Abstract
The present paper reviews the use of protein splicing for the biosynthesis of backbone cyclic polypeptides. This general method allows the in vivo and in vitro biosynthesis of cyclic polypeptides using recombinant DNA expression techniques. Biosynthetic access to backbone cyclic peptides opens the possibility to generate cell-based combinatorial libraries that can be screened inside living cells for their ability to attenuate or inhibit cellular processes thus providing a new way for finding therapeutic agents.
1. Introduction
A significant number of natural products with wide range of pharmacological activities are derived from cyclic polypeptides. In fact, peptide cyclization is widely used in medicinal chemistry to improve the biochemical and biophysical properties of peptide-based drug candidates [1, 2]. Cyclization rigidifies the polypeptide backbone structure, thereby minimizing the entropic cost of receptor binding and also improving the stability of the topologically constrained polypeptide. Among the different approaches used to cyclize polypeptides, backbone or head-to-tail cyclization remains one of the most extensively used to introduce structural constraints into biologically active peptides.
Despite the fact that the chemical synthesis of cyclic peptides has been well explored and a number different approaches involving solid-phase or liquid-phase exist [3–7], recent developments in the fields of molecular biology and protein engineering have now made possible the biosynthesis of cyclic peptides (Scheme 1). This progress has been made mainly in two areas, non-ribosomal peptide synthesis [8–10] and expressed protein ligation/protein trans-splicing [11–16]. The former strategy involves the use of genetically engineered non-ribosomal peptide synthetases and is reminiscent of more established technologies that yield novel polyketides. The later strategy relies on the heterologous expression of recombinant proteins fused to modified intein protein splicing/trans-splicing units [17].
Scheme 1.
Summary of the technologies used for the biosynthesis of backbone cyclized peptides. All these methods rely on the ribosomal synthesis of protein precursors that undergo protein splicing mediated by inteins, proteases or sortases. Three examples of naturally occurring backbone cyclized peptides with potential therapeutic value are also shown in the middle of the scheme. MCoTI-II is a naturally occurring cyclotide with trypsin inhibitory activity found in the seeds of tropical squash (Momordica conchichinensis) [74, 75] (PDB entry: 1IB9). SFTI-1 is a Bowman-Birk protease inhibitor found in the seeds of sunflower (Helianthus annuus) [76, 77] (PDB entry: 1JBN). Both peptides have been biosynthesized using EPL [49]. RTD-1 is a primate defensin with strong antibacterial and antiviral activity [78, 79] (PDB entry: 1HVZ).
The biosynthesis of cyclic polypeptides offers many advantages over purely synthetic methods. Using the tools of molecular biology, large combinatorial libraries of cyclic peptides, may be generated and screened in vivo. A typical chemical synthesis may generate 104 different molecules. It is not uncommon for a recombinant library to contain as many as 109 members. The molecular diversity generated by this approach is analogous to phage-display technology. Moreover, this approach takes advantage of the enhanced pharmacological properties of backbone-cyclized peptides as opposed to linear peptides or disulfide-stabilized polypeptides. Also, the approach differs from phage-display in that the backbone-cyclized polypeptides are not fused to or displayed by any viral particle or protein, but remain on the inside of the living cell where they can be further screened for biological activity in an analogous way as the yeast two hybrid technology works [18]. The complex cellular cytoplasm provides the appropriate environment to address the physiological relevance of potential leads.
Protein trans-splicing had been successfully used by Benkovic and co-workers to generate backbone cyclized or polypeptides in vivo [12]. In this approach, the peptide to be cyclized was nested between the two split intein fragments of the naturally occurring Ssp DnaE split intein [19] (usually referred as N- and C-inteins) in such way that the N-terminus of the peptide template is fused to C-intein fragment and vice versa. Protein splicing of this chimeric protein lead to the formation of the desired cyclic peptide inside E. coli cells. A potential limitation of this approach, however, was the requirement for specific N- and C-extein residues at the intein junction sites [20]. These amino acids were necessary for efficient protein splicing to occur, which restricts the sequence diversity within the sequence of the cyclic peptide.
An attractive alternative approach to the biosynthesis of circular polypeptides was the use of an intramolecular version of Native Chemical Ligation reaction [21–23]. The present paper reviews the use of these processes for the biosynthesis of circular polypeptides (i.e. peptides and proteins) and it will discuss also the potential of these methods for the biosynthesis of cyclic polypeptide libraries inside living cells as a complementary source for the rapid discovery of new therapeutics.
2. Native Chemical Ligation
Native Chemical Ligation (NCL) is an exquisitely specific ligation reaction that has been extensively used for the total synthesis, semi-synthesis and engineering of different proteins [22, 24–26]. In this reaction, two fully unprotected polypeptides, one containing a C-terminal α-thioester group and the other a N-terminal Cys residue, react chemoselectively under neutral aqueous conditions with the formation of a native peptide bond (Fig. 1A). The initial step in this ligation involves the formation of a thioester-linked intermediate, which is generated by a trans-thioesterification reaction involving the α-thioester moiety of one fragment and the N-terminal Cys thiol group of the other fragment. This intermediate then spontaneously rearranges to produce a peptide bond at the ligation site. This type of thioester-based chemistry was first discovered by Wieland in 1950’s for the synthesis of small Cys-containing peptides [27, 28].
Figure 1.
Backbone cyclization of polypeptides using native chemical Ligation. A. Principle of Native Chemical Ligation (NCL). B. Intramolecular NLC leads to the formation of a backbone cyclized polypeptide.
It is well established that when these two reactive groups, i.e. the C-terminal α-thioester group and the N-terminal Cys residue, are located in the same synthetic precursor, the chemical ligation proceeds in an intramolecular fashion thus resulting in the efficient formation of a circular polypeptide (Fig. 1B). This reaction has been successfully employed for the chemical synthesis of cyclic peptides and small protein domains [3, 5–7].
3. Expressed Protein Ligation
The discovery of protein splicing and advances in protein engineering have made also possible the introduction of the C-terminal α-thioester group and N-terminal Cys residue into recombinant proteins. These important developments made possible the use of NCL between synthetic and/or recombinant fragments. This technology, called Expressed Protein Ligation (EPL), has allowed access to a multitude of chemically engineered recombinant proteins including biosynthetic circular polypeptides [26].
3.1. Recombinant Polypeptide α-thioesters
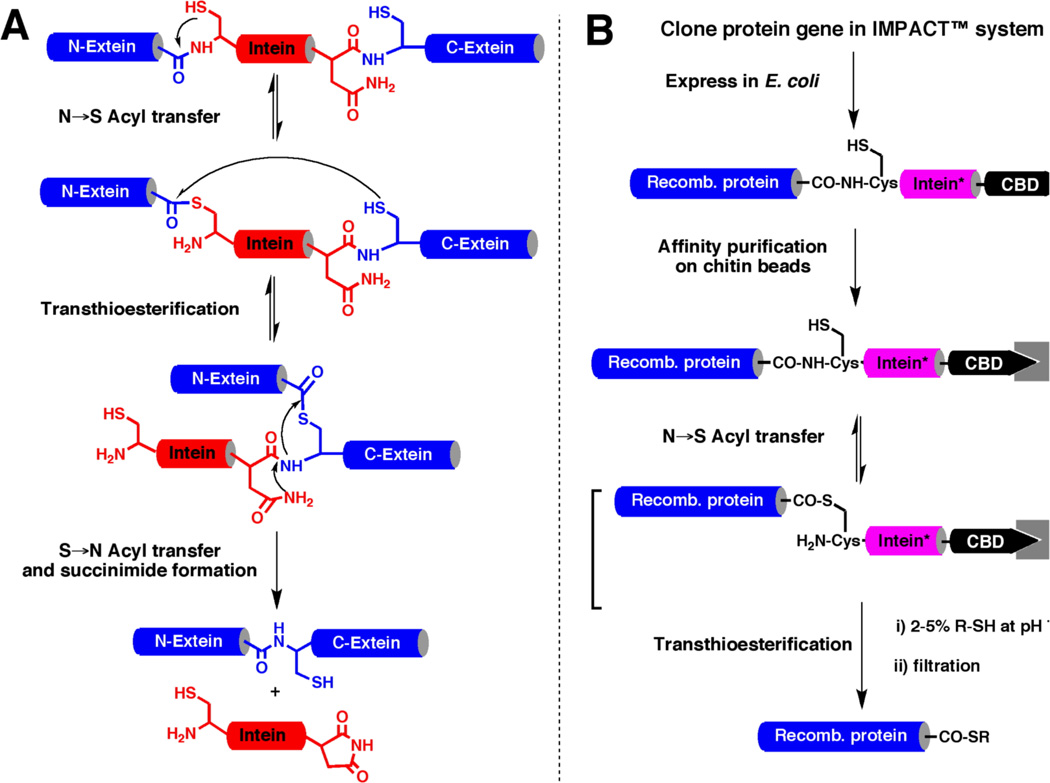
Recombinant protein α-thioesters can be obtained by using engineered inteins [25, 29–31]. Inteins are self-processing domains which mediate the naturally occurring process called protein splicing [32] (Fig. 2). Protein splicing is a cellular processing event that occurs post-translationally at the polypeptide level. In this multi-step process an internal polypeptide fragment, called intein, is self-excised from a precursor protein and in the process ligates the flanking protein sequences (N- and C-exteins) to give a different protein. The current understanding of the mechanism is summarized in Figure 2A and involves the formation of thioester/ester intermediates [32]. The first step in the splicing process involves an N→S or N→O acyl shift in which the N-extein is transferred to the thiol/alcohol group of the first residue of the intein. After the initial N→(S/O) acyl shift, a trans-esterification step occurs in which the N-extein is transferred to the side-chain of a second conserved Cys, Ser or Thr residue, this time located at the junction between the intein and the C-extein. The amide bond at this junction is then broken as a result of succinimide formation involving a conserved Asn residue within the intein. In the final step of the process, a peptide bond is formed between the N-extein and C-extein following an (S/O)→N acyl shift (similar to the last step of Native Chemical Ligation, see Fig. 1A). Mutation of the conserved Asn residue within the intein to Ala blocks the splicing process in midstream thus resulting in the formation of an α-thioester linkage between N-extein and the intein [32] (Fig. 2B). This thioester bond can be cleaved using an appropriate thiol through a trans-thioesterfication step to give the corresponding recombinant polypeptide α-thioester. The IMPACT expression system, commercially available from New England Biolabs [33, 34], allows the generation of recombinant α-thioester proteins by making use of such modified inteins in conjunction with a chitin binding domain (CBD) for easy purification by affinity chromatography (see Fig. 2B).
Figure 2.
Biosynthesis of recombinant polypeptide α-thioesters. A. Scheme representing the proposed canonical mechanism for protein splicing mediated by a Cys-intein. B. Expression and purification of recombinant polypeptide thioesters using a modified intein fusion protein. In the modified intein (represented with an asterisk) the last Asn residue of the intein has been mutated to Ala to prevent C-terminal cleavage and splicing. This mutation allows trapping a thioester intermediate that can be cleaved with a thiol to provide the corresponding thioester funtion.
3.2. Recombinant N-terminal Cys-containing polypeptides
The introduction of N-terminal Cys residues into expressed proteins can be readily accomplished by cleaving (by proteolysis or auto-proteolysis) the appropriate fusion proteins. The simplest way to generate a recombinant polypeptide containing a N-terminal Cys residue is to introduce a Cys downstream to the initiating Met residue. Once the translation step is completed, the endogeneous methionyl aminopeptidases (MAP) removes the Met residue, thereby generating in vivo a N-terminal Cys residue [14, 35–38]. Other approaches involve the use of exogenous proteases. Verdine and co-workers added a Factor Xa recognition sequence immediately in front of the N-terminal Cys residue of the protein of interest [39]. After purification, the fusion protein was treated with the protease Factor Xa which generated the corresponding N-terminal Cys protein. Tolbert and Wong also showed that the cysteine protease from tobacco etch virus (TEV) can also be used for the same purpose [40]. This protease is highly specific and it can be overexpressed in E. coli. Other proteases that cleave at the C-terminal side of their recognition site, like enterokinase and ubiquitin C-terminal hydrolase, could be also used for the generation of N-terminal Cys residues.
Protein splicing can also be engineered to produce recombinant N-terminal Cys-containing polypeptides. Several inteins have been already mutated in such a way that cleavage at the C-terminal splice junction (i.e. between the intein and the C-extein, see Fig. 2B) can be accomplished in a pH- and temperature-dependent fashion [41–43].
Proteins with N-terminal Cys can be also obtained by the convenient modification of vectors with the putative thrombin cleavage site LVPRG to LVPRC. Liu et al [44] successfully generated the Csk and Abl tyrosine kinase domains with N-terminal Cys using this method.
More recently, Hauser et al had used the N-terminal pelB leader sequence to direct newly synthesized fusion proteins to the E. coli periplasmic space where the corresponding endogenous leader peptidases [45, 46] can generate the desired N-terminal cysteine-containing protein fragment [47].
4. Biosynthesis of backbone cyclized peptides using Expressed Protein Ligation
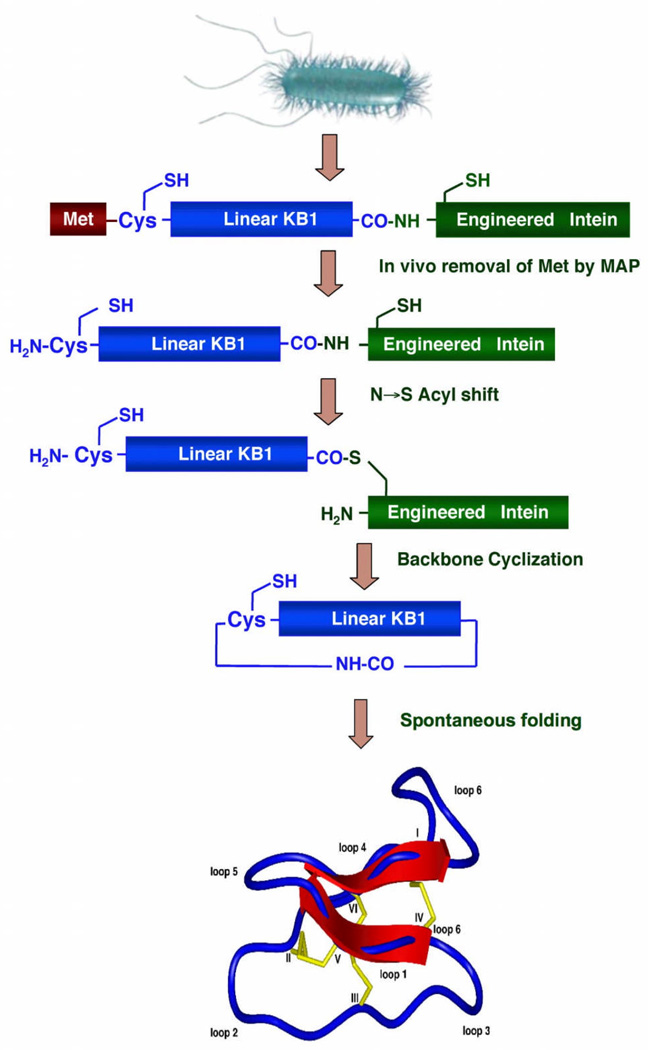
The approach employed for the biosynthesis of backbone cyclized polypeptides using EPL is depicted in Figure 3. The target polypeptide to be cyclized was fused at the N-terminus with a peptide leading sequence immediately followed by a Cys residue, and at the C-terminus with an engineered intein. The N-terminal leading sequence can be cleaved in vitro or in vivo by a proteolytic or self-proteolytic event thereby generating the required N-terminal Cys residue. This Cys residue then reacts in an intramolecular fashion with the α-thioester generated by the engineered intein at the C-terminus thus providing a recombinantly generated backbone cyclized polypeptide. This approach had been used for the in vitro and in vivo biosynthesis of different backbone cyclized polypeptides.
Figure 3.
Biosynthetic approach for the in vivo production of cyclotides inside live E. coli cells. Backbone cyclization of the linear cyclotide precursor is mediated by a modified protein splicing unit or intein. The cyclized product then folds spontaneously in the bacterial cytoplasm.
The demonstration of this biosynthetic cyclization strategy was first reported in vitro by Camarero and Muir in 1999 using the N-terminal SH3 domain of the c-Crk protein as model protein [11]. In this work, the SH3 domain was fused to a modified VMA intein at the C-terminus and to the MIEGRC motif (which contains a Factor Xa proteolysis site) at the N-terminus. After expression in E. coli and purification, the intein fusion protein was treated with Factor Xa protease. This proteolytic step afforded a N-terminal Cys-containing SH3-intein fusion protein, which spontaneously reacted in an intramolecular fashion to yield the corresponding cyclized SH3 domain. The cyclization process was extremely clean and fast, and the resulting cyclic SH3 protein domain was fully active [48]. Intriguingly, this intramolecular process did not require the presence of a thiol cofactor (absolutely necessary to facilitate intermolecular ligation reactions [25]). This interesting result was explained on basis of the close proximity of both reacting groups in the folded state of the SH3 domain (the N- and C-termini of the natively folded SH3 are located within 6Å), which was able to increase the local concentration of both reacting groups. This effect has been already reported in the cyclization of different small protein domains [5, 7].
Iwai and Pluckthum also reported the biosynthesis of a cyclized version of the β-lactamase protein using a similar approach [37]. In their case, the N-terminal Cys residue was generated in vivo by removal of the initiating Met residue by an endogenous Met amino peptidase. After purification of the N-terminal Cys-containing intein fusion protein at pH 8.0, the cyclization was triggered by addition of a thiol cofactor at pH 5.0. The resulting cyclized protein was found to be more stable against irreversible denaturation upon heating than the linear form.
Based on the high efficiency observed during the in vitro cyclization of the SH3 domain [11, 48], Camarero and Muir used a similar approach to test the possibility of carrying out the cyclization of the SH3 domain inside living cells. For this purpose, the Factor Xa recognition leading sequence in the SH3-VMA intein fusion protein was replaced by a Met residue. During the expression of the resulting fusion protein in E. coli cells, the Met residue was efficiently removed by an endogenous Met aminopeptidase [35]. This in vivo proteolytic event unmasked the N-terminal Cys residue which then reacted in an intramolecular fashion with the α-thioester group induced by the C-terminal engineered VMA intein [14]. Analysis by SDS-PAGE showed that most of the SH3-intein fusion protein (>90%) was cleaved in vivo. Remarkably, when the entire soluble cell fraction was analyzed by reverse-phase HPLC, the expected cyclic SH3 protein and the cleaved intein were found to be the major components in the mixture. It is worth noting that no linear SH3 domain was found in the cellular mixture, suggesting that in vivo hydrolysis of the α-thioester linkage present in the precursor protein was minimal. This work demonstrated the first example of a polypeptide chemical ligation reaction performed in the complex cytoplasmic environment of a living cell, and represents an important milestone in current efforts to generate and screen libraries of cyclic polypeptides inside living cells.
More recently, we have applied the same approach for the biosynthesis of cyclotides inside living bacterial cells (Fig. 3) [49]. Cyclotides are small globular microproteins with a unique head-to-tail cyclized backbone, which is stabilized by three disulfide bonds [50]. The number and positions of cysteine residues are conserved throughout the family, forming the cyclic cystine-knot motif (CCK) [50] that acts as a highly stable and versatile scaffold on which hyper-variable loops are arranged. This CCK framework gives the cyclotides exceptional resistance to thermal and chemical denaturation and enzymatic degradation. Moreover, several cyclotides had been found able to cross eukaryotic cell membranes [51]. All these unique properties make them ideal candidates for the development of peptide-based drugs [52]. Our group has recently developed and successfully used a bio-mimetic approach for the biosynthesis of several folded cyclotides inside cells by making use of intramolecular EPL in combination with modified protein splicing units [49, 53] (Fig. 3). Our important finding makes possible the generation of large libraries of cyclotides (≈109) for high throughput cell-based screening and selection of specific sequences able to recognize particular biomolecular targets [49, 53].
5. Biosynthesis of backbone cyclized peptides using protein trans-splicing
An alternative approach to EPL for the cell-based biosynthesis and screening of backbone cyclized polypeptides in vivo is the use of protein trans-splicing (Fig. 4). This approach was first reported by Benkovic and co-workers and makes use of Ssp DnaE split intein. Protein trans-splicing is a naturally occurring post-translational modification similar to protein splicing with the difference that the intein self-processing domain is split in two fragments (called N-intein and C-intein, respectively, Fig. 4A) [54, 55]. These two intein fragments are inactive individually, however, they can bind each other with high specificity under appropriate conditions to form a functional protein splicing domain (Fig. 4A). By rearranging the order of the elements of the intein (i.e. N-intein and C-intein, see Fig. 4) the result of the splicing produces a backbone cyclized polypeptide [12, 56]. This methodology, also denominated SICLOPPS (split intein circular ligation of proteins and peptides) has been used to generate several natural cyclic peptides [16] as well as large genetically-encoded libraries of small cyclic peptides [57].
Figure 4.
Production of backbone cyclized polypeptides using protein trans-splicing. A. Scheme representing the proposed canonical mechanism for protein trans-splicing mediated by a split Cys-intein. B. Cyclization of polypeptides using protein trans-splicing. To facilitate cyclization, the N-intein and C-intein moieties are fused the C- and N-terminus of the polypeptide to be cyclized, respectively.
It should be noted, however, that these systems require the presence of specific amino acid residues at both intein-extein junctions for efficient protein splicing to occur [12, 16, 58]. In contrast to protein trans-splicing, the only absolute sequence requirements for native chemical ligation is the presence of a N-terminal cysteine. Model studies had also shown that all 20 natural amino acids located at the C-terminus of a polypeptide α-thioester can support ligation [59]. It should be noted, however, the speed rate of the ligation reaction depends on the nature of the amino acid at the C-terminal of the thioester, being Gly the one that reacts faster, while β-branched amino acids react more slowly and in lower yields [60]. Furthermore, the engineered inteins commonly used to generate recombinant polypeptide α-thioesters had been also found to be compatible with most amino acids upstream of the cleavage site [17]. Thus, the EPL approach has been found to be quite general with respect to the sequence of the linear peptide precursor and therefore can generate a more diverse array of backbone cyclized polypeptides in vivo.
6. Protease catalyzed protein splicing for the biosynthesis of backbone cyclized peptides
Protease catalyzed protein splicing, also known as transpeptidation, is employed in prokaryotes to attach proteins to peptidoglycan in the cell-wall envelope [61]. For example, sortases are transpeptidase enzymes found in most Gram-positive bacteria that are specialized in this task. Among several isomorphs and homologues discovered so far, the Staphyloccocus aureus Sortase A (SrtA) [62] had been widely employed for protein engineering [63, 64]. SrtA recognizes substrates that contain an LPXTG sequence and catalyzes the cleavage of the amide bond between the Thr and Gly by means of an active-site cysteine (Cys184) residue (Fig. 5A). This process generates a covalent acyl-enzyme intermediate. The activated carboxyl group of the Thr residue then undergoes nucleophilic attack by an amino group of oligoglycine substrates (in S. aureus, a pentaglycine Gly5 cross-bridge on branch lipid II precursor) producing ligated products. In the absence of oligoglycine nucelophiles, the acyl-intermediate is hydrolyzed by a water molecule.
Figure 5.
Biosysnthesis of cyclic polypeptides using intramolecular sortase-mediated ligation. A. Principle of sortase-mediated ligation. Sortase A first recognizes an LPXTG sequence within polypeptide 1 and cleaves the amide bond between the Thr and the Gly with an active-site Cys184, generating a covalent acyl-enzyme intermediate. The thioester intermediate is then attacked by an amino group of the oligo Gly-containing polypeptide 2, which allows the ligation of the two polypeptides by a native peptide bond. B. Polypeptide cyclization of a dual tagged polypeptide by an intra-molecular transpeptidation reaction.
An intramolecular version of the sortase-mediated ligation (SML) can be used for the generation of backbone cyclized polypeptides (Fig. 5B). Bouder and co-workers reported the SrtA-mediated cyclization of EGFP containing the (Gly)3 and LPTEG peptides at the N- and C-terminus, respectively [65]. SML-based protein cyclization had been also reported by other groups for the cyclization of dyhydrolate reductase (DFHR) and pleckstrin homology (PH) domain [64].
Protease-catalyzed protein splicing has also recently found in animals and plants [66]. For example, recent studies have shown that the biosynthesis of cyclotides in plants (see above) involves an asparaginyl endopeptidase (AEP) catalyzed transpeptidation event [67]. Cyclotides are naturally processed from precursor proteins that have both N- and C-terminal pro-regions. Mutation of a highly conserved Asn residue at the C-terminus of the cyclotide domain eliminates cyclization, as does deletion of the C-terminal Pro-region following this residue [67]. Together, these findings indicate that an endopeptidase with specificity for asparagine residues is likely to be involved in the cyclization process [68]. The protease that cleaves the N-terminal pro-region, however, has yet to be identified. The identification of all the proteins required for the cyclization of cyclotides could provide an alternative method to intramolecular EPL for the biosynthesis of genetically-encoded libraries of cyclotides inside living cells. However, it should be emphasized the simplicity of the EPL-based biomimetic approach, which only uses one single self-processing protein that can easily expressed in any type of cell for rapid screening of genetically-encoded libraries.
7. Cell-based screening of genetically-encoded libraries of backbone cyclic polypeptides
The ability to create cyclic polypeptides in vivo opens up the possibility of generating large libraries of cyclic polypeptides. Using the tools of molecular biology, genetically encoded libraries of cyclic polypeptides containing billions of members can be readily generated. This tremendous molecular diversity forms the basis for selection strategies that model natural evolutionary processes. Also, since the cyclic polypeptides are generated inside living cells, these libraries can be directly screened for their ability to attenuate or inhibit cellular processes. In contrast to phage display, where the screening takes place in vitro, screening that takes place in the cytoplasm offers the advantages conferred by a native physiological environment where diverse biochemical events may be examined. In addition, problems resulting from the presence of a fusion tag (in this case the viral particle), in a phenomenon known as template effect, may be circumvented.
Backbone cyclized polypeptides are relatively more stable and more resistant to cellular catabolism than linear polypeptides or disulfide-based cyclic polypeptides. Naturally occurring cyclic peptides are often associated with diverse therapeutic activities ranging from immunosuppression to antimicrobial activity. The stability of backbone cyclized polypeptides displaying certain pharmacological properties suggests that they may be suitable scaffolds on which to graft the molecular diversity of an intracellular library [52, 58].
A number of advances in vivo library generation and screening have recently been reported. Scott and Benkovic used protein trans-splicing to generate the cyclic peptide Pseudostellarin F, an eight amino acid circular peptide with tyrosinase inhibitory activity [12]. The in vivo biosynthesized Pseudostellarin F was fully active and successfully screened in vivo for its tyrosinase inhibitory activity. More recently, cyclic peptide libraries based on the Pseudostellarin F scaffold demonstrated the structural requirements for this system. Apparently, several amino acids positions near the intein-extein junction were critical for expression and cyclization and the authors estimated that 70% of their library produced cyclic products.
Payan and coworkers also used a similar protein trans-splicing approach to generate random cyclic peptides in the cytoplasm of human cells using a retroviral expression vector [69]. Screening of the library for modulation of the IL-4 signaling pathway, led to the identification of several cyclic peptides that selectively inhibited the ε promoter activity. The library was based upon a five amino acid coding strategy, and the potential complexity of the library was about 160,000 members at the amino acids level. Of the 565 clones tested, twenty-three hits were identified as potential therapeutic agents against allergy and asthma, and they may serve in the future as leads for the development of more potent compounds. These results demonstrated for the first time an efficient functional screen for cyclic peptides in vivo in mammalian cells.
More recently, Cheng et al [70] have also used protein trans-splicing to select backbone cyclized peptides able to inhibit the bacterial ClpXP protease from genetically encoded of octapeptides, where only five residues were randomized. Screening of potential inhibitors was performed in E. coli cells using fluorescence activated cell sorting in combination with a genetically fluorescent reporter. The selected inhibitors had little shared sequence similarity and were able to interfere with unexpected steps in the ClpXP mechanism in vitro. One of the selected cyclic peptides showed antibacterial activity against Caulobacter crescentus, a model organism that requires ClpXP activity for viability.
Tavassoli et al [71] have also recently used protein trans-splicing for the screening and selection of genetically-encoded libraries of cyclized peptides able to interfere with the Gag-TSG101 interaction, which is required for the release of HIV particles from virus-infected cells. In this case the peptide library (composed of ≈ 106 different octamers) was screened in E. coli cells using a reverse bacterial two-hybrid system (RTHS) [72, 73]. The bacterial RTHS system used a combination of chimeric repressor fusion proteins and promoter sequences to link the disruption of targeted fusion protein heterodimers to the expression of several reported genes that allowed the selection of peptide inhibitors. This approach yielded several cyclic octapeptides that when linked to the cell penetrating peptide (CPP) TAT were able to inhibit the production of virus-like particles in human cells with a reported IC50 of 7 µM.
8. Conclusions and Remarks
The ability to biosynthesize backbone cyclic peptides using EPL, protein trans-splicing or SPL has important implications for drug-development efforts. The capability to screen for biochemical events in an environment as complex as the cell’s interior will result in valuable and unique information about potential leads identified by this method. Indeed, peptide-based libraries have been already shown to be effective in producing drug candidates in bacterial as well as mammalian systems [12, 69–71].
In summary, we have reviewed several approaches as well as recent developments on the use of different types of protein splicing for the biosynthesis of circular polypeptides. Protein trans-splicing has revealed itself as a powerful tool for the biosynthesis of circular polypeptides that include small peptides and large proteins [12, 15, 16, 20, 57]. It also has been shown that this approach can be used for the generation of large libraries of circular polypeptide inside living cells where they can be directly screened for biological activity [16, 69–71]. However, it has been shown that specific residues of the native N-extein and C-extein are required for efficient protein trans-splicing to occur with the naturally split DnaE intein [20]. It is conceivable, therefore, that this could restrict or bias the sequence diversity in the corresponding circular peptide libraries generated by this method. Similar sequence requirements are also found in Sortase-mediated intramolecular ligations, where 9 extra residues are required for the Sortase-catalyzed cyclization to take place.
Intramolecular EPL, on the other hand, had also been used successfully for the in vitro and in vivo biosynthesis of cyclic polypeptides [11, 13, 14, 37, 48, 49, 53]. In contrast with the protein trans-splicing approach, EPL is compatible with most of the amino acids at the cyclization site [17, 59], making this approach general with respect of the sequence of the linear polypeptide sequence.
Cyclic peptides represent an exciting new tool for deciphering complex biological processes. As the genomics era unfolds, there will be a continuing demand for innovative strategies to address difficult biological questions, and cyclic peptides offer biologists access to highly diverse molecular libraries for genetic experiments, approaches previously restricted to synthetic chemists. Moreover, methods are being developed for the application of cyclic peptides in a variety of experimental endeavors, providing a toolbox that should benefit evolving genomics-based approaches. In the coming years, scientists will continue unraveling the overwhelming amount of genomic data encoding complex biochemical pathways and protein networks, and cyclic peptides are an attractive complementary tool that can aid this challenging task.
Figure 6.
Shematic representation of the putative mechanism of protease-catalysed cyclotide cyclization. Cyclotides are a large family of plant defence proteins characterized by a cystine knot and cyclic backbone. Their prototypic linear precursor protein (top) comprises an endoplasmic reticulum (ER)-targeting sequence (dark blue), a pro-region (purple), an N-terminal repeat region (Ntr; green), a cyclotide domain (light grey) and a C-terminal tail (red). It features also a conserved asparagine (yellow) at the C-terminal cleavage point of the cyclotide domain. The precursor is processed in the ER and vacuole: disulfide bonds are formed (yellow) and a range of unidentified proteases (brown) trim the precursor. In the final stage the active-site cysteine of an AEP (yellow) displaces the C-terminal tail to form an enzyme-acyl intermediate (boxed). This intermediate is then attacked by the cyclotide N-terminal glycine to form the mature cyclic peptide. Figure adapted from reference [60].
Acknowledgments
JAC and HS are supported by funding from the School of Pharmacy at the University of Southern California.
References
- 1.Hruby VJ, Al-Obeidi F. Emerging approaches in the molecular design of receptor-selective peptide ligands: conformational, topographical and dynamic considerations. J. Biochem. 1990;268:249–262. doi: 10.1042/bj2680249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizo J, Gierasch LM. Constrained peptides: models of bioactive peptides and protein substructures. Ann. Rev. Biochem. 1992;61:387–418. doi: 10.1146/annurev.bi.61.070192.002131. [DOI] [PubMed] [Google Scholar]
- 3.Camarero JA, Muir TW. Chemoselective backbone cyclization of unprotected peptides. J. Chem. Soc., Chem. Comm. 1997;1997:1369–1370. [Google Scholar]
- 4.Zhang L, Tam JP. Synthesis and application of unprotected cyclic peptides as building blocks for peptide dendrimers. J. Am. Chem. Soc. 1997;119:2363–2370. [Google Scholar]
- 5.Camarero JA, Pavel J, Muir TW. Chemical Synthesis of a Circular Protein Domain: Evidence for Folding-Assisted Cyclization. Angew. Chem. Int. Ed. 1998;37:347–349. doi: 10.1002/(SICI)1521-3773(19980216)37:3<347::AID-ANIE347>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 6.Shao Y, Lu WY, Kent SBH. A novel method to synthesize cyclic peptides. Tetrahedron Lett. 1998;39:3911–3914. [Google Scholar]
- 7.Camarero JA, Cotton GJ, Adeva A, Muir TW. Chemical ligation of unprotected peptides directly form a solid support. J. Pept. Res. 1998;51:303–316. doi: 10.1111/j.1399-3011.1998.tb00428.x. [DOI] [PubMed] [Google Scholar]
- 8.Trauger JW, Kohli RM, Mootz HD, Marahiel MA, Walsh CT. Peptide cyclization catalysed by the thioesterase domain of tyrocidine synthetase. Nature. 2000;407:215–218. doi: 10.1038/35025116. [DOI] [PubMed] [Google Scholar]
- 9.Kohli RM, Walsh CT, Burkart MD. Biomimetic synthesis and optimization of cyclic peptide antibiotics. Nature. 2002;418:658–661. doi: 10.1038/nature00907. [DOI] [PubMed] [Google Scholar]
- 10.Walsh CT. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science. 2004;303:1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 11.Camarero JA, Muir TW. Biosynthesis of a Head-to-Tail Cyclized Protein with Improved Biological Activity. J. Am. Chem. Soc. 1999;121:5597–5598. [Google Scholar]
- 12.Scott CP, Abel-Santos E, Wall M, Wahnon D, Benkovic SJ. Production of cyclic peptides and proteins in vivo. Proc. Natl. Acad. Sci. USA. 1999;96:13638–13643. doi: 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans TC, Benner J, Xu M-Q. The cyclization and polymerization of bacterially expressed proteins using modified sef-splicing inteins. J. Biol. Chem. 1999;274:18359–18381. doi: 10.1074/jbc.274.26.18359. [DOI] [PubMed] [Google Scholar]
- 14.Camarero JA, Fushman D, Cowburn D, Muir TW. Peptide chemical ligation inside living cells: in vivo generation of a circular protein domain. Bioorg Med Chem. 2001;9:2479–2484. doi: 10.1016/s0968-0896(01)00217-6. [DOI] [PubMed] [Google Scholar]
- 15.Iwai H, Lingel A, Pluckthun A. Cyclic green fluorescent protein produced in vivo using an artificially split PI-PfuI intein from Pyrococcus furiosus. J. Biol. Chem. 2001;276:16548–16554. doi: 10.1074/jbc.M011639200. [DOI] [PubMed] [Google Scholar]
- 16.Abel-Santos E, Scott CP, Benkovic SJ. Use of inteins for the in vivo production of stable cyclic peptide libraries in E. coli. Methods Mol. Biol. 2003;205:281–294. doi: 10.1385/1-59259-301-1:281. [DOI] [PubMed] [Google Scholar]
- 17.Noren CJ, Wang JM, Perler FB. Dissecting the chemistry of protein splicing and its applications. Angew. Chem. Int. Ed. 2000;39:451–456. [PubMed] [Google Scholar]
- 18.Suter B, Auerbach D, Stagljar I. Yeast-based functional genomics and proteomics technologies: the first 15 years and beyond. Biotechniques. 2006;40:625–644. doi: 10.2144/000112151. [DOI] [PubMed] [Google Scholar]
- 19.Wu H, Hu Z, Liu XQ. Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. USA. 1998;95:9226–9231. doi: 10.1073/pnas.95.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans TC, Jr, Martin D, Kolly R, Panne D, Sun L, Ghosh I, Chen L, Benner J, Liu XQ, Xu MQ. Protein trans-splicing and cyclization by a naturally split intein from the dnaE gene of Synechocystis species PCC6803. J. Biol. Chem. 2000;275:9091–9094. doi: 10.1074/jbc.275.13.9091. [DOI] [PubMed] [Google Scholar]
- 21.Dawson PE, Muir TW, Clark-Lewis I, Kent SBH. Synthesis of Proteins by Native Chemical Ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 22.Dawson PE, Kent SB. Synthesis of native proteins by chemical ligation. Annu. Rev. Biochem. 2000;69:923–960. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 23.Tam JP, Lu YA, Liu CF, Shao J. Peptide synthesis using unprotected peptides through orthogonal coupling methods. Proc. Natl. Acad. Sci. U S A. 1995;92:12485–12489. doi: 10.1073/pnas.92.26.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans TC, Jr, Xu MQ. Intein-mediated protein ligation: harnessing nature's escape artists. Biopolymers. 1999;51:333–342. doi: 10.1002/(SICI)1097-0282(1999)51:5<333::AID-BIP3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 25.Camarero JA, Muir TW. Native Chemical Ligation of Polypeptides. Current Protocols in Protein Science. 1999:1–21. doi: 10.1002/0471140864.ps1804s15. [DOI] [PubMed] [Google Scholar]
- 26.Muir TW. Semisynthesis of proteins by expressed protein ligation. Annu. Rev. Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 27.Wieland T, Bokelmann E, Bauer L, Lang HU, Lau H. Polypeptide synthesis. VIII. Formation of sulfur containing peptides by intramolecular migration of aminoacyl groups. Liebigs Ann. Chem. 1953;583:129. [Google Scholar]
- 28.Wieland T. Sulfur in Biomimetic Peptide Synthesis. In: Kleinkauf vD, Jaeniche, editors. The Roots of Modern Biochemistry. Berlin, New York: Walter de Gruyter & Co.; 1988. pp. 213–221. [Google Scholar]
- 29.Muir TW, Sondhi D, Cole PA. Expressed protein ligation: a general method for protein engineering. Proc. Natl. Acad. Sci. U S A. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Severinov K, Muir TW. Expressed protein ligation, a novel method for studying protein-protein interactions in transcription. J. Biol. Chem. 1998;273:16205–16209. doi: 10.1074/jbc.273.26.16205. [DOI] [PubMed] [Google Scholar]
- 31.Evans TC, Benner J, Xu M-Q. Semisynthesis of cytotoxic proteins using a modified protein splicing element. Protein Sci. 1998;7:2256–2264. doi: 10.1002/pro.5560071103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu M-Q, Perler FB. The mechanism of protein splicing and its modulation by mutation. EMBO J. 1996;15:5146–5153. [PMC free article] [PubMed] [Google Scholar]
- 33.Chong S, Mersha FB, Comb DG, Scott ME, Landry D, Vence LM, Perler FB, Benner J, Kucera RB, Hirvonen CA, Pelletier JJ, Paulus H, Xu MQ. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 34.Chong S, Montenello GE, Zhang A, Cantor EJ, Liao W, Xu M-Q, Benner J. Utilizing the C-terminal cleavage activity of a protein splicing element to purify recombinant proteins in a single chromatographic step. Nucleic Acid Res. 1998;26:5109–5115. doi: 10.1093/nar/26.22.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirel PH, Schmitter MJ, Dessen P, Fayat G, Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. U S A. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dwyer MA, Lu W, Dwyer JJ, Kossiakoff AA. Biosynthetic phage display: a novel protein engineering tool combining chemical and genetic diversity. Chem. Biol. 2000;7:263–274. doi: 10.1016/s1074-5521(00)00102-2. [DOI] [PubMed] [Google Scholar]
- 37.Iwai H, Pluckthum A. Circular b-lactamase: stability enhancement by cyclizing the backbone. FEBS Lett. 1999:166–172. doi: 10.1016/s0014-5793(99)01220-x. [DOI] [PubMed] [Google Scholar]
- 38.Cotton GJ, Ayers B, Xu R, Muir TW. Insertion of a Synthetic Peptide into a Recombinant Protein Framework; A Protein Biosensor. J. Am. Chem. Soc. 1999;121:1100–1101. [Google Scholar]
- 39.Erlandson DA, Chytil M, Verdine GL. The leucine zipper domain controls the orientation of AP-1 in the NFAT·AP-1·DNA complex. Chem. Biol. 1996;3:981–991. doi: 10.1016/s1074-5521(96)90165-9. [DOI] [PubMed] [Google Scholar]
- 40.Tolbert TJ, Wong C-H. New methods for proteomic research: preparation of proteins with N-terminal cysteines for labeling and conjugation. Angew. Chem. Int. Ed. Engl. 2002;41:2171–2174. doi: 10.1002/1521-3773(20020617)41:12<2171::aid-anie2171>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 41.Evans TC, Benner J, Xu M-Q. The in Vitro Ligation of Bacterially Expressed Proteins Using an Intein from Metanobacterium thermoautotrophicum . J. Biol. Chem. 1999;274:3923–3926. doi: 10.1074/jbc.274.7.3923. [DOI] [PubMed] [Google Scholar]
- 42.Southworth MW, Amaya K, Evans TC, Xu MQ, Perler FB. Purification of proteins fused to either the amino or carboxy terminus of the Mycobacterium xenopi gyrase A intein. Biotechniques. 1999;27:110–114. doi: 10.2144/99271st04. 116, 118-120. [DOI] [PubMed] [Google Scholar]
- 43.Mathys S, Evans TC, Chute IC, Wu H, Chong S, Benner J, Liu XQ, Xu MQ. Characterization of a self-splicing mini-intein and its conversion into autocatalytic N- and C-terminal cleavage elements: facile production of protein building blocks for protein ligation. Gene. 1999;231:1–13. doi: 10.1016/s0378-1119(99)00103-1. [DOI] [PubMed] [Google Scholar]
- 44.Liu D, Xu R, Dutta K, Cowburn D. N-terminal cysteinyl proteins can be prepared using thrombin cleavage. FEBS Lett. 2008;582:1163–1167. doi: 10.1016/j.febslet.2008.02.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dalbey RE, Lively MO, Bron S, van Dijl JM. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 1997;6:1129–1138. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paetzel M, Dalbey RE, Strynadka NC. Crystal structure of a bacterial signal peptidase apoenzyme: implications for signal peptide binding and the Ser-Lys dyad mechanism. J Biol Chem. 2002;277:9512–9519. doi: 10.1074/jbc.M110983200. [DOI] [PubMed] [Google Scholar]
- 47.Hauser PS, Ryan RO. Expressed protein ligation using an N-terminal cysteine containing fragment generated in vivo from a pelB fusion protein. Protein Expr Purif. 2007;54:227–233. doi: 10.1016/j.pep.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Camarero JA, Fushman D, Sato S, Giriat I, Cowburn D, Raleigh DP, Muir TW. Rescuing a destabilized protein fold through backbone cyclization. J. Mol. Biol. 2001;308:1045–1062. doi: 10.1006/jmbi.2001.4631. [DOI] [PubMed] [Google Scholar]
- 49.Camarero JA, Kimura RH, Woo YH, Shekhtman A, Cantor J. Biosynthesis of a fully functional cyclotide inside living bacterial cells. Chembiochem. 2007;8:1363–1366. doi: 10.1002/cbic.200700183. [DOI] [PubMed] [Google Scholar]
- 50.Craik DJ, Daly NL, Bond T, Waine C. Plant cyclotides: A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J Mol Biol. 1999;294:1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- 51.Greenwood KP, Daly NL, Brown DL, Stow JL, Craik DJ. The cyclic cystine knot miniprotein MCoTI-II is internalized into cells by macropinocytosis. Int J Biochem Cell Biol. 2007;39:2252–2264. doi: 10.1016/j.biocel.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 52.Craik DJ, Simonsen S, Daly NL. The cyclotides: novel macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discov. Devel. 2002;5:251–260. [PubMed] [Google Scholar]
- 53.Kimura RH, Tran AT, Camarero JA. Biosynthesis of the cyclotide Kalata B1 by using protein splicing. Angew Chem Int Ed Engl. 2006;45:973–976. doi: 10.1002/anie.200503882. [DOI] [PubMed] [Google Scholar]
- 54.Saleh L, Perler FB. Protein splicing in cis and in trans. Chem Rec. 2006;6:183–193. doi: 10.1002/tcr.20082. [DOI] [PubMed] [Google Scholar]
- 55.Xu MQ, Evans TC., Jr Recent advances in protein splicing: manipulating proteins in vitro and in vivo. Curr Opin Biotechnol. 2005;16:440–446. doi: 10.1016/j.copbio.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 56.Naumann TA, Savinov SN, Benkovic SJ. Engineering an affinity tag for genetically encoded cyclic peptides. Biotechnol Bioeng. 2005;92:820–830. doi: 10.1002/bit.20644. [DOI] [PubMed] [Google Scholar]
- 57.Tavassoli A, Benkovic SJ. Split-intein mediated circular ligation used in the synthesis of cyclic peptide libraries in E. coli. Nat Protoc. 2007;2:1126–1133. doi: 10.1038/nprot.2007.152. [DOI] [PubMed] [Google Scholar]
- 58.Scott CP, Abel-Santos E, Jones AD, Benkovic SJ. Structural requirements for the biosynthesis of backbone cyclic peptide libraries. Chem. Biol. 2001;8:801–815. doi: 10.1016/s1074-5521(01)00052-7. [DOI] [PubMed] [Google Scholar]
- 59.Hackeng TM, Griffin JH, Dawson PE. Protein synthesis by native chemical ligation: expanded scope by using straightforward methodology. Proc. Natl. Acad. Sci. U S A. 1999;96:10063–10078. doi: 10.1073/pnas.96.18.10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hackeng TM, Griffin JH, Dawson PE. Protein synthesis by native chemical ligation: Expanded scope by using straightforward methodology. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:10068–10073. doi: 10.1073/pnas.96.18.10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 63.Mao H, Hart SA, Schink A, Pollok BA. Sortase-mediated protein ligation: a new method for protein engineering. J Am Chem Soc. 2004;126:2670–2671. doi: 10.1021/ja039915e. [DOI] [PubMed] [Google Scholar]
- 64.Tsukiji S, Nagamune T. Sortase-mediated ligation: a gift from Gram-positive bacteria to protein engineering. Chembiochem. 2009;10:787–798. doi: 10.1002/cbic.200800724. [DOI] [PubMed] [Google Scholar]
- 65.Parthasarathy R, Subramanian S, Boder ET. Sortase A as a novel molecular "stapler" for sequence-specific protein conjugation. Bioconjug Chem. 2007;18:469–476. doi: 10.1021/bc060339w. [DOI] [PubMed] [Google Scholar]
- 66.Saska I, Craik DJ. Protease-catalysed protein splicing: a new post-translational modification? Trends Biochem Sci. 2008;33:363–368. doi: 10.1016/j.tibs.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 67.Gillon AD, Saska I, Jennings CV, Guarino RF, Craik DJ, Anderson MA. Biosynthesis of circular proteins in plants. Plant J. 2008;53:505–515. doi: 10.1111/j.1365-313X.2007.03357.x. [DOI] [PubMed] [Google Scholar]
- 68.Saska I, Gillon AD, Hatsugai N, Dietzgen RG, Hara-Nishimura I, Anderson MA, Craik DJ. An asparaginyl endopeptidase mediates in vivo protein backbone cyclization. J Biol Chem. 2007;282:29721–29728. doi: 10.1074/jbc.M705185200. [DOI] [PubMed] [Google Scholar]
- 69.Kinsella TM, Ohashi CT, Harder AG, Yam GC, Li W, Peelle B, Pali ES, Bennett MK, Molineaux SM, Anderson DA, Masuda ES, Payan DG. Retrovirally delivered random cyclic Peptide libraries yield inhibitors of interleukin-4 signaling in human B cells. J. Biol. Chem. 2002;277:37512–37518. doi: 10.1074/jbc.M206162200. [DOI] [PubMed] [Google Scholar]
- 70.Cheng L, Naumann TA, Horswill AR, Hong SJ, Venters BJ, Tomsho JW, Benkovic SJ, Keiler KC. Discovery of antibacterial cyclic peptides that inhibit the ClpXP protease. Protein Sci. 2007;16:1535–1542. doi: 10.1110/ps.072933007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tavassoli A, Lu Q, Gam J, Pan H, Benkovic SJ, Cohen SN. Inhibition of HIV budding by a genetically selected cyclic peptide targeting the Gag-TSG101 interaction. ACS chemical biology. 2008;3:757–764. doi: 10.1021/cb800193n. [DOI] [PubMed] [Google Scholar]
- 72.Tavassoli A, Benkovic SJ. Genetically selected cyclic-peptide inhibitors of AICAR transformylase homodimerization. Angew Chem Int Ed Engl. 2005;44:2760–2763. doi: 10.1002/anie.200500417. [DOI] [PubMed] [Google Scholar]
- 73.Horswill AR, Savinov SN, Benkovic SJ. A systematic method for identifying small-molecule modulators of protein-protein interactions. Proc Natl Acad Sci U S A. 2004;101:15591–15596. doi: 10.1073/pnas.0406999101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hernandez JF, Gagnon J, Chiche L, Nguyen TM, Andrieu JP, Heitz A, Trinh Hong T, Pham TT, Le Nguyen D. Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry. 2000;39:5722–5730. doi: 10.1021/bi9929756. [DOI] [PubMed] [Google Scholar]
- 75.Felizmenio-Quimio ME, Daly NL, Craik DJ. Circular proteins in plants: solution structure of a novel macrocyclic trypsin inhibitor from Momordica cochinchinensis. J Biol Chem. 2001;276:22875–22882. doi: 10.1074/jbc.M101666200. [DOI] [PubMed] [Google Scholar]
- 76.Luckett S, Garcia RS, Barker JJ, Konarev AV, Shewry PR, Clarke AR, Brady RL. High-resolution structure of a potent, cyclic proteinase inhibitor from sunflower seeds. J Mol Biol. 1999;290:525–533. doi: 10.1006/jmbi.1999.2891. [DOI] [PubMed] [Google Scholar]
- 77.Korsinczky ML, Schirra HJ, Rosengren KJ, West J, Condie BA, Otvos L, Anderson MA, Craik DJ. Solution structures by 1H NMR of the novel cyclic trypsin inhibitor SFTI-1 from sunflower seeds and an acyclic permutant. J Mol Biol. 2001;311:579–591. doi: 10.1006/jmbi.2001.4887. [DOI] [PubMed] [Google Scholar]
- 78.Wang W, Mulakala C, Ward SC, Jung G, Luong H, Pham D, Waring AJ, Kaznessis Y, Lu W, Bradley KA, Lehrer RI. Retrocyclins kill bacilli and germinating spores of Bacillus anthracis and inactivate anthrax lethal toxin. J Biol Chem. 2006;281:32755–32764. doi: 10.1074/jbc.M603614200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gallo SA, Wang W, Rawat SS, Jung G, Waring AJ, Cole AM, Lu H, Yan X, Daly NL, Craik DJ, Jiang S, Lehrer RI, Blumenthal R. Theta-defensins prevent HIV-1 Env-mediated fusion by binding gp41 and blocking 6-helix bundle formation. J Biol Chem. 2006;281:18787–18792. doi: 10.1074/jbc.M602422200. [DOI] [PubMed] [Google Scholar]