Abstract
Aim: Functional in vivo studies on the mitochondrial thioredoxin system are hampered by the embryonic or larval lethal phenotypes displayed by murine or Drosophila knock-out models. Thus, the access to alternative metazoan knock-out models for the mitochondrial thioredoxin system is of critical importance. Results: We report here the characterization of the mitochondrial thioredoxin system of Caenorhabditis elegans that is composed of the genes trx-2 and trxr-2. We demonstrate that the proteins thioredoxin 2 (TRX-2) and thioredoxin reductase 2 (TRXR-2) localize to the mitochondria of several cells and tissues of the nematode and that trx-2 and trxr-2 are upregulated upon induction of the mitochondrial unfolded protein response. Surprisingly, C. elegans trx-2 (lof ) and trxr-2 (null) single and double mutants are viable and display similar growth rates as wild-type controls. Moreover, the lack of the mitochondrial thioredoxin system does not affect longevity, reactive oxygen species production or the apoptotic program. Interestingly, we found a protective role of TRXR-2 in a transgenic nematode model of Alzheimer's disease (AD) that expresses human β-amyloid peptide and causes an age-dependent progressive paralysis. Hence, trxr-2 downregulation enhanced the paralysis phenotype, while a strong decrease of β-amyloid peptide and amyloid deposits occurred when TRXR-2 was overexpressed. Innovation: C. elegans provides the first viable metazoan knock-out model for the mitochondrial thioredoxin system and identifies a novel role of this system in β-amyloid peptide toxicity and AD. Conclusion: The nematode strains characterized in this work make C. elegans an ideal model organism to study the pathophysiology of the mitochondrial thioredoxin system at the level of a complete organism. Antioxid. Redox Signal. 16, 1384–1400.
Introduction
Aerobic metabolism has the inescapable consequence of generating oxygen intermediates, collectively called reactive oxygen species (ROS), which were initially regarded as detrimental by-products of metabolism because uncontrolled ROS can be extremely harmful to all cellular components. Increasing evidence, however, demonstrates that ROS play a key role in many redox signaling pathways relevant for both healthy and pathological physiology (8). These two apparently contradictory aspects of ROS function have recently been reconciled in a new definition of oxidative stress as “an imbalance between oxidants and antioxidants in favor of the oxidants, leading to a disruption of redox signalling and control and/or molecular damage” (49). Hence, maintenance of redox homeostasis is critical for the survival of all organisms and this safeguarding function is largely accomplished by the thioredoxin and glutaredoxin systems, which are the two major redox regulatory enzymatic systems in most organisms (35).
Innovation.
The in vivo study of the metazoan mitochondrial thioredoxin system function(s) is severely hampered by the lack of an appropriate model at the level of a complete organism. This critical limitation is illustrated by both the murine and Drosophila knock-out models that display embryonic or larval lethal phenotypes, respectively. The present work describes the functional characterization of the mitochondrial thioredoxin system of Caenorhabditis elegans. Our findings show that this nematode model is the first reported metazoan in which the absence of the mitochondrial thioredoxin system does not cause any lethal phenotype. Thus, the availability of a viable metazoan model lacking a functional mitochondrial system combined with the genetic amenability of C. elegans provide an ideal scenario to further investigate the in vivo functions of this system as well as to identify other redundant enzymatic systems functionally overlapping with the mitochondrial thioredoxin system.
Mitochondrial dysfunction and alterations in redox homeostasis have been unequivocally linked to the progression of many neurodegenerative diseases. Using Caenorhabditis elegans, we have uncovered a protective role of the mitochondrial thioredoxin reductase 2 against the paralysis phenotype caused by the aggregation of the β-amyloid peptide in the muscle cells of the nematode, a well-established invertebrate model of Alzheimer's disease (AD). This unexpected finding paves the way to use the strains generated in this work in pharmacological screens and other studies of clinical significance aimed at evaluating the role of mitochondria, and more specifically, that of the mitochondrial thioredoxin system, in the pathophysiology of AD.
Being an important source of ROS (4, 28), mitochondria are well protected against the deleterious effects of ROS by a set of antioxidant defenses such as superoxide dismutases, glutathione peroxidases, glutathione transferases, peroxiredoxins and specific mitochondrial thioredoxin and glutaredoxin systems (27). In addition to its key role as antioxidant defense (5, 44, 60), the mitochondrial thioredoxin system has been shown to be an important mediator of many processes where the mitochondrial function is crucial, such as apoptosis (55), angiogenesis (9), transcription factor DNA binding activity (46) or cell growth and differentiation (25, 38), among others. Interestingly, the mitochondrial thioredoxin system has been shown to functionally interact with the mitochondrial glutaredoxin system by directly reducing mitochondrial glutaredoxin (22). Furthermore, both mitochondrial systems display overlapping functions as, for example, in the reduction of the mitochondrial peroxiredoxin (18).
Mitochondrial thioredoxin systems are, in most cases, composed of dedicated thioredoxin reductase and thioredoxin proteins encoded by nuclear genes, different from those that encode the cytoplasmic thioredoxin system (36). An exception to this generality is found in Drosophila melanogaster, where the same gene (trxr-1) encodes both the cytoplasmic and the mitochondrial thioredoxin reductase isoforms by alternative splicing (37). Surprisingly, a mitochondria-specific thioredoxin has not yet been identified in this organism. In yeast, null mutants of the mitochondrial thioredoxin system (composed of trxr-2 and trx-3 genes) are viable but show enhanced sensitivity to oxidative stress (43). In contrast, inactivation of the mouse mitochondrial thioredoxin system trxr-2 and trx-2 genes results in early embryonic lethality (7, 40) and null mutants of the Drosophila trxr-1 gene die at the second instar larval stage (37). These lethal phenotypes in several model organisms entail important difficulties to genetically dissect the pathways in which the eukaryotic mitochondrial thioredoxin system participates.
In this context, we focused our attention on C. elegans, a well-established, genetically tractable model, in order to study the functions of the mitochondrial thioredoxin system in vivo and in the framework of a complete organism. Surprisingly, C. elegans mutants of the mitochondrial thioredoxin system are viable and do not show any apparent phenotype even under stress conditions. However, we have uncovered an unexpected protective role of the mitochondrial thioredoxin reductase trxr-2 gene in a worm model of Alzheimer's disease (AD) that shows an age-dependent progressive paralysis phenotype upon overexpression of the human β-amyloid peptide (31).
Results
C. elegans trx-2 and trxr-2 genes form a functional thioredoxin system
Previous in silico genomic analyses have demonstrated the presence of two different thioredoxin reductases in C. elegans encoded by the trxr-1 and trxr-2 genes, respectively (16). We have recently demonstrated that thioredoxin reductase 1 (TRXR-1) has a cytoplasmic localization (51), while TRXR-2 is assumed to be a mitochondrial protein based on the presence of a putative mitochondrial targeting sequence (MTS) at its N-terminus (Supplementary Fig. S1A; Supplementary Data are available online at www.liebertonline.com/ars). Although C. elegans TRXR-2 has been demonstrated to function as thioredoxin reductase in enzymatic assays (29), its subcellular localization has not yet been addressed experimentally in vivo. To identify potential candidates for the mitochondrial thioredoxin protein, we screened the C. elegans proteome searching for proteins harboring the conserved thioredoxin active sequence WCGPC plus any potential N-terminal sequence with characteristics of an MTS. We found the product of the worm gene B0024.9 as the only one meeting both criteria and accordingly named it trx-2. The putative thioredoxin 2 (TRX-2) MTS is shown in Supplementary Figure S1B. Both C. elegans TRXR-2 and TRX-2 predicted proteins display a high homology with worm and human TRXR-1 and TRX-1 proteins, respectively [(16) and Supplementary Fig. S1C], with the exception that C. elegans TRXR-2 is not a selenocysteine-containing protein.
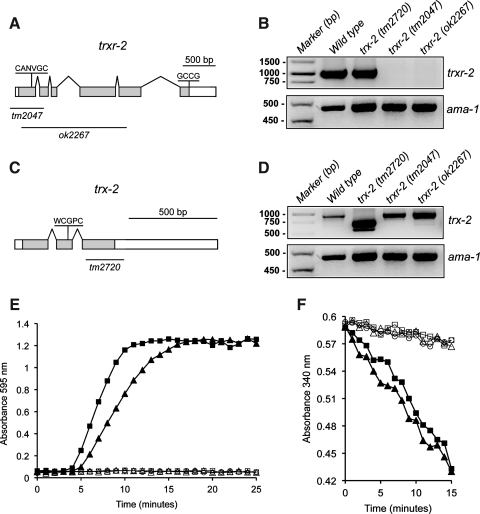
Two deletion alleles, tm2047 and ok2267, are available for the trxr-2 gene, which is composed of 6 exons and spans about 3 kb on linkage group III (www.wormbase.org; Fig. 1A). The tm2047 allele is a 507 bp deletion that removes part of the proximal trxr-2 promoter, the first exon, which contains the ATG codon and the N-terminal redox active site (CANVGC), plus part of the second exon. Likewise, the ok2267 allele is a 1649 bp deletion spanning from exon 1 to 5, thus removing most of the trxr-2 gene (Fig. 1A). Reverse–transcriptase–polymerase chain reaction (RT-PCR) failed to identify any messenger RNA (mRNA) species, demonstrating that both tm2047 and ok2267 are null trxr-2 alleles (Fig. 1B).
FIG. 1.
Genomic organization, cDNA expression of Caenorhabditis elegans trxr-2 and trx-2 genes and enzymatic activity of recombinant TRX-2. Exon-intron structure of the trxr-2 (A) and trx-2 (C) genes where boxes represent exons and lines introns. White boxes indicate 5′- and 3′-UTRs respectively, while gray boxes indicate the ORF. The sequence and position of their respective redox-active sites are shown and the lines underneath denote the regions deleted in the alleles used in this study. RT-PCR expression of trxr-2 (B) and trx-2 (D) genes from N2 wild-type, trx-2 and trxr-2 single mutants. While both trxr-2 (tm2047) and (ok2267) are null alleles (no mRNA produced), two shorter bands, as compared with the one of wild-type control, are produced from the trx-2 (tm2720) allele. The ama-1 gene was used as a housekeeping gene loading control. Recombinant C. elegans histidine-tagged TRX-2 (His-CeTRX-2) and a truncated variant from the conceptual translation of the tm2720 allele (His-CeΔTRX-2) were assayed for their capability of reducing insulin disulfide bonds using DTT (E) or NADPH and rat thioredoxin reductase-1 (F) as electron donor (20). ▪ His-hTRX-1; ▴ His-CeTRX-2; □ His-CeΔTRX-2; ▵ His-CeTRX-2 without DTT (E) or thioredoxin reductase (F); ○, reaction mix only. An increase in absorbance at OD595 measures insulin precipitation on reduction, while a decrease in absorbance at OD340 measures NADPH consumption. Recombinant human histidine-tagged TRX-1 (His-hTRX-1) was used as positive control. UTR, untranslated region; RT-PCR, reverse transcriptase–polymerase chain reaction; ORF, open reading frame; OD, optical density; DTT, 1,4-dithio-d-threitol; NADPH, reduced nicotinamide adenine dinucleotide phosphate.
Similarly, two deletion alleles, tm2720 and ok1526, are also available for the trx-2 gene, which is composed of 3 exons and spans about 1.1 kb on linkage group V (www.wormbase.org; Fig. 1C). The tm2720 allele is a 218 bp deletion that eliminates part of the third exon of the trx-2 gene (Fig. 1C), while the ok1526 deletion completely removes the trx-2 gene plus part of a neighboring gene (www.wormbase.org) and was not further considered for our study. RT-PCR analysis on the tm2720 allele identifies two shorter trx-2 mRNA variants (Fig. 1D) that differ by an additional splicing event in the 3′-UTR of the trx-2 mRNA (data not shown). The conceptual translation of the tm2720 mRNA variant produces a truncated protein (ΔTRX-2) that lacks the C-terminal half of the mature wild-type TRX-2 protein, which contains amino acid residues critical for protein-protein interaction and redox active-site stabilization (11). Consistent with this analysis, recombinant ΔTRX-2 is devoid of enzymatic activity in both the 1,4-dithio-d-threitol (DTT) and reduced nicotinamide adenine dinucleotide phosphate (NADPH)/thioredoxin reductase assay as compared with that of mature full-length TRX-2 (Fig. 1E, F), thus supporting the notion that the tm2720 allele is a loss-of-function allele. The TRX-2 enzymatic activity described here together with that reported by Hondal's group on TRXR-2 (29) suggests that TRXR-2 and TRX-2 constitute a functional thioredoxin system.
TRX-2 and TRXR-2 are mitochondrial proteins
To demonstrate TRX-2 and TRXR-2 subcellular localization in mitochondria as well as to identify the cells and tissues expressing these proteins within the nematode, we generated transgenic animals expressing transcriptional (promoter only) and translational (promoter plus gene) green fluorescent protein (GFP) fusion reporters. Noteworthy, trxr-2 is the third gene in an operon composed of the unc-32, tpk-1 and trxr-2 genes (www.wormbase.org). It has been recently reported that downstream genes within an operon can be additionally transcribed from internal promoters (21). Therefore, to evaluate the possibility that C. elegans trxr-2 could also be transcribed from an internal promoter, we made transcriptional and translational GFP fusions using both the unc-32 promoter and the putative internal trxr-2 promoter (which spans 343 bp from the end of tpk-1 gene to the ATG codon of trxr-2 gene).
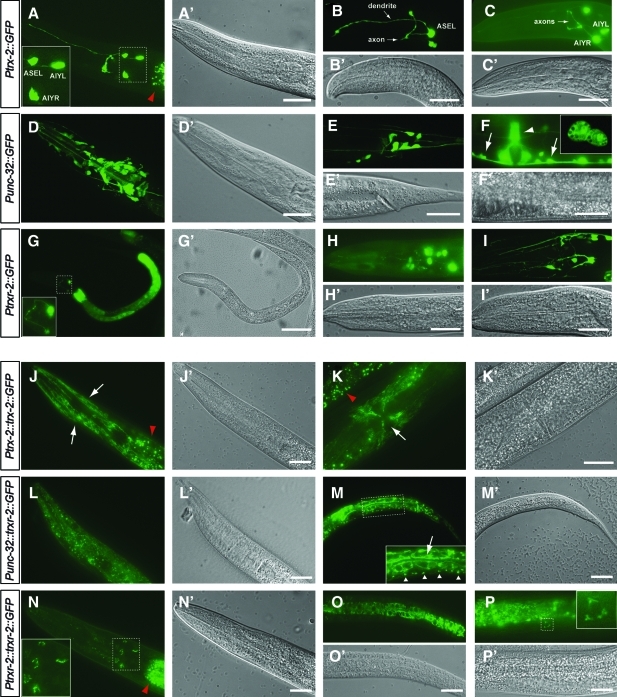
A transcriptional construct Ptrx-2::GFP was expressed only in AIYL/R and ASEL neurons as determined by the position of their cell bodies, morphology of their axons and dendrites and colocalization experiments with specific markers for these neurons (Fig. 2A–C and Supplementary Fig. S2). In some, but not all, transgenic animals, a fourth neuron identified as ASER also displayed fluorescent signals (data not shown). The unc-32 gene, the first gene of the trxr-2 operon, has been previously reported to be expressed in several cells and tissues such as the ventral nerve cord, nerve ring and spermathecal-uterine valve, among others (41). We confirmed most of these expression patterns in transgenic animals harboring a Punc-32::GFP construct and, in addition, we also detected expression in muscle cells, several neurons in the tail, intestinal cells and coelomocytes (Fig. 2D–F). This expression pattern was complemented with that of the trxr-2 internal promoter where a transcriptional construct Ptrxr-2::GFP was found to be expressed in the intestine plus additional neurons in the head (Fig. 2G–I). Interestingly, in L1 animals, only two (unidentified) neurons are marked, but the number of neurons showing fluorescence increases as the animal grows to adulthood (Fig. 2G–I). As a whole, the combined expression pattern of unc-32 and trxr-2 internal promoters suggests a basically ubiquitous expression of TRXR-2, which strongly contrasts the restricted expression for TRX-2 in only three to four head neurons.
FIG. 2.
Expression patterns of trx-2 and trxr-2 GFP reporters. Transgenic worms carrying different expression constructs were visualized with fluorescence (A–P) or DIC (A′-P′) optics. A transcriptional Ptrx-2::GFP fusion shows fluorescence in AIYL/R and ASEL neurons cell bodies and processes (A–C). Transgenic worms expressing the transcriptional fusion Punc-32::GFP displayed fluorescence in many cells in the head area, including several neurons (D), tail neurons (E) ventral nerve cord (F, arrows), vulva muscles (F, arrowhead) and coelomocytes (F, inset). A transcriptional Ptrxr-2::GFP construct was expressed in two neurons in the head area and intestine of L1 animals (G), while in older animals, GFP appears in additional head neurons (H, I). All transgenic worms expressing translational fusions show a fluorescent pattern compatible with mitochondrial localization. Thus, worms expressing the Ptrx-2::trx-2::GFP construct display fluorescence in muscle cell tubular mitochondria mainly in the head area (J, arrows), precluding the identification of the AIYL/R and ASEL neurons and in the vulva muscles mitochondria (K, arrow). Punc-32::trxr-2::GFP translational construct is expressed in mitochondria in many cells of the head (L), reticular mitochondria in the intestine (M, arrow) and mitochondria of ventral nerve cord (M, arrowheads). When expressed from the trxr-2 internal promoter, TRXR-2::GFP is found in a perinuclear location in several head neurons (N), reticular mitochondria in the intestine (O) and, in a few transgenic animals, a single unidentified cell located ventrally to the intestine also expresses GFP in perinuclear mitochondria (P). Red arrowheads indicate the intestinal autofluorescent granules. Insets (except in F) show amplification of the respective dashed areas. Bar 20 μm. (To see this illustration in color, the reader is referred to the web version of this article at www.liebertonline.com/ars). GFP, green fluorescent protein; DIC, differential interference contrast; TRXR, thioredoxin reductase.
Transgenic animals expressing translational Ptrx-2::trx-2::GFP, Punc-32::trxr-2::GFP and Ptrxr-2::trxr-2::GFP constructs all showed an expression pattern consistent with mitochondrial localization (Fig. 2J–P). Thus, Ptrx-2::trx-2::GFP was clearly expressed in muscle cells in the head, vulva or body wall where mitochondria display their typical tubular morphology (Fig. 2J, K). Indeed, the expression in head muscle cells precluded in most cases the identification of the AIYL/R and ASEL neurons labeling (Fig. 2J). The fact that muscle cells are marked only when using the translational construct Ptrx-2::trx-2::GFP but not the transcriptional construct Ptrx-2::GFP indicates that additional regulatory sequences directing muscle expression are present within the trx-2 genomic open reading frame. The translational Punc-32::trxr-2::GFP construct was found to be expressed in a punctate pattern in the head neurons and ventral nerve cord, while in body wall muscle cells and intestine, the mitochondria distribution was found in a tubular/reticular pattern excluding the nucleus (Fig. 2L, M). Finally, the Ptrxr-2::trxr-2::GFP translational construct expression demonstrated perinuclear punctate fluorescence in several head neurons and reticular labeling in the intestinal cells when transcribed from its internal promoter (Fig. 2N–P).
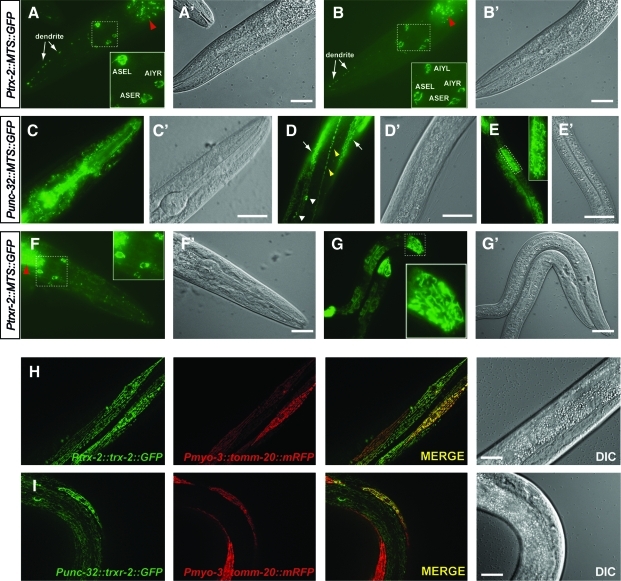
To unequivocally demonstrate in vivo that the patterns obtained with the translational fusion constructs do indeed pertain to mitochondria, we first generated expression constructs in which the putative TRXR-2 and TRX-2 MTS (Supplementary Fig. S1A and B, respectively) were directly fused to GFP. Transgenic animals harboring Ptrx-2::MTS::GFP, Punc-32::MTS::GFP and Ptrxr-2::MTS::GFP all confirmed that the punctate/reticular patterns obtained with the translational GFP constructs are indeed driven by their respective putative MTS (Fig. 3A–G). Next, we performed mitochondrial colocalization experiments in the worm body wall muscle cells, because this tissue expresses both TRX-2::GFP and TRXR-2::GFP fusion proteins from the trx-2 and unc-32 promoters (Fig. 2J–L). As mitochondrial marker, we used a fusion protein of C. elegans mitochondrial import receptor subunit TOMM-20 and monomeric red fluorescent protein (mRFP), whose expression is driven in body wall muscle cells by the myo-3 promoter (Supplementary Table S1, a kind gift of Dr. Amir Sapir and Dr. Paul Sternberg). As shown in Figure 3H and I, the mitochondrial reporter TOMM-20::mRFP colocalizes with both TRX-2::GFP and TRXR-2::GFP fusion proteins expressed in body wall muscle cells from their respective trx-2 and unc-32 promoters. Taken together, all these experiments demonstrate that both trx-2 and trxr-2 genes encode the C. elegans mitochondrial thioredoxin system.
FIG. 3.
TRX-2 and TRXR-2 N-terminal MTS direct GFP translocation into mitochondria. Transgenic worms carrying expression constructs with TRX-2 and TRXR-2 MTS were visualized with fluorescence (A–G) or DIC (A′-G′) optics. When GFP was fused to the TRX-2 MTS, perinuclear mitochondria of AIYL/R and ASEL neurons (and in some cases, also ASER) were marked and GFP puncta were also found along ASEL dendrites (A, B). When GFP fused to the TRXR-2 MTS was expressed under the control of the unc-32 promoter, a strong punctated pattern was obtained in most cells of the head area (C), seam cells (D, yellow arrowheads), muscle cell (D, arrows), coelomocytes (D, white arrowheads) and intestinal tubular mitochondria (E) while when expressed under the control of the trxr-2 internal promoter expression is restricted to perinuclear mitochondria of several head neurons (F) and tubular intestinal mitochondria (G). Transgenic animals overexpressing simultaneously in body wall muscle cells the fusion proteins TOMM-20::mRFP (driven by the myo-3 promoter) and either TRX-2::GFP (driven by the trx-2 promoter) (H) or TRXR-2::GFP (driven by the unc-32 promoter) (I) demonstrate mitochondrial colocalization by the merged yellow color. Note that only in those cells where the expression levels of both fusion proteins are similar, the colocalization is evident. Red arrowheads indicate the intestinal autofluorescent granules. Insets show amplification of the respective dashed areas. Bar 20 μm. (To see this illustration in color, the reader is referred to the web version of this article at www.liebertonline.com/ars). TRX, thioredoxin; MTS, mitochondrial targeting sequence; mRFP, monomeric red fluorescent protein.
The mitochondrial thioredoxin system is dispensable for C. elegans development
Next, we set out to evaluate the phenotypic effect of the lack of trx-2 and trxr-2 genes during C. elegans development. To our surprise, and in sharp contrast with the early embryonic lethal phenotypes displayed by their mammalian orthologs (7, 40), C. elegans trxr-2 and trx-2 single mutants were viable, grew normally to adulthood with no apparent embryonic or postembryonic defects, were fertile and had normal mitochondrial morphology (Supplementary Table S2 and Supplementary Fig. S3). Similarly, double mutants trx-2 (tm2720); trxr-2 (ok2267) and trx-2 (tm2720); trxr-2 (tm2047) did not display any obvious phenotype.
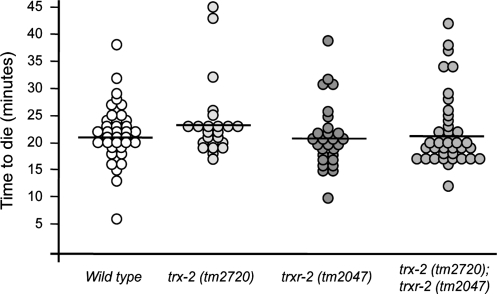
Apoptosis is a developmental process where the mammalian mitochondrial thioredoxin system has been shown to play a key role (10, 42). In fact, trx-2 knock-out mice embryos die by massive apoptosis, while liver apoptosis has been shown to partially underlie the lethal embryonic phenotype of trxr-2 knock-out mice (7, 40). During C. elegans embryogenesis, the apoptotic program is invariant with 113 cells undergoing apoptosis out of 671 cells generated from zygote to newly hatched larva (54). During postembryonic development, 18 additional cells die by programmed cell death, thus making up a total of 131 somatic cells undergoing apoptosis in C. elegans (53). Furthermore, in the adult nematode germ line, another round of cell death takes place to reduce the number of cells that complete oogenesis (17). Given the important role of the mitochondrial thioredoxin system in the apoptotic process in other organisms, we investigated whether C. elegans trx-2 and/or trxr-2 mutants exhibited defective regulation of apoptosis. To this purpose, we used four-dimensional differential interference contrast (DIC) timelapse video microscopy in trx-2 and/or trxr-2 mutant embryos to determine the kinetics of apoptosis of the first 13 programmed cell death events that occur in the AB lineage (39). As shown in Figure 4, we found no significant alteration of cell death kinetics of the mitochondrial thioredoxin system single and double mutants compared with wild-type control embryos. We also measured the penetrance of germ line cell death in adults of the same set of mutants and again found no significant difference in the kinetics of apoptosis in this tissue [wild type 1.00±1.17; trx-2 (tm2720) 0.90±1.07; trxr-2 (tm2047) 0.95±0.88 and trx-2 (tm2720); trxr-2 (tm2047) 1.05±0.94 (Numbers indicate the average±standard deviation of apoptotic corpses per gonad arm, n=20, p>0.05 in all cases by unpaired two-tailed t-test)]. Together, these results demonstrate that in C. elegans, the mitochondrial thioredoxin system is dispensable for normal development under standard growth conditions and does not regulate apoptosis.
FIG. 4.
Loss of trx-2 and trxr-2 does not affect embryonic apoptosis kinetics. The first 13 apoptotic cell deaths in the AB lineage were followed by 4D microscopy and the time to die (defined as the time from cell birth till the onset of apoptosis) was measured. Each circle represents a single cell, the position of the circle along the Y axis indicates the time that the cell takes to die, and the horizontal line represents the mean time to die for each genotype. The time to die for apoptotic cells is distributed in a Gaussian manner with the values of 21.8±5.4 (mean±SD) min after their birth for wild-type embryos, 23.8±7.1 min for trx-2 (tm2720) embryos, 21.3±6.1 min for trxr-2 (tm2047) embryos and 22.1±6.7 min for trx-2 (tm2720); trxr-2 (tm2047) embryos. Analyses were performed in three independent embryos of each genetic background (n=23–39 cells). Unpaired two-tailed t-tests were performed and differences were found to be not significant in all cases (p>0.05). SD, standard deviation.
The mitochondrial thioredoxin system is not a major defense system against stress but might play a role in the mitochondrial unfolded protein response
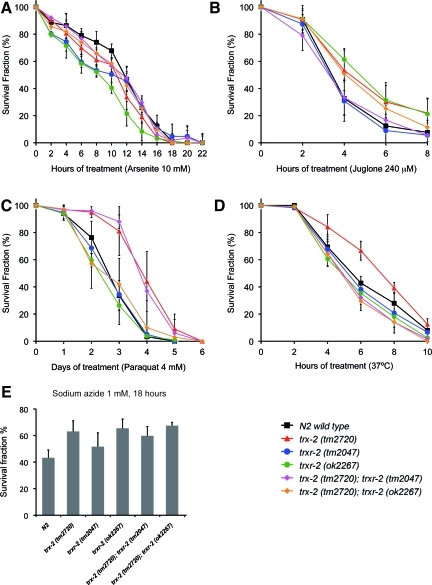
The mammalian mitochondrial thioredoxin system functions as a key defense system against oxidative stress and other cellular insults either directly (19, 50) or by reducing other antioxidant enzymes such as peroxiredoxins (18). To determine whether this protective function is also conserved in worms, we evaluated the sensitivity of trx-2 and trxr-2 mutants exposed to different agents and treatments that induce oxidative stress. No significant differences in sensitivity were found with arsenite, juglone, sodium azide, paraquat or thermal stress treatments (Fig. 5A–E), although the trx-2 (tm2720) mutant was slightly more resistant to thermal stress and trx-2 (tm2720) single and trx-2 (tm2720); trxr-2 (tm2047) double mutant showed a slightly higher resistance to paraquat treatment. Consistent with a dispensable role as a major defense system against oxidative stress, trx-2 and trxr-2 single and double mutants do not show increased ROS production either under basal unstressed conditions or in the presence of 1 mM sodium azide or 0.8 mM paraquat (Supplementary Fig. S4). Besides, efficient RNA interference (RNAi) downregulation of C. elegans trx-2 and trxr-2 gene expression (Supplementary Fig. S5A) in animals carrying integrated GFP fusion transgene markers of different types of cellular stress such as DAF-16, SKN-1, SOD-3, GST-4, HSP-16.2, HSP-4 and HSP-6 (Supplementary Table S1) did not modify the expression levels or subcellular localization of these stress markers (data not shown).
FIG. 5.
Sensitivity of the mitochondrial thioredoxin system mutants to stress treatments. N2 wild-type and single and double mutant combinations of the mitochondrial thioredoxin system alleles were assayed for their resistance to different stress treatments such as arsenite 10 mM (A), juglone 240 μM (B), paraquat 4 mM (C), heat shock at 37°C (D), and sodium azide 1 mM (E). Graphs represent the average of three to five independent experiments. All treatments (except heat shock) were carried out at 20°C. Error bars indicate the standard error of the mean (SEM). For A–D, One-way analysis of variance (ANOVA) test were performed and differences were found to be not significant in all cases (p>0.05). For (E), differences were found to be not significant in all cases by unpaired two-tailed t-test (p>0.05). (To see this illustration in color, the reader is referred to the web version of this article at www.liebertonline.com/ars).
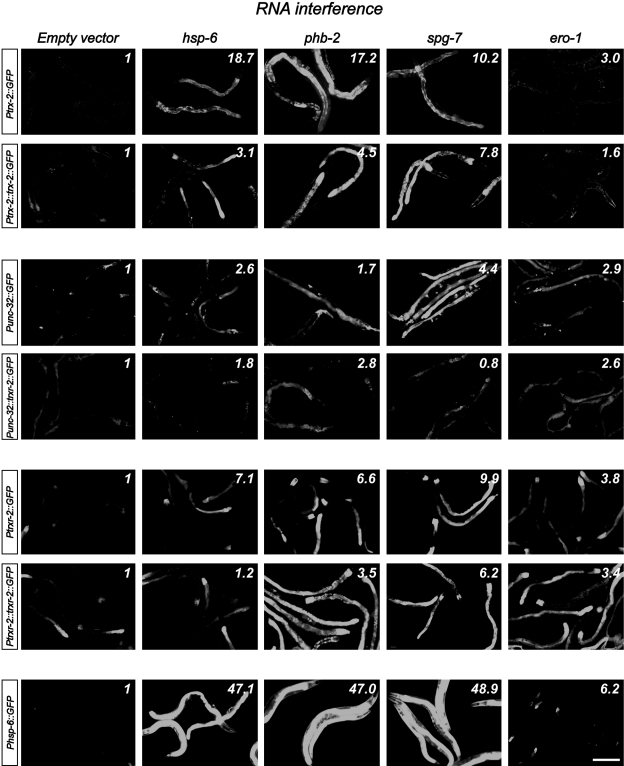
Next, we asked whether the expression levels of trx-2 and trxr-2 GFP reporters were modified under different stress conditions such as food deprivation, heat shock, sodium azide or paraquat treatments as well as induction of the mitochondrial unfolded protein response (UPRmito) resulting from spg-7 RNAi (59). Interestingly, out of all these treatments, we only found a clear induction of GFP expression in transgenic worms grown in spg-7 RNAi (Fig. 6). RNAi downregulation of other genes known to induce UPRmito such as hsp-6 and phb-2 (1, 59) confirmed that the trx-2 and trxr-2 GFP reporters were upregulated under conditions that activate UPRmito (Fig. 6). Unexpectedly, we also found some induction of trxr-2, trx-2 and hsp-6 GFP mitochondrial reporters when using ero-1 RNAi as negative control for UPRmito [ero-1 RNAi activates UPRer (59)] (Fig. 6) This result adds further support to the increasing evidence of an important crosstalk between ER and mitochondria (45). Despite trx-2 and trxr-2 genes being upregulated on UPRmito induction, trx-2 and trxr-2 mutants did not activate the UPRmito reporter HSP-6::GFP nor further enhanced the UPRmito-dependent developmental arrest phenotypes caused by spg-7, phb-2 and hsp-6 RNAi (59). Consistent with these results, overexpression of TRX-2 and TRXR-2 did not alleviate the developmental arrest phenotypes just mentioned that are caused by UPRmito induction (data not shown).
FIG. 6.
The mitochondrial thioredoxin system is upregulated upon mitochondrial UPR induction. Transgenic worms carrying GFP reporter fusions for trx-2, unc-32 and trxr-2 genes were subjected to RNAi of genes inducing either UPRmito (hsp-6, phb-2 and spg-7) or UPRer (ero-1). Transgenic worms expressing the Phsp-6::GFP construct (59) were used as controls. All micrographs were taken with identical image capture settings and, when needed, brightness and contrast were equally modified in all images of the same transgenic strain. Quantification of GFP expression was performed using the ImageJ Software and it is denoted in the upper right corner of each RNAi condition. The fluorescence mean of 5–10 worms per RNAi condition was divided by the selected area (complete intestinal region) and normalized by the background adjacent to the selected worm in the same image. Fluorescence of the respective controls expressing the empty vector was set to value=1 and the different RNAi treatment values indicate the fold induction compared with the corresponding empty vector control. Bar 200 μm. UPR, unfolded protein response; RNAi, RNA interference.
Collectively, these data indicate that the mitochondrial thioredoxin system is not required to maintain general redox homeostasis in C. elegans even under different stress conditions, but it might play a role when mitochondrial protein folding homeostasis (mitochondrial proteostasis) is compromised.
C. elegans trx-2 and trxr-2 mutants have normal longevity
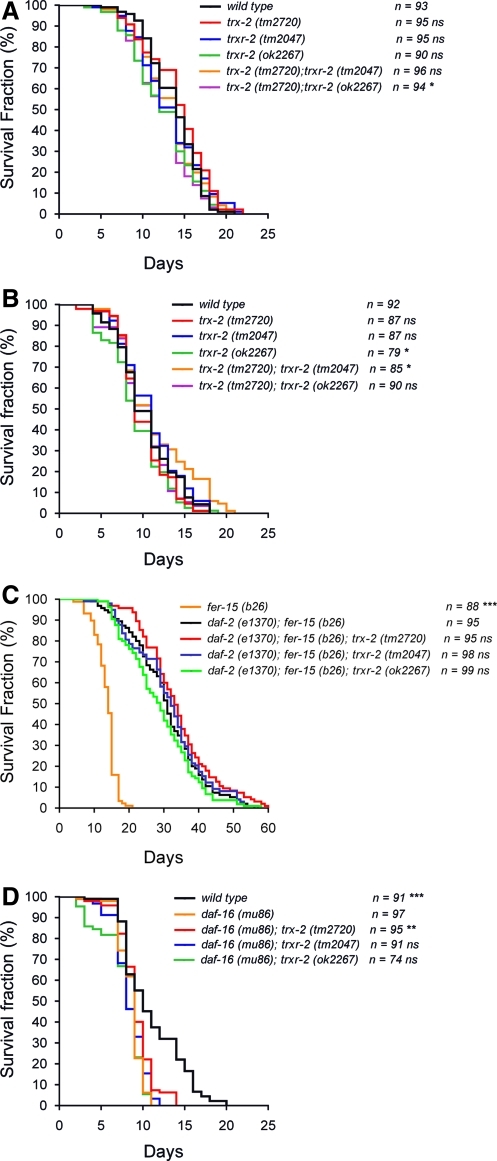
Proteostasis is critical for survival and, therefore, eukaryotic organisms integrate a complex network of chaperones, proteases and assembly factors across all cellular compartments, known as UPR, to ensure a proper balance between protein folding, assembly and degradation (3). In this context, it has been recently proposed that mitochondrial function in cellular proteostasis is critical for worm longevity (26). The finding that trx-2 and trxr-2 gene expression is induced when UPRmito is activated prompted us to investigate their role in C. elegans longevity. First, we determined C. elegans trx-2 and trxr-2 single and double mutants lifespan at both 20°C and 25°C (Fig. 7A, B) and found no major differences in mean and maximum lifespan when compared with N2 wild-type control animals. Since the insulin pathway is a major regulator of longevity (24) and because reduced insulin signaling protects against proteotoxicity (6), we next asked whether the mitochondrial thioredoxin system regulates the lifespan of mutants in the insulin pathway. For this purpose, we made double mutants combining the trx-2 and trxr-2 deletions with mutations in the genes for the insulin receptor daf-2 or the FOXO transcription factor daf-16, which are key determinants of insulin-pathway-dependent longevity (24). Again, no significant differences were observed in any mutant combination with either daf-2 or daf-16 mutants at 25°C, except for a slightly higher maximum lifespan of the trx-2 mutant in the daf-16 background (Fig. 7C, D). Overall, these data indicate that the mitochondrial thioredoxin system is not involved in regulating C. elegans longevity.
FIG. 7.
Lifespan analysis of the mitochondrial thioredoxin system mutants. The effect on longevity of trx-2 and trxr-2 single and double mutants was assayed at 20°C (A) or 25°C (B). The effect of daf-2 (e1370) (C) and daf-16 (mu86) (D) backgrounds on the longevity of trx-2 and trxr-2 mutants was assayed at 25°C. The fer-15 (b26) mutation was included in all the daf-2 (e1370) combinations to prevent internal hatching of the progeny. Kaplan–Meier plots were used to show the fraction of animals that survive over time. Longevity assays were performed twice, yielding similar results and one representative experiment is shown. The survival rate of each population was compared with that of the control [wild type in A and B; daf-2 (e1370); fer-15 (b26) in C and daf-16 (mu86) in D] using the log-rank [Mantel-Cox] test (ns, not significant or p>0.05; *p<0.05; **p<0.01; ***p<0.001). (To see this illustration in color, the reader is referred to the web version of this article at www.liebertonline.com/ars).
Mitochondrial TRXR-2 protects against the paralysis phenotype of a transgenic worm model of AD expressing human β-amyloid peptide
Given the increased expression of trx-2 and trxr-2 genes upon UPRmito activation, we decided to study whether these genes play a role in protein aggregation and proteotoxicity. For this purpose, we used different transgenic worms that overexpress human β-amyloid peptide in body wall muscle cells (hereafter called Aβ worms), resulting in muscular dysfunction and age-dependent progressive paralysis as a consequence of intracellular Aβ aggregation (31, 34). These Aβ worms are commonly considered a C. elegans model of AD (32).
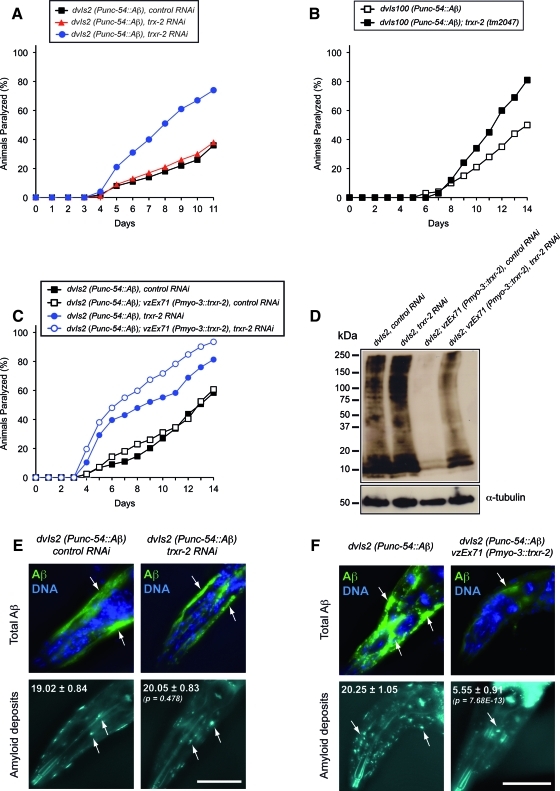
First, we evaluated the impact of trx-2 and trxr-2 RNAi downregulation on the paralysis phenotype of the CL2006 strain, which overexpresses the Aβ peptide in a constitutive manner from the dvIs2 (Punc-54::Aβ) integrated array (31). As shown in Figure 8A, decreased levels of trxr-2 resulted in increased paralysis onset, while, in turn, trx-2 downregulation did not affect paralysis. To validate the RNAi results, we aimed at generating derivatives of dvIs2 Aβ worms in trx-2 and trxr-2 mutant backgrounds. trx-2 (tm2720); dvIs2 (Punc-54::Aβ) animals reproduced the trx-2 RNAi result and had similar paralysis onset as the dvIs2 control (data not shown). However, to our surprise, we were unable to generate animals carrying the dvIs2 integrated array in a trxr-2 mutant background. This unexpected synthetic lethal interaction could be explained by either a positional effect of the dvIs2 array insertion locus or, given the enhanced paralysis caused upon trxr-2 downregulation, by worms being unable to withstand Aβ production in the complete absence of trxr-2. To discriminate between these two alternatives, we made use of the strain CL2750 (C.D. Link, unpublished results), which, similar to CL2006, produces the Aβ peptide constitutively in muscle cells from the integrated array dvIs100 (Supplementary Table S1). Interestingly, unlike dvIs2 worms, we managed to generate Aβ dvIs100 animals in a trxr-2 (tm2047) mutant background and confirmed their increased paralysis when compared with its respective control (Fig. 8B). To rule out that variations in Aβ content might underlie the differences between the CL2006 and CL2750 strains, we examined the total Aβ levels in these two strains and found that CL2006 has a little higher amount of Aβ compared with that of CL2750 (Supplementary Fig. S5B). Based on these results, we support the notion that the failure to obtain animals expressing the dvIs2 array in a trxr-2 mutant background is due to a positional effect of the locus where the dvIs2 array is inserted (which we have independently mapped at LG II) rather than to the higher Aβ levels in CL2006 as compared with those of CL2750, although we cannot completely rule out that this slight difference might play a role.
FIG. 8.
TRXR-2 alleviates Aβ-dependent paralysis and modulates Aβ aggregation and β-amyloid deposit formation. (A) Progressive paralysis of CL2006, dvIs2 [Punc-54::Aβ3–42::unc-54 3′-UTR; rol-6 (su1006)] worms growing on HT-115 bacteria expressing either no dsRNA (▪), trx-2 dsRNA (▴) or trxr-2 dsRNA (•). The graph shows one representative experiment out of three independent trials with similar results. (B) Progressive paralysis of CL2750, dvIs100 [Punc-54::Aβ1–42::unc-54 3′-UTR; Pmtl-2::GFP] worms (□) and its trxr-2 (tm2047) derivative VZ223 (▪) growing on OP50 bacteria. The graph shows one representative experiment out of two independent trials with similar results. (C) Progressive paralysis of CL2006 worms and its derivative transgenic strain VZ209 carrying the extrachromosomal array vzEx71 [Pmyo-3::trxr-2::trxr-2 3′-UTR; Punc-122::GFP] growing on HT-115 bacteria expressing either no dsRNA (▪, □) or trxr-2 dsRNA (•, ○). The graph shows one representative experiment out of three independent trials with similar results. All paralysis assays were scored at 20°C. (D) Aβ immunoblot with the 6E10 monoclonal antibody. All lanes were loaded with total protein extract from 100 one-day adult synchronized CL2006 and VZ209 worms grown on HT-115 bacteria expressing either no dsRNA or trxr-2 dsRNA. A strong decrease in Aβ content is found in TRXR-2 overexpressing animals, an effect that is abolished when trxr-2 overexpression is downregulated by RNAi. α-tubulin is used as loading control. One blot is shown out of three independent trials with similar results. dvIs2, control RNAi (3.33±0.09); dvIs2, trxr-2 RNAi (3.52±0.91, p=0.37); dvIs2, vzEx71, control RNAi (0.66±0.10, p=2.5×10−6); dvIs2, vzEx71, trxr-2 RNAi (3.03±0.53, p=0.20) (mean Aβ content±SD, p-value by Student's unpaired t-test with a two-tailed distribution, compared with dvIs2, control RNAi). The quantification of the blots and the statistical analyses are included in Supplementary Figure S6A. (E) Immunohistochemistry of total Aβ content (upper panels, arrows) and amyloid deposits (lower panels, arrows) in the muscles of the head region of 1-day adult CL2006 synchronized worms grown on HT-115 bacteria expressing either no dsRNA or trxr-2 dsRNA. Total Aβ content was determined using the 6E10 monoclonal antibody (green) counterstained with DAPI (blue) for identification of nuclei, while amyloid deposits detection was performed using the fluorescent amyloid dye X-34. The quantification of amyloid deposits was performed on a total of 20 worms per condition and the results are shown in the corresponding panel (number of amyloid deposits±SD). Unpaired two-tailed t-test was performed and the difference was found to be not significant (p=0.478). (F) Immunohistochemistry of total Aβ content (upper panels, arrows) and amyloid deposits (lower panels, arrows) in the muscles of the head region of 1-day adult CL2006 and VZ209 synchronized worms grown on OP50 bacteria. Total Aβ content determination and amyloid deposits quantification was performed as described in (E). Unpaired two-tailed t-test was performed and the difference was found to be highly significant (p=7.68E-13). Note that animals used for experiments shown in panels A, C, D and E were grown on the corresponding RNAi bacteria for two generations before performing the experiment. Bar 50 μm. (To see this illustration in color, the reader is referred to the web version of this article at www.liebertonline.com/ars). DAPI, 4′,6-diamidino-2-phenylindole.
We next asked whether the protective effect of trxr-2 on the paralysis of Aβ (constitutive) worms is a consequence of its requirement during embryogenesis and early development, when body wall muscle cells are generated (53, 54), or it can also be implemented at later developmental stages, when muscle cells are fully formed. For this purpose, we used the CL4176 strain that overproduces the Aβ peptide from the dvIs27 integrated array in an inducible manner after temperature upshift at the L3 larval stage (34). Our results show that trxr-2 also protects against the paralysis even when Aβ is produced in an inducible fashion (Supplementary Fig. S5C), indicating that the protective effect of trxr-2 in Aβ-dependent paralysis is associated to muscle function but not to muscle biogenesis.
Since the lack of trxr-2 enhances the Aβ-dependent paralysis phenotype, we wondered whether high levels of TRXR-2 would improve the paralysis onset. To this aim, we generated transgenic strains expressing high levels of TRXR-2 in both constitutive dvIs2 and inducible dvIs27 Aβ worms (Supplementary Fig. S5F). Surprisingly, while decreased levels of TRXR-2 worsen paralysis, transgenic Aβ worms expressing high levels of TRXR-2 did not improve the paralysis phenotype when compared with their corresponding nonoverexpressing control siblings (Fig. 8C and Supplementary Fig. S5D). These data indicate that restoring endogenous TRXR-2 levels is sufficient to rescue the enhanced paralysis of Aβ worms caused by dowregulation of trxr-2 and that increased amount of TRXR-2 does not provide additional protection against Aβ-dependent paralysis.
To determine whether the effect of TRXR-2 on paralysis onset was associated with differences in Aβ aggregation or oligomerization, we first performed Aβ immunoblotting on constitutive dvIs2 Aβ worms using the Aβ-peptide specific antibody 6E10. This antibody detects all forms of Aβ (amyloid deposits, amorphous Aβ aggregates and soluble Aβ oligomers and monomers) (14). As shown in Figure 8D, and except for a slight increase in Aβ oligomeric species (range 75–150 kDa), no major differences were found in Aβ worms when trxr-2 was downregulated by RNAi. Unexpectedly, we found a dramatic decrease in total Aβ amount in TRXR-2 overexpressing Aβ worms, an effect that was abolished upon trxr-2 RNAi when Aβ oligomeric species reappeared (Fig. 8D). Similar results were obtained with inducible dvIs27 Aβ worms, although the reduction of total Aβ in TRXR-2 overexpressing worms was not as strong as in the constitutive Aβ strain. (Supplementary Fig. S5E). This difference in Aβ reduction between the constitutive and inducible strains most likely reflects saturation of the (unknown) clearance mechanism as a consequence of the rapid and robust Aβ production upon temperature upshift in the inducible strain.
Immunohistochemistry with the 6E10 antibody or the fluorescent amyloid vital dye X-34 (33) on dvIs2 Aβ worms corroborated the immunoblot data, as no differences were found in either the total Aβ load or amyloid deposits formation between control and trxr-2 downregulated animals (Fig. 8E). On the other hand, dvIs2 Aβ worms overexpressing TRXR-2 showed a robust reduction in both total Aβ species and amyloid deposits (Fig. 8F).
Together, our data demonstrate an in vivo role of TRXR-2 on Aβ-dependent paralysis and aggregation in C. elegans, suggesting a possible protective function of TRXR-2 in AD.
Discussion
Given the difficulty of studying the function of the mitochondrial thioredoxin system in the context of a complete animal using the mouse or Drosophila models (due to embryonic or larval lethal phenotypes, respectively), we turned our interest to the nematode C. elegans, an excellent genetically tractable model where the mitochondrial thioredoxin system is highly homologous to that of mammals.
We first demonstrated that C. elegans trx-2 and trxr-2 genes encode proteins that are targeted to mitochondria in vivo, driven by putative MTS at their respective N-terminus. Interestingly, the rather ubiquitous expression pattern of the trxr-2 gene deeply contrasts with the highly restricted one of the trx-2 gene in AIYL/R and ASEL neurons and muscle cells, which suggests that TRXR-2 might have additional substrates other than TRX-2. We identified TRX-2 on the basis of its conserved active site sequence WCGPC and it cannot be ruled out that other thioredoxin family members with a more divergent active site sequence are also present in mitochondria. Furthermore, it has been reported that human mitochondrial glutaredoxin 2 is a substrate for mitochondrial thioredoxin reductase (22), a function that could also be conserved in worms (at least four glutaredoxin genes have been reported in C. elegans, see below). Likely, other TRXR-2 substrates not functionally or structurally related to thioredoxins and glutaredoxins might also exist in mitochondria. It is worth noting that TRX-2 is not detected in the intestine under unstressed conditions (Fig. 2A–C and J, K), but it is expressed in this organ upon UPRmito induction (Fig. 6), indicating that TRX-2 expression might be turned on in additional tissues depending on stress conditions.
C. elegans is, to date, the only metazoan in which the mitochondrial thioredoxin system is not essential for survival. More surprisingly, trx-2 and trxr-2 single and double mutants do not show enhanced sensitivity to different stressors. Collectively, this lack of phenotype supports the idea that a redundant system exists to counterbalance the absence of the mitochondrial thioredoxin system under both unstressed and stressed conditions. One alternative could be that the C. elegans cytosolic thioredoxin reductase gene trxr-1 might produce an isoform targeted to mitochondria as it happens in other organisms (48). However, we have ruled out this possibility, as trxr-1; trxr-2 double mutants are also fully viable (51). The viability of the trxr-1; trxr-2 double mutants raises the interesting question of which enzyme(s) maintain the different worm thioredoxins in their reduced active state in the absence of both thioredoxin reductases. An obvious candidate is the glutaredoxin system based on the numerous reports of functional redundancy of both thioredoxin and glutaredoxin systems (either cytoplasmic or mitochondrial) in several organisms (12, 58). We have recently uncovered an example of such functional redundancy in C. elegans as trxr-1 animals feeding on glutathione reductase (gsr-1) RNAi bacteria display a highly penetrant larval arrest phenotype due to defective cuticle ecdysis (51). However, RNAi downregulation of gsr-1 or any of the four worm glutaredoxins (glrx-5, glrx-10, glrx-21 and glrx-22) in trx-2 and trxr-2 single and double mutant backgrounds did not result in any obvious synthetic phenotype (Supplementary Table S3). Since RNAi feeding penetrance can be highly variable depending on genetic backgrounds and tissue expression, combinations of glutaredoxin system mutants with those of trx-2 and trxr-2 will be needed to unequivocally identify the redundant system for the mitochondrial thioredoxin system. Other plausible explanations for the lack of phenotype of the mitochondrial thioredoxin system mutants could be that the system is dispensable under normal laboratory growth conditions while being essential in the wild or that, alternatively, the mitochondrial thioredoxin system plays a modulatory role in nonessential ROS-mediated signaling mechanisms.
The induction of trx-2 and trxr-2 upon UPRmito activation suggests that the mitochondrial thioredoxin system is a part of the chaperone machinery required to cope with the load of unfolded/misfolded proteins that accumulate and aggregate when mitochondrial proteostasis is compromised. Most likely, the function of the mitochondrial thioredoxin system under UPRmito is aimed at reducing incorrect disulfide bonds in these misfolded proteins to facilitate either native refolding or their export from mitochondria and subsequent degradation by proteasome (56). The maintenance of cellular and subcellular proteostasis is essential for organismal survival and its imbalance dramatically affects the function of all cellular organelles, including mitochondria, eventually leading to enhanced progression of aging and development of neurodegenerative diseases such as Alzheimer's, Parkinson's or Huntington's disease (26).
In this work, we have found a protective role of the trxr-2 gene on the aging-dependent progressive paralysis phenotype caused by Aβ aggregation in worm muscle cells, a well-established model for AD (31). This protective role is not shared by the trx-2 gene despite it is also expressed in muscle cells, reinforcing the notion that additional substrates for TRXR-2 might mediate such a protective function. We have shown here that while trxr-2 downregulation clearly enhances paralysis, increased TRXR-2 expression does not alleviate the paralysis onset. On the other hand, trxr-2 downregulation does not substantially modify the total levels of Aβ species nor amyloid deposits, which, in turn, are dramatically diminished in TRXR-2 overexpressing animals. The lack of correlation between total amyloid loads and the paralysis rate has been previously reported in this AD worm model (15, 32), although no molecular mechanism has been proposed yet to clarify this fact. A plausible explanation relates to the recent finding that muscle mitochondria of Aβ worms are highly fragmented (C.D. Link, unpublished data). Our preliminary unpublished results indicate that decreased TRXR-2 levels further enhance the mitochondrial fragmentation of Aβ worms, which might account for the increased paralysis phenotype in trxr-2 mutants or trxr-2 RNAi treated Aβ worms without provoking noticeable changes in total Aβ load or amyloid deposits. Alternatively, despite the fact that higher levels of TRXR-2 strongly reduce both total Aβ load and amyloid deposits, it is possible that the residual amount of Aβ species detected in both immunoblots and immunostainings of overproducing TRXR-2 Aβ worms might still be sufficient to induce paralysis, thus explaining why overexpressing TRXR-2 does not improve paralysis onset.
Elucidating the mechanism(s) by which TRXR-2 overexpression degrades most Aβ load is of enormous interest. For instance, it is conceivable that TRXR-2 might interact with proteins implicated in the specific degradation of the Aβ peptide and amyloid deposits in mammals, such as angiotensin-converting enzyme, insulin-degrading enzyme or neprilysin (2), all of which have close orthologs in worms. Alternatively, an interesting possibility relates to the finding that reduced insulin signaling improves paralysis of Aβ worms by promoting autophagic degradation of Aβ (13). Thus, it could be possible that TRXR-2 overexpression triggers a decrease of insulin signaling, resulting in Aβ autophagic-dependent degradation. As a whole, it will be very important to gain a deeper insight into the molecular mechanisms underlying the dual role of TRXR-2 in alleviating paralysis and decreasing amyloid deposits in Aβ worms and the potential application of this knowledge to the human scenario in AD.
In conclusion, the characterization of the C. elegans mitochondrial thioredoxin system reported in this work provides a new tool to further investigate the function of this redox system in metazoan physiology and pathology. The genetic amenability of C. elegans paves the way to use the mitochondrial thioredoxin mutants in genetic screens to identify other systems involved in the maintenance of organismal redox homeostasis and, particularly, opens new possibilities to understand the role of mitochondrial redox systems in the pathology of neurodegenerative diseases.
Materials and Methods
C. elegans strains and culture conditions
The standard methods used for culturing and maintenance of C. elegans were as described previously (52). The strains used in this work are described in Supplementary Table S1. All experiments were performed at 20°C unless otherwise noted. All VZ strains were 6× backcrossed with N2 wild type.
RNA extraction and RT-PCR analysis
RNA extraction (from embryos of the different genotypes) and cDNA generation were performed by standard methods using commercial kits. Details on the nested RT-PCR procedures are found in Supplementary Materials and Methods.
Thioredoxin activity assays
The enzymatic activity of the recombinant His-CeTRX-2 and His-CeΔTRX-2 proteins was tested by their ability to reduce bovine insulin A and B chains (Sigma) using either DTT or NADPH (Sigma) and rat thioredoxin reductase-1 (IMCO) as electron donors, as previously described, with slight modifications (20). Details on recombinant protein purification and enzymatic assays are found in Supplementary Materials and Methods.
GFP expression constructs and transgenesis
All transcriptional and translational GFP fusion constructs were generated using the pPD95.77 vector backbone. Information on primer sequences, cloning sites and the size of the respective inserts will be provided upon request. Details on transgenic strains generation are found in Supplementary Materials and Methods.
Expression pattern analysis
GFP transgenic animals were mounted in a 5 μL drop of 10 mM levamisole (Sigma) on a 3% agarose pad covered with a 24×24 mm coverslip. DIC and fluorescence imaging was performed on a Zeiss AxioImager M2 ApoTome fluorescence microscope equipped with an AxioCam MRn (Zeiss). Images were captured with the AxioVision 4.8 Software (Zeiss) and equal adjustment of brightness and contrast on control and matched experimental images was done using Adobe Photoshop 10 Software (Adobe Systems).
Analysis of embryonic and germ line apoptosis
Apoptotic corpses were detected by their refractile shape using a Leica DM6000 microscope equipped with DIC optics and scored as described (47). For embryonic apoptosis, three embryos of each genotype were mounted at a very early stage (2–4 cell stage) and the recording of their development was carried out at 25°C. The 4-D microscopic analysis was carried out by recording 30 focal planes of the embryos for 10 h. As a result, four-dimensional (4D) movies (3D of the embryo+time) were obtained for each genotype. The SIMI Biocell software (SIMI GmbH) allowed tracing each and every cell of the embryo in time and space as described (39). Time of birth and onset of apoptosis were measured to define the kinetics of cell death. Time to die is defined as the time period from which a cell is born until it begins to show morphological characteristics of apoptosis, such as rounding of the cell, cytoplasmic condensation or nuclear swelling (23). For germ line apoptosis, 20 gonad arms per genotype were analyzed and the apoptotic corpses detection and scoring were performed as described above.
Stress assays
For paraquat, sodium arsenite, sodium azide (Sigma) and heat-shock treatments, L4 larval stage hermaphrodites were used to initiate the assays. For juglone treatment, young adult hermaphrodites were utilized. The viability of each animal was assessed at the indicated times by microscopic examination of pharyngeal pumping and movement in response to prodding with a pick. Dead worms were counted and removed from the plates. Details are found in Supplementary Materials and Methods. The percentage of surviving animals for each treatment was calculated from three or more independent experiments.
RNAi and UPRmito induction quantification
Feeding RNAi was performed as previously described (57). Details are found in Supplementary Materials and Methods. Fluorescence quantification of UPRmito induction was done on images (acquired under identical settings) of 5–10 worms for each RNAi condition, using the ImageJ Software (National Institutes of Health).
Longevity assays
Life-span assays were performed at 20°C or 25°C as previously described (30) with slight modifications. Details are found in Supplementary Materials and Methods.
Paralysis phenotype, Aβ peptide expression and β-amyloid deposits quantification
For strains CL2006, CL2750 and derivatives, synchronous populations of Aβ worms were generated by time-limited (2 h) egg lay at 16°C. Parents were removed and progeny was grown continuously at 20°C. Paralysis scoring was initiated at the first day of adulthood and determined daily, whereby paralyzed worms were removed from plates. For strains CL4176 and VZ296, synchronous populations of Aβ worms were generated by time-limited (2 h) egg lay at 16°C. Parents were removed and progeny was grown continuously at 16°C for 48 hours followed by a temperature upshift to 23°C or 25°C to induce Aβ production. Paralysis scoring was initiated 24 h after a temperature upshift and determined every hour, whereby paralyzed worms were removed from plates. Aβ immunoblotting and immunostaining was carried out using the 6E10 monoclonal antibody (Covance Inc.). The quantification of β-amyloid blots was performed using the ImageJ Software, while the quantification of β-amyloid deposits using the fluorescent amyloid dye X-34 (a kind gift from Prof. William Klunk) was performed as described, with brief modifications (15, 33). Details are found in Supplementary Materials and Methods.
Supplementary Material
Abbreviations Used
- 4D
four-dimensional
- Aβ
beta amyloid peptide
- AD
Alzheimer's disease
- CGC
Caenorhabditis Genetics Center
- DAPI
4′,6-diamidino-2-phenylindole
- DIC
differential interference contrast
- DTT
1,4-dithio-d-threitol
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- H2-DCF-DA
2,7-dichlorodihydrofluorescein-diacetate
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- LB
Luria-Bertani
- mRFP
monomeric red fluorescent protein
- mRNA
messenger RNA
- MTS
mitochondrial targeting sequence
- NA
not applicable
- NADPH
reduced nicotinamide adenine dinucleotide phosphate
- NGM
nematode growth medium
- OD
optical density
- ORF
open reading frame
- PVDF
polyvinylidene fluoride
- RNAi
RNA interference
- ROS
reactive oxygen species
- RT-PCR
reverse transcriptase–polymerase chain reaction
- SD
standard deviation
- SDS-PAGE
sodium dodecyl sulphate-polyacrylamide gel electrophoresis
- SEM
standard error of the mean
- TRX
thioredoxin
- TRXR
thioredoxin reductase
- UPR
unfolded protein response
- UTR
untranslated region
Acknowledgments
We thank the Caenorhabditis Genetics Center, the Japanese National Bioresource Project, Ellen A. Nollen, Piali Sengupta, Amir Sapir, Paul Sternberg, Simon Tuck, Oliver Hobert, Alexander van der Bliek, Andrew Fire, Cesar Santiago and Julian Cerón for sharing worm strains and plasmids. We are grateful to Pilar Alarcón, María Jesús Rodríguez and Francisco José Naranjo for excellent technical assistance, Lizett Rodríguez for help with enzymatic assays and Fernando Calahorro for his expert assistance with sequence analysis. We also thank Elias Arnér for critical reading of the article. Continuous support from Peter Askjaer's and Manuel Muñoz's groups is deeply acknowledged. A.M.-V. was supported by the Instituto de Salud Carlos III (Projects PI050065 and PI080557, co-financed by the Fondo Social Europeo, FEDER) and Junta de Andalucía (Projects P07-CVI-02697 and P08-CVI-03629), Spain. B.C.-V. was supported by a fellowship from the Consejo Nacional de Ciencia y Tecnología (CONACYT) from the Government of Mexico. P.N. was supported by the Instituto de Salud Carlos III (project PI080500, co-financed by the Fondo Social Europeo, FEDER) and Junta de Andalucía (project P08-CTS-03988). Work in the laboratory of P.S., a member of the NordForsk Nordic C. elegans network, was supported by a grant from the Swedish Research Council. J.C. was supported by the Spanish Ministry of Science and Innovation Grant BFU2010-21794 and the RiojaSalud Foundation.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Artal-Sanz M. Tsang WY. Willems EM. Grivell LA. Lemire BD. van der Spek H. Nijtmans LG. The mitochondrial prohibitin complex is essential for embryonic viability and germline function in Caenorhabditis elegans. J Biol Chem. 2003;278:32091–32099. doi: 10.1074/jbc.M304877200. [DOI] [PubMed] [Google Scholar]
- 2.Bates KA. Verdile G. Li QX. Ames D. Hudson P. Masters CL. Martins RN. Clearance mechanisms of Alzheimer's amyloid-beta peptide: implications for therapeutic design and diagnostic tests. Mol Psychiatry. 2009;14:469–486. doi: 10.1038/mp.2008.96. [DOI] [PubMed] [Google Scholar]
- 3.Bukau B. Weissman J. Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 4.Chen Q. Vazquez EJ. Moghaddas S. Hoppel CL. Lesnefsky EJ. Production of reactive oxygen species by mitochondria: central role of complex III. J Biol Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y. Cai J. Jones DP. Mitochondrial thioredoxin in regulation of oxidant-induced cell death. FEBS Lett. 2006;580:6596–6602. doi: 10.1016/j.febslet.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen E. Du D. Joyce D. Kapernick EA. Volovik Y. Kelly JW. Dillin A. Temporal requirements of insulin/IGF-1 signaling for proteotoxicity protection. Aging Cell. 2010;9:126–134. doi: 10.1111/j.1474-9726.2009.00541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conrad M. Jakupoglu C. Moreno SG. Lippl S. Banjac A. Schneider M. Beck H. Hatzopoulos AK. Just U. Sinowatz F. Schmahl W. Chien KR. Wurst W. Bornkamm GW. Brielmeier M. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol Cell Biol. 2004;24:9414–9423. doi: 10.1128/MCB.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Autreaux B. Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 9.Dai S. He Y. Zhang H. Yu L. Wan T. Xu Z. Jones D. Chen H. Min W. Endothelial-specific expression of mitochondrial thioredoxin promotes ischemia-mediated arteriogenesis and angiogenesis. Arterioscler Thromb Vasc Biol. 2009;29:495–502. doi: 10.1161/ATVBAHA.108.180349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damdimopoulos AE. Miranda-Vizuete A. Pelto-Huikko M. Gustafsson JA. Spyrou G. Human mitochondrial thioredoxin. Involvement in mitochondrial membrane potential and cell death. J Biol Chem. 2002;277:33249–33257. doi: 10.1074/jbc.M203036200. [DOI] [PubMed] [Google Scholar]
- 11.Eklund H. Gleason FK. Holmgren A. Structural and functional relations among thioredoxins of different species. Proteins. 1991;11:13–28. doi: 10.1002/prot.340110103. [DOI] [PubMed] [Google Scholar]
- 12.Fernando MR. Lechner JM. Lofgren S. Gladyshev VN. Lou MF. Mitochondrial thioltransferase (glutaredoxin 2) has GSH-dependent and thioredoxin reductase-dependent peroxidase activities in vitro and in lens epithelial cells. FASEB J. 2006;20:2645–2647. doi: 10.1096/fj.06-5919fje. [DOI] [PubMed] [Google Scholar]
- 13.Florez-McClure ML. Hohsfield LA. Fonte G. Bealor MT. Link CD. Decreased insulin-receptor signaling promotes the autophagic degradation of beta-amyloid peptide in C. elegans. Autophagy. 2007;3:569–580. doi: 10.4161/auto.4776. [DOI] [PubMed] [Google Scholar]
- 14.Fonte V. Dostal V. Roberts CM. Gonzales P. Lacor P. Magrane J. Dingwell N. Fan EY. Silverman MA. Stein GH. Link CD. A glycine zipper motif mediates the formation of toxic beta-amyloid oligomers in vitro and in vivo. Mol Neurodegener. 2011;6:61. doi: 10.1186/1750-1326-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fonte V. Kipp DR. Yerg J., 3rd Merin D. Forrestal M. Wagner E. Roberts CM. Link CD. Suppression of in vivo beta-amyloid peptide toxicity by overexpression of the HSP-16.2 small chaperone protein. J Biol Chem. 2008;283:784–791. doi: 10.1074/jbc.M703339200. [DOI] [PubMed] [Google Scholar]
- 16.Gladyshev VN. Krause M. Xu XM. Korotkov KV. Kryukov GV. Sun QA. Lee BJ. Wootton JC. Hatfield DL. Selenocysteine-containing thioredoxin reductase in C. elegans. Biochem Biophys Res Commun. 1999;259:244–249. doi: 10.1006/bbrc.1999.0765. [DOI] [PubMed] [Google Scholar]
- 17.Gumienny TL. Lambie E. Hartwieg E. Horvitz HR. Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- 18.Hanschmann EM. Lonn ME. Schutte LD. Funke M. Godoy JR. Eitner S. Hudemann C. Lillig CH. Both thioredoxin 2 and glutaredoxin 2 contribute to the reduction of the mitochondrial 2-Cys peroxiredoxin Prx3. J Biol Chem. 2010;285:40699–40705. doi: 10.1074/jbc.M110.185827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen JM. Zhang H. Jones DP. Mitochondrial thioredoxin-2 has a key role in determining tumor necrosis factor-alpha-induced reactive oxygen species generation, NF-kappaB activation, and apoptosis. Toxicol Sci. 2006;91:643–650. doi: 10.1093/toxsci/kfj175. [DOI] [PubMed] [Google Scholar]
- 20.Holmgren A. Bjornstedt M. Thioredoxin and thioredoxin reductase. Methods Enzymol. 1995;252:199–208. doi: 10.1016/0076-6879(95)52023-6. [DOI] [PubMed] [Google Scholar]
- 21.Huang P. Pleasance ED. Maydan JS. Hunt-Newbury R. O'Neil NJ. Mah A. Baillie DL. Marra MA. Moerman DG. Jones SJ. Identification and analysis of internal promoters in Caenorhabditis elegans operons. Genome Res. 2007;17:1478–1485. doi: 10.1101/gr.6824707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansson C. Lillig CH. Holmgren A. Human mitochondrial glutaredoxin reduces S-glutathionylated proteins with high affinity accepting electrons from either glutathione or thioredoxin reductase. J Biol Chem. 2004;279:7537–7543. doi: 10.1074/jbc.M312719200. [DOI] [PubMed] [Google Scholar]
- 23.Kao AW. Eisenhut RJ. Martens LH. Nakamura A. Huang A. Bagley JA. Zhou P. de Luis A. Neukomm LJ. Cabello J. Farese RV., Jr. Kenyon C. A neurodegenerative disease mutation that accelerates the clearance of apoptotic cells. Proc Natl Acad Sci U S A. 2011;108:4441–4446. doi: 10.1073/pnas.1100650108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 25.Kim MR. Chang HS. Kim BH. Kim S. Baek SH. Kim JH. Lee SR. Kim JR. Involvements of mitochondrial thioredoxin reductase (TrxR2) in cell proliferation. Biochem Biophys Res Commun. 2003;304:119–124. doi: 10.1016/s0006-291x(03)00547-3. [DOI] [PubMed] [Google Scholar]
- 26.Kirstein-Miles J. Morimoto RI. Caenorhabditis elegans as a model system to study intercompartmental proteostasis: interrelation of mitochondrial function, longevity, and neurodegenerative diseases. Dev Dyn. 2010;239:1529–1538. doi: 10.1002/dvdy.22292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koehler CM. Beverly KN. Leverich EP. Redox pathways of the mitochondrion. Antioxid Redox Signal. 2006;8:813–822. doi: 10.1089/ars.2006.8.813. [DOI] [PubMed] [Google Scholar]
- 28.Kowaltowski AJ. de Souza-Pinto NC. Castilho RF. Vercesi AE. Mitochondria and reactive oxygen species. Free Radic Biol Med. 2009;47:333–343. doi: 10.1016/j.freeradbiomed.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Lacey BM. Hondal RJ. Characterization of mitochondrial thioredoxin reductase from C. elegans. Biochem Biophys Res Commun. 2006;346:629–636. doi: 10.1016/j.bbrc.2006.05.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsen PL. Albert PS. Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Link CD. Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1995;92:9368–9372. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Link CD. C. elegans models of age-associated neurodegenerative diseases: lessons from transgenic worm models of Alzheimer's disease. Exp Gerontol. 2006;41:1007–1013. doi: 10.1016/j.exger.2006.06.059. [DOI] [PubMed] [Google Scholar]
- 33.Link CD. Johnson CJ. Fonte V. Paupard M. Hall DH. Styren S. Mathis CA. Klunk WE. Visualization of fibrillar amyloid deposits in living, transgenic Caenorhabditis elegans animals using the sensitive amyloid dye, X-34. Neurobiol Aging. 2001;22:217–226. doi: 10.1016/s0197-4580(00)00237-2. [DOI] [PubMed] [Google Scholar]
- 34.Link CD. Taft A. Kapulkin V. Duke K. Kim S. Fei Q. Wood DE. Sahagan BG. Gene expression analysis in a transgenic Caenorhabditis elegans Alzheimer's disease model. Neurobiol Aging. 2003;24:397–413. doi: 10.1016/s0197-4580(02)00224-5. [DOI] [PubMed] [Google Scholar]
- 35.Meyer Y. Buchanan BB. Vignols F. Reichheld JP. Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu Rev Genet. 2009;43:335–367. doi: 10.1146/annurev-genet-102108-134201. [DOI] [PubMed] [Google Scholar]
- 36.Miranda-Vizuete A. Damdimopoulos AE. Spyrou G. The mitochondrial thioredoxin system. Antioxid Redox Signal. 2000;2:801–810. doi: 10.1089/ars.2000.2.4-801. [DOI] [PubMed] [Google Scholar]
- 37.Missirlis F. Ulschmid JK. Hirosawa-Takamori M. Gronke S. Schafer U. Becker K. Phillips JP. Jackle H. Mitochondrial and cytoplasmic thioredoxin reductase variants encoded by a single Drosophila gene are both essential for viability. J Biol Chem. 2002;277:11521–11526. doi: 10.1074/jbc.M111692200. [DOI] [PubMed] [Google Scholar]
- 38.Nalvarte I. Damdimopoulos AE. Nystom C. Nordman T. Miranda-Vizuete A. Olsson JM. Eriksson L. Bjornstedt M. Arner ES. Spyrou G. Overexpression of enzymatically active human cytosolic and mitochondrial thioredoxin reductase in HEK-293 cells. Effect on cell growth and differentiation. J Biol Chem. 2004;279:54510–54517. doi: 10.1074/jbc.M408494200. [DOI] [PubMed] [Google Scholar]
- 39.Nieto C. Almendinger J. Gysi S. Gomez-Orte E. Kaech A. Hengartner MO. Schnabel R. Moreno S. Cabello J. ccz-1 mediates the digestion of apoptotic corpses in C. elegans. J Cell Sci. 2010;123:2001–2007. doi: 10.1242/jcs.062331. [DOI] [PubMed] [Google Scholar]
- 40.Nonn L. Williams RR. Erickson RP. Powis G. The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol Cell Biol. 2003;23:916–922. doi: 10.1128/MCB.23.3.916-922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oka T. Toyomura T. Honjo K. Wada Y. Futai M. Four subunit a isoforms of Caenorhabditis elegans vacuolar H+-ATPase. Cell-specific expression during development. J Biol Chem. 2001;276:33079–33085. doi: 10.1074/jbc.M101652200. [DOI] [PubMed] [Google Scholar]
- 42.Patenaude A. Ven Murthy MR. Mirault ME. Mitochondrial thioredoxin system: effects of TrxR2 overexpression on redox balance, cell growth, and apoptosis. J Biol Chem. 2004;279:27302–27314. doi: 10.1074/jbc.M402496200. [DOI] [PubMed] [Google Scholar]
- 43.Pedrajas JR. Kosmidou E. Miranda-Vizuete A. Gustafsson JA. Wright AP. Spyrou G. Identification and functional characterization of a novel mitochondrial thioredoxin system in Saccharomyces cerevisiae. J Biol Chem. 1999;274:6366–6373. doi: 10.1074/jbc.274.10.6366. [DOI] [PubMed] [Google Scholar]
- 44.Perez VI. Lew CM. Cortez LA. Webb CR. Rodriguez M. Liu Y. Qi W. Li Y. Chaudhuri A. Van Remmen H. Richardson A. Ikeno Y. Thioredoxin 2 haploinsufficiency in mice results in impaired mitochondrial function and increased oxidative stress. Free Radic Biol Med. 2008;44:882–892. doi: 10.1016/j.freeradbiomed.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 45.Pizzo P. Pozzan T. Mitochondria-endoplasmic reticulum choreography: structure and signaling dynamics. Trends Cell Biol. 2007;17:511–517. doi: 10.1016/j.tcb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 46.Psarra AM. Hermann S. Panayotou G. Spyrou G. Interaction of mitochondrial thioredoxin with glucocorticoid receptor and NF-kappaB modulates glucocorticoid receptor and NF-kappaB signalling in HEK-293 cells. Biochem J. 2009;422:521–531. doi: 10.1042/BJ20090107. [DOI] [PubMed] [Google Scholar]
- 47.Reddien PW. Horvitz HR. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat Cell Biol. 2000;2:131–136. doi: 10.1038/35004000. [DOI] [PubMed] [Google Scholar]
- 48.Rundlöf AK. Janard M. Miranda-Vizuete A. Arner ESJ. Evidence for intriguinly complex transcription of human thioredoxin reductase 1. Free Radic Biol Med. 2004;36:641–656. doi: 10.1016/j.freeradbiomed.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 49.Sies H. Jones DP. Oxidative stress. In: Fink G, editor. Encyclopedia of Stress. San Diego, CA: Elsevier; 2007. pp. 45–48. [Google Scholar]
- 50.Stanley BA. Sivakumaran V. Shi S. McDonald I. Lloyd D. Watson WH. Aon MA. Paolocci N. Thioredoxin reductase-2 is essential for keeping low levels of H2O2 emission from isolated heart mitochondria. J Biol Chem. 2011;286:33669–33677. doi: 10.1074/jbc.M111.284612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stenvall J. Fierro-Gonzalez JC. Swoboda P. Saamarthy K. Cheng Q. Cacho-Valadez B. Arner ES. Persson OP. Miranda-Vizuete A. Tuck S. Selenoprotein TRXR-1 and GSR-1 are essential for removal of old cuticle during molting in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2011;108:1064–1069. doi: 10.1073/pnas.1006328108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sulston J. Hodgkin J. Methods. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1988. pp. 587–606. [Google Scholar]
- 53.Sulston JE. Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 54.Sulston JE. Schierenberg E. White JG. Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka T. Hosoi F. Yamaguchi-Iwai Y. Nakamura H. Masutani H. Ueda S. Nishiyama A. Takeda S. Wada H. Spyrou G. Yodoi J. Thioredoxin-2 (TRX-2) is an essential gene regulating mitochondria-dependent apoptosis. EMBO J. 2002;21:1695–1703. doi: 10.1093/emboj/21.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor EB. Rutter J. Mitochondrial quality control by the ubiquitin-proteasome system. Biochem Soc Trans. 2011;39:1509–1513. doi: 10.1042/BST0391509. [DOI] [PubMed] [Google Scholar]
- 57.Timmons L. Court DL. Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 58.Trotter EW. Grant CM. Overlapping roles of the cytoplasmic and mitochondrial redox regulatory systems in the yeast Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:392–400. doi: 10.1128/EC.4.2.392-400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoneda T. Benedetti C. Urano F. Clark SG. Harding HP. Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 60.Zhang H. Go YM. Jones DP. Mitochondrial thioredoxin-2/peroxiredoxin-3 system functions in parallel with mitochondrial GSH system in protection against oxidative stress. Arch Biochem Biophys. 2007;465:119–126. doi: 10.1016/j.abb.2007.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.