signals obtained with IgG and anti-PolII, anti-TBP, and anti-Dll. For these activators, a strong signal was obtained for LT/304, I2, I4, and M. An amplicon close to the Ubx promoter served as a positive control for PolII and TBP binding, but showed no binding to Dll, as expected.
Results and Discussion
To dissect the regulation of Dll beyond the characterization of CRMs, we initially carried out standard ChIP experiments with whole embryos using antibodies directed against several factors known to regulate Dll. In these ChIP experiments, we typically surveyed the LT/304 region, the Dll promoter (M) region, as well as three to four intermediate regions (I1 to I4) in between LT/304 and M (Figure 1A). We carried out ChIPs for both abdominal repressors (the Hox proteins Ubx and AbdA), known activators (Mad (Mothers against Dpp, a transcriptional effector in the Dpp pathway), Armadillo (Arm, a coactivator in the Wg pathway) and Dll) as well as two components of the basal transcription machinery (TATA-binding protein, TBP, and RNA Polymerase II (PolII)) (Figure 1B). Curiously, we found that all three activators, TBP, and PolII behaved differently in these ChIP experiments compared to the repressors. When ChIPing for Ubx or AbdA only the LT/304 region, but not any of the intermediate or M regions, was robustly detected compared to control ChIPs (Figure 1D,E). In contrast, all of these regions, even sequences far from the known CRMs and promoter, were detected in ChIPs for any of the activators (Mad, Arm, Dll), TBP, or PolII (Figure 1C,D,F).
Two scenarios can account for the different abilities of activators and repressors to ChIP Dll DNA sequences. In one, the activators and basal transcriptional machinery are bound, directly or indirectly, to binding sites scattered throughout the 12 kb 5’ Dll DNA, while the repressors are bound only to the LT/304 region. Alternatively, the configuration of the chromatin may be different in cells where the activators are bound compared to cells in which the repressors are bound. According to this idea, in cells where the activators are bound the chromatin may be configured such that multiple regions of the 12 kb 5’ DNA are close to each other, within a cross-linkable distance to the promoter. In contrast, in cells where the repressors are bound, the LT/304 region, which contains known binding sites for these factors, would not be in close proximity to the rest of the 5’ Dll DNA and promoter. These two configurations may correspond to cells that express or repress Dll, respectively.
Standard ChIP experiments with whole embryos, including 3C and its derivatives (Gavrilov et al., 2009), cannot discriminate between these two scenarios because they do not distinguish cells that express Dll from cells where Dll is repressed. Existing methods also have limited resolution and sensitivity, especially for genes such as Dll that are expressed transiently and in only a small subset of total embryonic cells. To overcome these obstacles, we established a method, called cell and gene-specific ChIP (cgChIP), in which one can monitor the chromatin structures of specific DNA sequences in specific cell types. We used this approach to characterize the 14 kb 5’ Dll region in both Dll-expressing and -nonexpressing cell types. cgChIP is a two-component system that relies on an interaction between the E. coli DNA binding protein LacI and its binding site, lacO. The first component of cgChIP consists of cell type specific expression of a flag-tagged version of LacI. To study Dll, we generated two genotypes that differ only in the expression pattern of flag-LacI: (1) thorax>lacI, (Dll304-Gal4; UAS-flag-lacI) in which LacI is expressed in the Dll-expressing cells of the thoracic appendage primordia; and (2) abdomen>lacI, (DMEAct-Gal4, Dll304-Gal80; UAS-flag-lacI) in which LacI is expressed in the homologous cells of the abdomen (Figure 2A and see Experimental Procedures for details). Notably, although they do not express Dll, abdomen>lacI-expressing cells receive the same positive inputs (e.g. Wg and Dpp signaling) as thorax>lacI-expressing cells. In a second component of cgChIP we generated lacO-tagged, lacZ-expressing transgenes under the control of ~14 kb of DNA 5’ to the start of Dll transcription, which includes Dll304, LT and the native Dll promoter (Figure 2B,C). In one (lacO:M) eight copies of lacO were inserted adjacent to the M element, close to the Dll promoter. In a second (lacO:LT/304) eight copies of lacO were inserted into a non-conserved region at LT/304. Importantly, both lacO:LT/304 and lacO:M drove expression of lacZ in a pattern that was indistinguishable from Dll, in the presence or absence of LacI, suggesting that the 14 kb region is sufficient to drive accurate Dll-like expression, and that binding of LacI to the lacO sequences did not interfere with the normal activities of the Dll CRMs or promoter (Figure 2A-C). By combining these tools, we expressed Flag-LacI in the Dll-expressing or non-expressing cells in flies that also contained either the lacO:LT/304 or lacO:M transgenes. Flag-lacI-bound chromatin was immunoprecipitated with anti-Flag antibody and analyzed by PCR (Figure 2D). The cell type specific expression of Flag-lacI, coupled with the lacO-tagged Dll transgenes (lacO:M or lacO:LT/304), allowed us to ask questions about the state of Dll regulatory sequences in specific cell types that cannot be answered by conventional ChIP experiments.
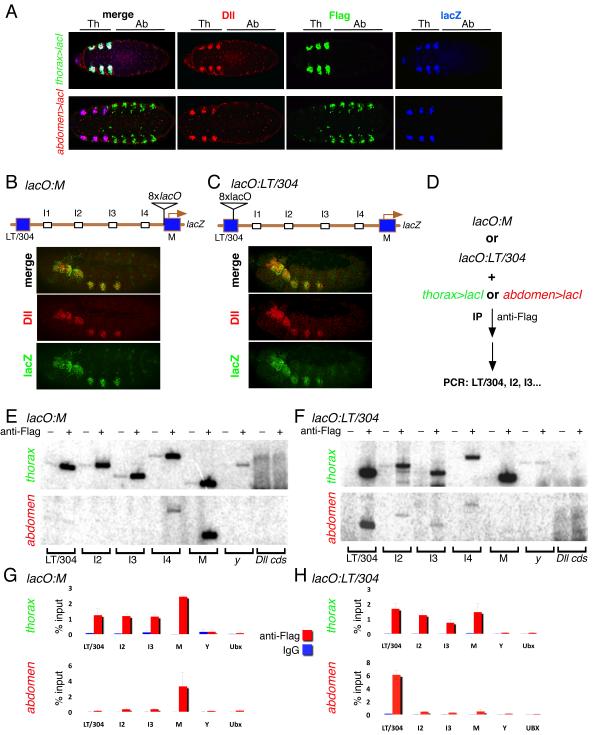
Figure 2. Cell type specific CRM-promoter interactions at Dll.
(A) Ventral views of stage 14 embryos stained for Dll (red), βgal (blue), and Flag-lacI (green). Top row: lacO:M-lacZ; thorax>lacI (thorax-Gal4; UAS-flag-lacI); bottom row: lacO:M-lacZ; abdomen>lacI (abdomen-Gal4; UAS-flag-lacI). The positions of the thoracic and abdominal segments are indicated above the images. Wild type, Dll-like expression of lacZ is observed despite the presence of lacO binding sites and expression of LacI. Note that although abdomen>lacI is expressed in some non-abdominal cells, they do not express Dll (see Methods for more details).
(B,C) Lateral views of stage 14 embryos containing the lacO:M (B) and lacO:LT/304 (C) transgenes, stained for Dll (red) and βgal (green). Schematic diagrams of these two lacZ-expressing transgenes are shown above the images. The expression patterns of Dll and lacZ are indistinguishable.
(D) Outline of cgChIP experiments for monitoring cell type specific interactions between LT/304 and M using the tools defined in (A-C).
(E) 32P PCRs of cgChIPs from lacO:M embryos expressing either thorax>lacI (thorax) or abdomen>lacI (abdomen) as indicated. When Flag-lacI was expressed in the thorax multiple Dll 5’ sequences, but not those from y or the Dll coding sequence (Dll cds), were amplified. In contrast, when Flag-lacI was expressed in the abdomen, only the M element (close to the lacO sites) was amplified. ‘-‘ and ‘+’ above the gels indicate IPs with IgG or anti-Flag, respectively.
(F) 32P PCRs of cgChIPs from lacO:LT/304 embryos expressing either thorax>lacI (thorax) or abdomen>lacI (abdomen) as indicated. When Flag-lacI was expressed in the thorax multiple Dll 5’ sequences, but not those from y or the Dll coding sequence (Dll cds), were amplified. In contrast, when Flag-lacI was expressed in the abdomen, only the LT/304 region (close to the lacO sites) was amplified. ‘-‘ and ‘+’ above the gels indicate IPs with IgG or anti-Flag, respectively.
(G) qPCR results of cgChIP experiments with lacO:M and thorax>lacI or abdomen>lacI as indicated. The results confirm the 32P PCR results shown in (E).
(H) qPCR results for cgChIP experiments with lacO:LT/304 and thorax>lacI or abdomen>lacI as indicated. The results confirm the 32P PCR results shown in (F).
The first set of results, shown in Figure 2E-H by both 32P-labelled and real-time qPCRs, demonstrates that the 14 kb of 5’ Dll DNA is in a distinct configuration in Dll-expressing cells in the thorax compared to Dll-non-expressing cells in the abdomen. When Flag-lacI was expressed in the thorax in embryos containing lacO:M, the M, LT/304, I2, I3, and I4 regions were all efficiently immunoprecipitated compared to control (IgG) ChIPs and negative control sequences in the yellow (y) gene and Dll exons (Figure 2E). In contrast, when Flag-lacI was expressed in the abdomen in lacO:M embryos, only the M element was immunoprecipitated compared to the same negative controls (Figure 2E). Analogous results were obtained when Flag-lacI was expressed in the thorax or abdomen in embryos containing lacO:LT/304: LT/304, M, I2, I3, and I4 were all immunoprecipitated from thoracic cells, while only the LT/304 region was immunoprecipitated from abdominal cells (Figure 2F). These results were confirmed and quantified by carrying out real time qPCR experiments (Figure 2G,H). We conclude that there is no detectable interaction between the LT/304 region and the Dll promoter in abdominal cells, where Dll is repressed by Ubx and AbdA. In contrast, such an interaction is readily observed in thoracic cells that express Dll. Interestingly, in Dll-expressing cells this interaction is not limited to the LT/304 and promoter regions. Instead, the entire 12 kb, including the sequences in between LT/304 and the promoter, are in close proximity to each other in Dll-expressing thoracic cells. The alternative scenario, that LacI “spreads” from its binding site into nearby DNA, is argued against because LacI is a highly specific DNA binding protein and the version used here does not have its self-associating tetramerization domain (Robinett et al., 1996). Nevertheless, because our LacI cgChIPs show clear tissue-specific differences, both the spreading and interaction models argue that the local chromatin structure of the Dll 5’ region is different in Dll-expressing and non-expressing cells. Together, these results suggest that abdominal Hox proteins repress Dll by modifying chromatin structure, in part by interfering with CRM-promoter communication.
We next used cgChIP to study the distribution of transcription factors in 5’ Dll sequences in thoracic and abdominal cell types. In these experiments two consecutive immunopreciptiations were carried out: a primary IP using anti-Flag was used to pull-down Flag-lacI bound to lacO-tagged chromatin, followed by a secondary ChIP using an antibody directed against a protein of interest (Figure 3A). In parallel to the secondary ChIP, we carried out two control IPs: a negative control with unprogrammed IgG and a positive control with an antibody directed against LacI. Obtaining a strong signal (relative to IgG) with anti-LacI confirmed that both rounds of precipitation were successful. In addition, we confirmed that primary anti-Flag cgChIPs using thorax>lacI embryos pulled-down multiple Dll sequences (M, LT/304, and I3), while anti-Flag cgChIPs using abdomen>lacI embryos only detected sequences close to the lacO binding sites (Figure 3B). We again employed both 32P-labelled and real-time qPCRs to quantify ChIP signals. Given the increased number of controls and the limiting quantities of material available for these sequential-ChIP experiments, we limited this analysis to amplicons that detected the LT/304, M, and I3 regions.
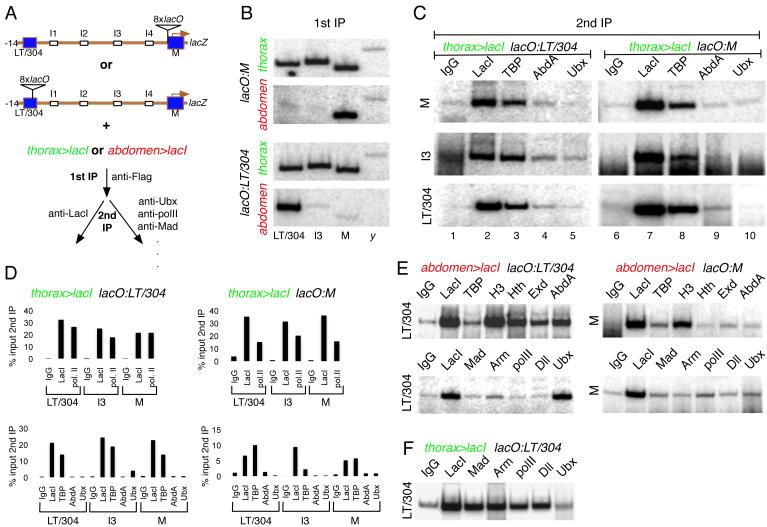
Figure 3. Cell type specific binding of activators and repressors at Dll.
(A) Outline of cgChIP experiments for monitoring the presence of factors bound to Dll regulatory regions in thoracic and abdominal cells.
(B) 32P PCRs confirming the thoracic-specific interaction between Dll regulatory elements after the primary anti-Flag IP. These data served as a quality control for the primary anti-Flag IP before carrying out any secondary ChIPs as in panels C-F. Independent experiments are shown for both lacO:M and lacO:LT/304. thorax (green) and abdomen (red) refer to thorax>lacI and abdomen>lacI, respectively.
(C) 32P PCRs of cgChIPs from thorax>lacI; lacO:LT/304 (left) and thorax>lacI; lacO:M (right) embryos. These PCRs assess the presence of M, I3, and LT/304 sequences following a secondary IP using the antibodies indicated above the gels (IgG, anti-LacI, anti-TBP, anti-AbdA, and anti-Ubx). The results confirm that IPs for LacI and TBP, but not abdominal Hox proteins, pull-down multiple Dll 5’ regions in Dll-expressing cells in the thorax.
(D) qPCR measurements of cgChIP experiments for chromatin isolated from thorax>lacI; lacO:LT/304 (left) and thorax>lacI; lacO:M (right). Measurements are for the three Dll sequences (LT/304, I3, and M) after secondary IPs with the antibodies indicated (top gels: IgG, anti-LacI, anti-PolII; bottom gels: IgG, anti-LacI, anti-TBP, anti-AbdA, anti-Ubx). Quantifications are presented as percentages (%) of the qPCR signals obtained from PCRs for the same amplicons after the primary, anti-Flag IP (i.e.; % input 2nd IP). See also Supplementary Figure 1.
(E) 32P PCRs of cgChIPs from abdomen>lacI; lacO:LT/304 (left) and abdomen>lacI; lacO:M (right). These PCRs assess the presence of the M or LT/304 sequences following a secondary IP using the antibodies indicated above each gel. IPs for repressors (e.g. Hth, Exd, AbdA, and Ubx) pull-down LT/304 sequences but not M sequences; IPs for activators (TBP, Mad, Arm, PolII, and Dll) fail to pull-down any Dll sequences from abdominal cells.
(F) 32P PCRs of cgChIPs from thorax>lacI; lacO:LT/304 embryos. IPs for activators (Mad, Arm, Dll and PolII), but not repressors (Ubx), pull-down these sequences from thoracic cells.
In general, these cgChIP experiments revealed that factors involved in Dll activation, including PolII, TBP, Mad, Tcf (a transcription factor in the Wg pathway), Arm, and Dll, bind to Dll in Dll-expressing thoracic cells, but not in Dll-non-expressing abdominal cells (Figure 3C-E and Supplementary Figure 1). Moreover, thorax>lacI cgChIPs for these factors pulled-down LT/304, the Dll promoter, and DNA sequences in between these two elements, regardless of where the lacO sequences were inserted. In contrast, cgChIPs for activators and RNA PolII failed to pull-down any Dll sequences when abdomen>lacI was used to examine the Dll-nonexpressing cells in the abdomen (Figure 3E). These results suggest that these activators are bound to the structurally compact 5’ Dll sequences in thoracic Dll-expressing cells, but are not bound to these sequences when they are in a more extended state in Dll-nonexpressing cells in the abdomen.
A different picture emerged when we examined factors known to be important for Dll repression, including the Hox proteins Ubx and AbdA and their cofactors Hth and Exd. In cgChIP experiments using thorax>lacI embryos, Dll sequences were not detected above background with anti-Ubx or anti-AbdA, consistent with the abdominal-specific expression of Ubx and AbdA (Figure 3C,D,F). In contrast, when abdomen>lacI was used to examine Dll-nonexpressing cells in lacO:LT/304 embryos, cgChIPs for repressors pulled-down the LT/304 region, which contains essential binding sites for these factors (Figure 3E, left). Further, consistent with the results shown in Figure 2, M sequences were not detected above background in abdomen>lacI lacO:M cgChIPs (Figure 3E, right). Thus, in the abdomen factors used for Dll repression are bound only to the LT/304 region, which is not in close proximity to other regions of the 5’ Dll regulatory DNA.
To gain insight into the factors contributing to the observed tissue-specific chromatin configurations, we examined the distributions of two proteins previously implicated in establishing distinct chromatin structures, GAGA factor (GAF) and the histone variant H2Av. GAF, encoded by the Trithorax-like (Trl) gene in Drosophila, has been shown to mediate long range and even trans-interactions between DNA elements in vivo (Mahmoudi et al., 2002; Petrascheck et al., 2005), making it a good candidate for promoting CRM-promoter communication at Dll. Supporting this idea, whole embryo ChIPs using an anti-GAF antibody were able to pull-down multiple regions of the Dll 5’ regulatory DNA including LT/304, M, and all four intermediate regions (I1 to I4) (Figure 4A-C). A robust signal of GAF binding was also detected at the Ubx promoter (Negre et al., 2006). The distribution of GAF at Dll is identical to that observed for Dll activators (Figure 1), suggesting that GAF is also used to promote Dll expression. Due to its ability to self interact via its BTB/POZ domain (Katsani et al., 1999), these observations suggest that GAF may play a role in promoting the compact chromatin structure present in Dll-expressing thoracic cells.
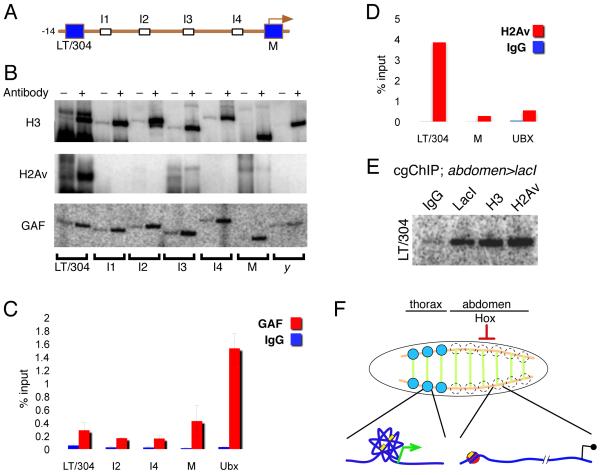
Figure 4. GAF and H2Av have distinct patterns of binding at Dll.
(A) Schematic of the −14 kb 5’ Dll regulatory region.
(B) Whole embryo ChIPs using anti-H3, anti-H2Av, and anti-GAF as indicated. H2Av, like other repressors, is bound to LT/304, but not other 5’ Dll regions. In contrast, binding of GAF appears to be widespread in the Dll 5’ region.
(C) qPCRs of whole embryo ChIPs with anti-GAF, showing widespread binding to the Dll 5’ region, similar to the distribution of other activators (Figure 1).
(D) qPCRs of whole embryo ChIPs with anti-H2Av, showing that it is bound to the LT/304 region, but not to the M region.
(E) 32P PCR of a cgChIP experiment from abdomen>lacI; lacO:LT/304 embryos, showing that H2Av is bound to the LT/304 region in abdominal cells.
(F) Summary of observed cell type specific chromatin configurations in Dll-expressing (thorax) and Dll-non-expressing (abdominal) cells. Thoracic Dll-expressing domains are indicated by the blue circles and occur close to the intersections of Wg expression (green) and Dpp expression (orange). Although Wg and Dpp are present in the same positions in abdominal segments, Dll is repressed in these segments by the abdominal Hox proteins. Our data suggest that in thoracic Dll-expressing cells the entire 5’ region of Dll (with its regulatory elements; yellow boxes) is in a compact state, while in abdominal segments the chromatin structure is more extended and the LT/304 region has H2Av-containing nucleosomes.
In contrast to the broad distribution of GAF, binding of H2Av, a histone variant implicated in both gene activation and repression (Clarkson et al., 1999; Hanai et al., 2008; Swaminathan et al., 2005), was only observed at LT/304, but not at any of the I regions or at the Dll promoter (Figure 4B,D). This polarized distribution of H2Av at Dll is similar to the binding pattern of Ubx and AbdA, implying that H2Av is present at LT/304 in abdominal cells, where Dll is repressed. This conclusion was confirmed by carrying out cgChIP experiments for H2Av using abdomen>lacI; lacO:LT/304 embryos (Figure 4E).
Together, these findings suggest that activation of Dll in thoracic cells may be mediated by GAF’s ability to facilitate long-range interactions between distant regulatory elements and that abdominal Hox factors block these long-range interactions (Figure 4F). The association of H2Av with LT/304 suggests that Hox-mediated recruitment of this histone variant may contribute to the lack of CRM-promoter interaction in abdominal cells. Indeed H2A.Z, the yeast homolog of H2Av, has been implicated in blocking fiber-fiber interactions in in vitro chromatin reconstitution experiments (Fan et al., 2004). Attempts to further test the proposed roles of GAF or H2Av at Dll using genetic approaches were unsuccessful, likely because of the pleiotropic requirement for these factors at many genes and in many cells during Drosophila development. Therefore, we cannot exclude that the presence of GAF or H2Av is a consequence, rather than a cause, of the distinct chromatin configurations present in abdominal and thoracic cells.
In summary, the local chromatin conformation at Dll varies in a developmentally relevant manner: its 5’ regulatory DNA is present in different states depending on whether it is expressed or repressed by abdominal Hox proteins (Figure 4F). In contrast to previous studies where 3D chromatin organization was compared in very different tissues (e.g. forebrain versus limb; Noordermeer et al., 2011), our experiments compared a small group of Dll-expressing cells in the thorax that are fated to give rise to the appendages with the homologous groups of cells in the abdomen. The fates of these two populations of cells differ only due to the expression of Hox selector proteins. As we observed long-distance interactions only in the thorax, our results suggest that abdominal Hox proteins suppress limb development at least in part by preventing distant enhancer elements from being brought into proximity with the Dll promoter. We further speculate that abdominal Hox proteins block these long-range interactions by interfering with the binding of GAF and other activators, perhaps by promoting the assembly of H2Av-containing nucleosomes.
It is also noteworthy that the interactions we observe in Dll-expressing cells are not limited to communication between individual enhancers and the promoter. Instead, the entire 5’ Dll regulatory region appears to be in a more compact state, as many of these sequences are in close proximity to each other and to the Dll promoter. These observations suggest that the entire 5’ 12 kb region functions as a single unit, consistent with the presence of additional Dll CRMs within this region (Estella et al., 2008). Thus, while isolated CRMs and shadow enhancers (Hong et al., 2008) are often sufficient to drive accurate reporter gene expression, multiple CRMs may be integrated within larger functional regulons when in their native context.
Finally, our observations raise the question of whether other genes also have distinct chromatin conformations when activated. Consistent with this view, there are many examples of ChIP experiments that show broad transcription factor binding (>5 kb) that are reminiscent of what we observe for Dll activators (e.g. Li et al., 2011; MacArthur et al., 2009), and broad binding of the circadian rhythm factors Clock and Period was observed at some of their targets (Menet et al., 2010). As we suggest for Dll, these examples may represent the chromatin conformations of large regulons that contain multiple functionally related CRMs. In contrast to these examples, other transcription factor ChIPs typically pull-down short (<1 kb) DNA fragments. However, because many of these experiments were carried out using heterogeneous populations of cells, such as whole embryos, cell-type specific chromatin conformations may be difficult to detect. In addition, chromatin interactions may occur between non-adjacent CRMs that function together to drive gene expression, leading to what appears to be independently ChIPed DNA sequences. It follows that some fraction of the widespread binding observed in conventional ChIP experiments (Biggin, 2011; Li et al., 2011) may be an indirect consequence of interactions between regulatory elements. The recent identification of large chromatin interactomes, in which specific genomic regions interact with each other, is consistent with this view (Fullwood et al., 2009; Handoko et al., 2011; Schoenfelder et al., 2010). In addition to cell type specific chromatin conformations, cell type specific differences in transcription factor binding (e.g. Mad and Tcf binding to Dll in the thorax but not in the abdomen) may also be missed when heterogeneous populations of cells are examined. Only by carrying out cell type specific analyses, such as the cgChIP experiments described here, can such questions be fully resolved.
Experimental Procedures
Antibodies
Immunostaining embryos was performed as in (McKay et al., 2009) with minor modifications: 1) Blocking was carried out overnight in PBST with 5% BSA at 4°C. 2) Both the primary and the secondary antibody incubations were 12 hours at 4°C. The antibodies used for immunostaining were: anti-pMad (gift of G. Morata); anti-AbdA (gift of K. White); anti-Dll (Estella et al., 2008); anti-Wg (Drosophila Hybridoma Bank); anti-β-gal (MP Biomedicals); anti-Flag (Sigma, M2); anti-Ubx (Drosophila Hybridoma Bank). The antibodies used for ChIPs were: anti-Ubx (modEncode; gift of K. White); anti-AbdA (Santa Cruz, SC-27063); anti-Mad (Santa Cruz, SC-25760); anti Arm (Santa Cruz, SC-133180); anti-Dll (Santa Cruz, SC-15858); anti-Hth (Santa Cruz, SC-26187); anti-Exd (Santa Cruz, SC-26190); anti-GAF (Santa Cruz, SC-98263); anti-Flag (Sigma, M2); anti-LacI (Rockland, 600-401-B04); anti-PolII (Abcam, ab5408); anti-TBP (Abcam; ab61411); anti Histone3 (Abcam, ab1791); anti-Histone2Av (Abcam, ab18263).
Whole embryo ChIPs
Performed as in (Orlando et al., 1997) with the below minor modifications: 1) Ultracentrifugation step was carried out for 30 hours. 2) 6 μg of primary antibody were used in an incubation step of 16 hours at 4 degrees. 3. Instead of agarose beads, magnetic beads (Invitrogen) were used and the coupling procedure we carried out for 1 hour at room temperature.
cgChip
The cgChIP experiments included several controls to assess any possible contamination. For one, we routinely carried out anti-abdominal Hox ChIPs side by side with ChIPs for activators and basal factors from thorax>lacI embryos. Because abdominal Hox proteins are not expressed in the thorax, we did not continue with experiments in which these factors were detected in thorax>lacI-derived chromatin. Conversely, an anti-Dll ChIP was carried out in parallel with abdominal>lacI embryos. As Dll is not expressed in the abdomen we did not continue with experiments in which Dll binding was observed in abdomen>lacI-derived chromatin. In addition, for both thorax>lacI and abdomen>lacI experiments, anti-LacI ChIPs were used as a positive control for both the primary and secondary IPs.
Genotypes
thorax>lacI flies were generated by combining Dll304-Gal4 with UAS-3Xflag-lacI (simplified as flag-lacI). abdomen>lacI flies were generated by combining a Dll304-Gal80 transgene and a DMEAct-Gal4 transgene with UAS-3Xflag-lacI. DMEAct is a mutant version of Dll304 that is derepressed in the abdominal segments because the Hox, Exd, and Hth binding sites have been deleted (Gebelein et al., 2004) and Dll304-Gal80 blocks Gal4 activity in thoracic Dll-expressing cells. The result is predominant expression in cells of the abdominal segments that have the potential to express Dll (i.e. they receive the necessary positive inputs) in the absence of Hox repression. Because DMEAct is active in a slightly broader domain than Dll304, some non-Dll-expressing thoracic cells express flag-lacI in the abdomen>lacI embryos. UAS-3Xflag-lacI was generated from a lacI cDNA plasmid obtained from A. Belmont and expresses a form of LacI that has its tetramerization domain removed to avoid the formation of higher-order complexes and an NLS inserted at the N-terminus (Robinett et al., 1996).
Collection and fixing
Embryos ranging in age from 6 to 9 hours were grown at room temperature to ensure Gal80 (when present) was active. About 8 g of embryos were collected and dechorionated using standard procedures. Embryos were washed to remove any nonembryonic structures and fixed at room temperature for 30 min with 3:1 heptane:fix solutions. After washing, the embryos were transferred to Falcon tubes and placed at −80 °C at least for 4 hours.
Chromatin isolation
Embryos were pulverized and incubated twice in buffer A (0.25% Triton X-100, 10 mM Na-EDTA, 0.5 mM Na-EGTA,10mM Hepes pH7.9) for 10 min at room temperature and then twice with buffer B (0.2 M NaCl, 1 mM Na-EDTA, 0.5 mM Na-EGTA, 10 mM Hepes pH 7.9) for 15 min at 4 degrees. Sonication was on ice for at least 7 times 40 sec at maximum power. Upon centrifugation in 4.000 rpm for 10 minutes the supernatant was separated to 1.5 ml volumes followed by centrifugation for 20 min at 12000 rpm at 4 degrees. Sheared isolated chromatin was stored at −80 upon addition of glycerol (5% final).
Looping experiments
500 μg chromatin was pre-cleared by incubation with 10 μl of magnetic beads for 1 hour at 4 degrees in 1x Ripa buffer. The reaction was divided into two tubes and 2.5 μg of anti-Flag antibody or IgG were added, respectively. For the looping experiments the above reaction was at a final volume of 800 μl and incubated at 4 degrees for 4 hours with rocking. 2 μl of beads was added for 1 hour at room temperature, followed by 2 rounds of incubation with 10 mM Hepes pH 7.9, 0.5% Triton X-100, 140mM Nacl, 0.14% DOC, 0.2%SDS. A final wash step was carried out before proteinase K treatment and phenol/chlorophorm extraction and precipitations (Agelopoulos and Thanos, 2006). Formaldehyde cross-linking was reversed and the extracted/precipitated DNA fragments were used as a template for the PCR amplification in which multiple domains of Dll 5’ DNA and control sequences were scanned. An equally divided sample was analyzed side by side with individual pairs of primers. Sequences of the primers are available upon request.
Double cgChIP experiments
8 gr of embryos were used in experiments with 5 antibodies in secondary IPs. Staged embryos were collected, harvested, and IPd for Flag-lacI as described above. Precipitated Dll chromatin was eluted by the addition of 600 μl elution buffer and incubation at 4 degrees for 4 hours. The eluted material was pre-cleared for a second time before further use. A small fraction of the eluted material was treated with proteinase K and after reversal of the cross-links and extraction, the DNA was amplified with primers inside and outside of the transgene that contains the lacO binding sites. Thus, the purity of the first IP was tested before the second IP. PCR with primers that amplify lacZ sequences or sequences outside of the tagged transgene at irrelevant chromosomes were used to ensure the absence of any contamination of non-specific chromatin.
The second round of IP was carried out at 4 degrees. At this stage, two controls (IgG, a negative control, and anti-LacI, a positive control) were performed side by side to insure that the first IP was successful. If confirmed, the eluted chromatin was divided into equal samples and tested with 2 μg of a primary antibody in a total reaction of 300 μl. After 12 hours of incubation chromatin/antibody complex was bound to magnetic beads as above. The reactions were washed twice with 1xWash Ripa buffer and then treated with proteinase K and cross-links reversed. Finally the extracted/precipitated DNA was analyzed with gene specific primers in 32P (Agelopoulos and Thanos, 2006) or SYBR Green based qPCR (Applied Biosystems).
Supplementary Material
Highlights.
Transcriptional repressors and activators have distinct binding patterns at Dll
cgChIP is a new method to analyze cell-type specific chromatin conformations in vivo
Dll regulatory DNA is in distinct conformations in expressing and nonexpressing cells
GAF and H2Av bind to Dll in expressing and nonexpressing cells, respectively
Acknowledgements
We are grateful to A. Belmont, G. Morata, V. Ranade, G. Struhl, and K. White for sharing reagents and/or technical assistance; R. Axel, S. Kalogirou, V. Ranade, G. Struhl, D. Thanos, and members of the Mann lab for feedback during the course of this project; and O. Hobert and C. Desplan for comments on the manuscript. This work was supported by GM058575 and GM054510 awarded to R.S.M. D.J.M. was supported by 5T32DK07328 and M.A. was supported by a long-term EMBO fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Marios Agelopoulos, Department of Biochemistry and Molecular Biophysics, Columbia University, 701 W. 168th St., HHSC 1104, New York, NY 10032, USA.
Daniel J. McKay, Department of Biochemistry and Molecular Biophysics, Columbia University, 701 W. 168th St., HHSC 1104, New York, NY 10032, USA.
Richard S. Mann, Department of Biochemistry and Molecular Biophysics, Columbia University, 701 W. 168th St., HHSC 1104, New York, NY 10032, USA
References
- Agelopoulos M, Thanos D. Epigenetic determination of a cell-specific gene expression program by ATF-2 and the histone variant macroH2A. EMBO J. 2006;25:4843–4853. doi: 10.1038/sj.emboj.7601364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev Cell. 2009;16:47–57. doi: 10.1016/j.devcel.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Apostolou E, Thanos D. Virus Infection Induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- Bartkuhn M, Renkawitz R. Long range chromatin interactions involved in gene regulation. Biochim Biophys Acta. 2008;1783:2161–2166. doi: 10.1016/j.bbamcr.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Biggin MD. Animal transcription networks as highly connected, quantitative continua. Dev Cell. 2011;21:611–626. doi: 10.1016/j.devcel.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Cavalli G. Chromosome kissing. Curr Opin Genet Dev. 2007;17:443–450. doi: 10.1016/j.gde.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Clarkson MJ, Wells JR, Gibson F, Saint R, Tremethick DJ. Regions of variant histone His2AvD required for Drosophila development. Nature. 1999;399:694–697. doi: 10.1038/21436. [DOI] [PubMed] [Google Scholar]
- Cleard F, Moshkin Y, Karch F, Maeda RK. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat Genet. 2006;38:931–935. doi: 10.1038/ng1833. [DOI] [PubMed] [Google Scholar]
- Cohen SM, Bronner G, Kuttner F, Jurgens G, Jackle H. Distal-less encodes a homoeodomain protein required for limb development in Drosophila. Nature. 1989;338:432–434. doi: 10.1038/338432a0. [DOI] [PubMed] [Google Scholar]
- Cohen SM, Jurgens G. Proximal-distal pattern formation in Drosophila: cell autonomous requirement for Disal-less gene activity in limb development. EMBO J. 1989;8:2045–2055. doi: 10.1002/j.1460-2075.1989.tb03613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. Gene regulation in the third dimension. Science. 2008;319:1793–1794. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Estella C, McKay DJ, Mann RS. Molecular integration of wingless, decapentaplegic, and autoregulatory inputs into Distalless during Drosophila leg development. Dev Cell. 2008;14:86–96. doi: 10.1016/j.devcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estella C, Rieckhof G, Calleja M, Morata G. The role of buttonhead and Sp1 in the development of the ventral imaginal discs of Drosophila. Development. 2003;130:5929–5941. doi: 10.1242/dev.00832. [DOI] [PubMed] [Google Scholar]
- Estella C, Voutev R, Mann RS. A dynamic network of morphogens and transcription factors patterns the fly leg. Curr Top Dev Biol. 2012;98:173–198. doi: 10.1016/B978-0-12-386499-4.00007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JY, Rangasamy D, Luger K, Tremethick DJ. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Mol Cell. 2004;16:655–661. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo MI, Fernandez-Garza D, Phillips R, Couso JP. Control of Distalless expression in the Drosophila appendages by functional 3′ enhancers. Dev Biol. 2011;353:396–410. doi: 10.1016/j.ydbio.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilov A, Eivazova E, Priozhkova I, Lipinski M, Razin S, Vassetzky Y. Chromosome conformation capture (from 3C to 5C) and its ChIP-based modification. Methods Mol Biol. 2009;567:171–188. doi: 10.1007/978-1-60327-414-2_12. [DOI] [PubMed] [Google Scholar]
- Gebelein B, Culi J, Ryoo HD, Zhang W, Mann RS. Specificity of Distalless repression and limb primordia development by abdominal Hox proteins. Dev Cell. 2002;3:487–498. doi: 10.1016/s1534-5807(02)00257-5. [DOI] [PubMed] [Google Scholar]
- Gebelein B, McKay DJ, Mann RS. Direct integration of Hox and segmentation gene inputs during Drosophila development. Nature. 2004;431:653–659. doi: 10.1038/nature02946. [DOI] [PubMed] [Google Scholar]
- Gothard LQ, Hibbard JC, Seyfred MA. Estrogen-mediated induction of rat prolactin gene transcription requires the formation of a chromatin loop between the distal enhancer and proximal promoter regions. Mol Endocrinol. 1996;10:185–195. doi: 10.1210/mend.10.2.8825558. [DOI] [PubMed] [Google Scholar]
- Hanai K, Furuhashi H, Yamamoto T, Akasaka K, Hirose S. RSF governs silent chromatin formation via histone H2Av replacement. PLoS Genet. 2008;4:e1000011. doi: 10.1371/journal.pgen.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19:541–549. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istrail S, Davidson EH. Logic functions of the genomic cis-regulatory code. Proc Natl Acad Sci U S A. 2005;102:4954–4959. doi: 10.1073/pnas.0409624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadauke S, Blobel GA. Chromatin loops in gene regulation. Biochim Biophys Acta. 2009;1789:17–25. doi: 10.1016/j.bbagrm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsani KR, Hajibagheri MA, Verrijzer CP. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 1999;18:698–708. doi: 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Thomas S, Sabo PJ, Eisen MB, Stamatoyannopoulos JA, Biggin MD. The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome Biol. 2011;12:R34. doi: 10.1186/gb-2011-12-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Liu Z, Garrard WT. Long-range interactions between three transcriptional enhancers, active Vkappa gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol Cell Biol. 2005;25:3220–3231. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- MacArthur S, Li XY, Li J, Brown JB, Chu HC, Zeng L, Grondona BP, Hechmer A, Simirenko L, Keranen SV, et al. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 2009;10:R80. doi: 10.1186/gb-2009-10-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi T, Katsani KR, Verrijzer CP. GAGA can mediate enhancer function in trans by linking two separate DNA molecules. EMBO J. 2002;21:1775–1781. doi: 10.1093/emboj/21.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay DJ, Estella C, Mann RS. The origins of the Drosophila leg revealed by the cis-regulatory architecture of the Distalless gene. Development. 2009;136:61–71. doi: 10.1242/dev.029975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev. 2010;24:358–367. doi: 10.1101/gad.1883910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- Negre N, Hennetin J, Sun LV, Lavrov S, Bellis M, White KP, Cavalli G. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006;4:e170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolis IK, McKay DJ, Mantouvalou E, Lomvardas S, Merika M, Thanos D. Transcription factors mediate long-range enhancer-promoter interactions. Proc Natl Acad Sci U S A. 2009;106:20222–20227. doi: 10.1073/pnas.0902454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. The dynamic architecture of Hox gene clusters. Science. 2011;334:222–225. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- Ohtsuki S, Levine M. GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev. 1998;12:3325–3330. doi: 10.1101/gad.12.21.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- Petrascheck M, Escher D, Mahmoudi T, Verrijzer CP, Schaffner W, Barberis A. DNA looping induced by a transcriptional enhancer in vivo. Nucleic Acids Res. 2005;33:3743–3750. doi: 10.1093/nar/gki689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Murrell A, Lewis A, Mitsuya K, Umlauf D, Dean W, Higgins M, Feil R. Chromosome loops, insulators, and histone methylation: new insights into regulation of imprinting in clusters. Cold Spring Harb Symp Quant Biol. 2004;69:29–37. doi: 10.1101/sqb.2004.69.29. [DOI] [PubMed] [Google Scholar]
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007;21:3027–3043. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Swaminathan J, Baxter EM, Corces VG. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 2005;19:65–76. doi: 10.1101/gad.1259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachon G, Cohen B, Pfeifle C, McGuffin ME, Botas J, Cohen SM. Homeotic genes of the Bithorax complex repress limb development in the abdomen of the Drosophila embryo through the target gene Distal-less. Cell. 1992;71:437–450. doi: 10.1016/0092-8674(92)90513-c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.