A fully automated high-throughput solution X-ray scattering data collection system, developed for protein structure studies at beamline 4-2 of the Stanford Synchrotron Radiation Lightsource, is described.
Keywords: proteomics, structural genomics, X-ray scattering, laboratory automation, SAXS
Abstract
A fully automated high-throughput solution X-ray scattering data collection system has been developed for protein structure studies at beamline 4-2 of the Stanford Synchrotron Radiation Lightsource. It is composed of a thin-wall quartz capillary cell, a syringe needle assembly on an XYZ positioning arm for sample delivery, a water-cooled sample rack and a computer-controlled fluid dispenser. It is controlled by a specifically developed software component built into the standard beamline control program Blu-Ice/DCS. The integrated system is intuitive and very simple to use, and enables experimenters to customize data collection strategy in a timely fashion in concert with an automated data processing program. The system also allows spectrophotometric determination of protein concentration for each sample aliquot in the beam via an in situ UV absorption spectrometer. A single set of solution scattering measurements requires a 20–30 µl sample aliquot and takes typically 3.5 min, including an extensive capillary cleaning cycle. Over 98.5% of measurements are valid and free from artefacts commonly caused by air-bubble contamination. The sample changer, which is compact and light, facilitates effortless switching with other sample-handling devices required for other types of non-crystalline X-ray scattering experiments.
1. Introduction
Structural biologists are increasingly interested in utilizing complementary structural techniques to overcome difficulties encountered in high-resolution structural methods such as macromolecular crystallography and nuclear magnetic resonance. Solution small-angle X-ray scattering (SAXS) is one such technique experiencing a renaissance, aided by advanced synchrotron instrumentation developments and sophisticated computational tools for structural rendering at nanometer resolution (Tsuruta & Irving, 2008 ▶; Putnam et al., 2007 ▶; Petoukhov et al., 2007 ▶; Konarev et al., 2006 ▶; Hura et al., 2009 ▶). It is particularly useful for obtaining whole three-dimensional structures of difficult-to-crystallize systems such as large protein complexes and assemblies from individual high-resolution subcomponent structures, and evaluating crystallographic tertiary and quaternary structures in the context of the physiologically relevant solution environment. Every major synchrotron radiation facility either hosts or is in a process of developing at least one SAXS station suitable for these studies to meet rapidly growing beam time demand. The structural genomics and proteomics communities are seriously considering incorporating solution SAXS into their portfolio of high-throughput structural techniques to complement high-resolution studies, creating additional demand for developing reliable high-throughput solution SAXS data collection systems. As we have witnessed in macromolecular crystallography, automation minimizes human error and leads to efficient use of synchrotron beam time. It also allows high-throughput studies which would hardly be feasible by manual sample handling. In the past few years a few SAXS beamlines have developed or acquired robotic systems for automatic solution sample handling (Round et al., 2008 ▶; Hura et al., 2009 ▶). We have taken a different approach to overcome some of the technical challenges encountered by others while incorporating advanced data collection features, and built a high-throughput solution scattering data collection system at beamline 4-2 (BL4-2), the dedicated structural biology X-ray scattering station at the Stanford Synchrotron Radiation Lightsource (Smolsky et al., 2007 ▶).
2. The automated sample changer
X-ray scattering studies of proteins and macromolecular complexes in solution constitute the majority of experimental activities on SSRL BL4-2 while studies of partially oriented systems such as lipids and fibres are another important class of studies at the beamline. The rapidly configurable instrument on BL4-2 allows us to accommodate these different types of experiments, some of which require additional features for anomalous scattering and time-resolved studies. Several different sample-handling devices are still required to effectively conduct these studies, thus requiring our automated solution scattering system to be rapidly exchangeable and compatible with the existing hardware and software in order to minimize reconfiguration effort while maximizing available beam time. We developed an automated sample changer which is light and compact, and fits on the standard motorized sample stage used for all experiments on BL4-2 (Fig. 1 ▶). It consists of a temperature-controlled sample rack in the standard 96-well microplate format, an XYZ translation arm for positioning a syringe needle, a computer-controlled fluid dispenser (MicroLab 560; Hamilton Company, Reno, NV, USA), an electromagnetic air valve, an open-end quartz capillary (10 µm-thick walls, 1.5 mm diameter) vertically held in the X-ray beam, and a capillary holder which interfaces the capillary, the air valve and the fluid dispenser via a thin Tygon tube. The capillary is housed in an aluminium sheath which makes direct contact with the sample rack for temperature control. The fluid dispenser carries two independently operated glass syringes: a smaller sample syringe (100 or 250 µl) for precise sample aliquot delivery and data collection, and a larger cleaning syringe (1 ml) for flushing and cleaning of the capillary. Each syringe is coupled with a four- or eight-port valve to select a syringe input among water and a few different cleaning solutions. The output of the sample syringe is always set to the syringe needle via a short piece of Tygon tubing, which is filled with purified water for precise positioning of the sample in the capillary. The cleaning syringe is connected to the outlet immediately above the quartz capillary via a short piece of PTEF tubing, which is also filled with water or one of the cleaning solutions. Compressed air is introduced to dry the capillary by opening the air valve just above the quartz capillary.
Figure 1.
Schematic view of the key hardware components of the sample changer (a). It consists of a motorized XYZ translation arm (A), a syringe needle (B), a temperature-controlled sample rack in the 96-well microplate format (C), a temperature-controlled capillary holder assembly (D) and a quartz capillary. The capillary holder assembly (D), made of anodized aluminium, interfaces the capillary to compressed air via a valve for drying, as well as two syringes, for capillary cleaning and sample aliquot handling. The gentle compression of the syringe needle holder onto the capillary holder assembly makes an air-tight O-ring seal, enabling precise positional control of the sample aliquot within the quartz capillary. The inset (b) depicts the capillary holder assembly in detail. It consists of three parts: an air valve (D1) that switches the compressed air flow for drying the capillary, an interface part (D2) with internal channels (as outlined in the drawing) connecting the capillary to the cleaning syringe, the air valve and the syringe needle, and a capillary holder (D3) into which the quartz capillary is glued. The O-rings sealing the connections between (D2) and (D3) as well as between (D2) and the syringe needle holder are also depicted. Samples are placed on the sample rack in two different patterns (c) for dilution series (columns 1 and 2) or for titration series (columns 4 and 5): the white wells represent buffer solutions, the coloured wells protein samples and the blue wells empty.
A typical set of measurements proceeds as follows. The XYZ arm positions the syringe needle into a microtube containing a sample aliquot on the sample rack. The sample syringe aspirates the sample (typically 20–30 µl) into the syringe needle. Then the XYZ arm positions the needle to the quartz capillary in which the sample is released. The rubber gasket at the bottom of the needle and the O-ring at the top of the capillary holder assembly together achieve an air-tight seal, allowing precise control of the sample in the capillary. Immediately after triggering the X-ray data acquisition system the sample syringe oscillates the sample up and down at a typical rate of 1 µl s−1 in the X-ray beam during X-ray exposures. Once a set of exposures have been completed, the sample syringe injects water through the needle to flush its interior and the capillary to eliminate the irradiated sample. A second cleaning solution can be used to clean the needle if desired. Additional fluids, such as water, detergent and ethanol solutions, are then sent successively by the cleaning syringe to thoroughly clean the capillary. This is followed by final water flushing and air drying of the capillary by opening the air valve directly above the capillary. Finally, the needle moves out of the capillary and aspirates 10 µl of air into it, separating the next sample aliquot from the water filling the Tygon tubing.
The aluminium sheath surrounding the quartz capillary is equipped with a pair of standard SMA 905 optic fiber fittings a few millimeters below the X-ray exposure point for UV/vis spectroscopy measurements using a fiber optic spectrometer (S2000, Ocean Optics, Dunedin, FL, USA), which usually follow each set of X-ray exposures, providing an accurate means of recording protein concentration practically in situ (Fig. 2 ▶). The system can be programmed to recover each sample aliquot back into the microtube prior to cleaning the capillary; for instance, to check physiological activities after X-ray exposures. The system provides an option to translate the sample only in one direction in the beam to minimize physical disturbance which can harm highly sensitive samples instead of repetitively oscillating up and down.
Figure 2.
Photograph of the sample changer hardware. Depicted are the motorized XYZ translation arm (A), the syringe needle (B), the temperature-controlled sample rack (C), and the temperature-controlled capillary holder assembly (D). The X-ray capillary is housed within a protective aluminium sheath (E) with two pairs of X-ray and UV/vis ports, perpendicular to the capillary long axis. A piece of plastic tubing is connected to the bottom opening of the capillary for drain collection (not depicted).
2.1. Blu-Ice software
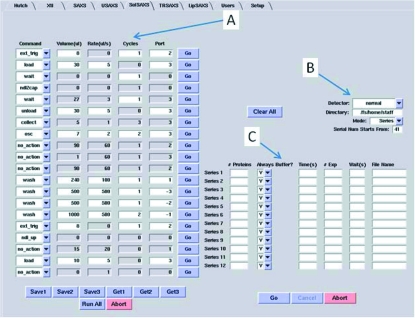
The sample changer is controlled by a graphical user interface SolSAXS ‘tab’ (Fig. 3 ▶) in a version of Blu-Ice software (McPhillips et al., 2002 ▶) specifically developed for non-crystalline X-ray scattering studies (Liu et al., 2012 ▶). The left panel of this tab defines a sequence of commands that the sample changer performs for each sample. These commands, requiring up to four parameters, control sample delivery, capillary cleaning, data collection and external triggering of auxiliary equipment such as the UV spectrophotometer. The upper-right panel defines the data acquisition parameters common to all sample series entered below it. Each of the 12 columns on the sample rack is designated as a series, involving at least one buffer and one protein sample. Sample aliquots need to follow one of the two different patterns which are depicted in Fig. 1(c) ▶ for dilution/concentration series (columns 1 and 2) or titration pairs (columns 4 and 5). The option between these two patterns enables maximizing the number of samples in the rack. Each series can be measured either in the dilution/concentration series or in the titration series by selecting ‘Series’ or ‘Pairs’ (for titration series) in the field ‘Mode’. Each series in the lower-right panel (C) corresponds to a column, numbered from 1 through 12 on the sample rack. The user defines how many protein samples are placed in each series as well as the exposure parameters for X-ray data acquisition. In the case of a dilution series the field ‘Always Buffer?’ enables the user to choose whether the buffer solution is measured for each protein solution (Y) or only twice, at the beginning and the end of each series (N).
Figure 3.
The SolSAXS Blu-Ice tab showing a set of sample handling and exposure command sequences (A) and global data acquisition parameters (B). A separate set of data acquisition parameters, for example frequency of blank (buffer) measurement and number/length of X-ray exposures, can be defined for each column of the sample rack (C).
There are currently 22 commands in four categories: the commands acting on the main data collection system (e.g. wait, collect, ext_trig), those controlling the XYZ arm and therefore the position of the needle (e.g. ndl_up/to/down/by, ndl2cap, home), those for controlling the fluid dispenser (e.g. init, load, unload, flush, wash), and the combination commands operating multiple hardware components in a specific order (osc, recover). Each command takes up to four parameters. For instance, the ‘ext_trig’ command generates a TTL (transistor–transistor logic) level signal to one of three outputs for controlling the air valve and triggering the spectrophotometer, requiring two parameters: the duration of the signal in seconds and the output port. The ‘wash’ command combines a syringe aspiration and injection. It takes four parameters: solution volume in µl to aspirate/expel, flow rate in µl s−1, number of repetitions and the input port through which a fluid is aspirated. The ‘osc’ command pushes and pulls the sample syringe plunger repeatedly to oscillate a defined volume of the sample at a defined rate during X-ray exposures. The Blu-Ice software sets the number of sample oscillation required according to exposure parameters for each series. Up to three sets of command sequences can be saved and recalled for later use.
3. System performance
A typical sequence of automated sample loading, recording ten 3 s exposures, a series of capillary cleaning and drying takes 3.5 min, which can be shortened to less than 3 min with a simplified cleaning sequence. The system has been highly reliable, but very occasionally recorded erroneous diffraction images caused by the X-ray beam hitting air bubbles or aliquot meniscus. A streak of parasitic scattering near the beam stop and/or an abnormally high level of beam transmission through the sample aliquot was observed in 1.5% of the total 6750 images recorded during the five separate sets of measurements (8–20 exposures per sample aliquot) on 262 protein samples over a five-month period in the early stage of commissioning the system in 2009. Some of the invalid images were due to the empty water reservoir for capillary cleaning. The failure rate, ranging from 0 to 5.85% in each of five sets of measurements, appears dependent upon physical properties such as viscosity and surface tension of the sample solutions and the amount of minuscule protein deposits on the quartz capillary. UV spectra were simultaneously recorded for a subset of these data using the integrated UV/vis spectrophotometer, and showed a satisfactory linearity in the protein concentration range from 0.25 to 10 mg ml−1 for most proteins.
4. Discussion
The automated solution scattering system presented here achieves high reliability of sample handling and data acquisition by directly pipetting out a sample aliquot into a quartz capillary. This approach eliminates problems associated with transferring the sample aliquot via long plastic tubing into the sample cell. In an earlier version of our sample changer we indeed tested this approach but often experienced the loss of protein aliquots owing to air bubbles breaking an aliquot apart into tiny portions too small to position them in the X-ray beam. Furthermore, moving a sample aliquot through long lines of tubing often leads to accidental dilution or contamination of precious samples. Our current approach avoids the problems associated with the long tubing.
The sample changer system presented here also maintains compatibility with the flow-through chromatography approach (Mathew et al., 2004 ▶; David & Pérez, 2009 ▶). This is accomplished by connecting the syringe needle, placed in the quartz capillary, directly to the chromatography column outlet.
The vertical orientation of the capillary imposes some limits on the maximum width of the beam to avoid reflections from the capillary walls. We typically use a slit-collimated X-ray beam of 0.3 mm × 0.5 mm FWHM (vertical × horizontal) at the sample position. At this beam size the photon flux at the sample is approximately 2 × 1012 photons s−1 at 11 keV. The necessary reduction of the horizontal width lowers the available flux at the sample by about 20% compared with a set-up using a horizontal capillary. This reduction in intensity is compensated for by a small increase in exposure time. Gentle oscillation or one-way translation of a protein solution aliquot in the quartz capillary during each X-ray exposure at a typical flow rate of 1–2 µl s−1 can be used to limit radiation dosage below the level discussed by Kuwamoto et al. (2004 ▶). The sample changer is compatible with all commonly used solution conditions and poses no additional constraints on selection of experimental condition. For instance, we have successfully measured protein solutions containing 40% w/v glycerol at 288 K. Flushing the quartz capillary with several cleaning solutions prior to each set of X-ray exposures ensures that the capillary is always clean, minimizing air-bubble contamination. The sample volume necessary for a measurement is satisfactorily small for all but the most precious protein systems. Improvements are under way to minimize the sample volume further. Currently the sample changer is operated with the X-ray capillary in air using a pair of thin mica windows separating the capillary from the vacuum up- and downstream of the sample position. This out-of-vacuum design slightly increases the background signal owing to the scattering from the mica windows and the small air gap around the capillary, but it greatly facilitates the maintenance of the device, which was particularly important during the development of the system. However, our sample changer design can be easily adapted to an in-vacuum capillary and we are currently in the process of developing this.
The automated solution scattering system has quite significantly facilitated solution SAXS measurements for structural biology studies on BL4-2. The high level of automation relieves experimenters from manual sample handling while minimizing sample handling errors in data collection. The system is very user-friendly and intuitive to use owing to its Blu-Ice graphical interface. Furthermore, two-dimensional SAXS patterns are immediately reduced to fully processed one-dimensional scattering curves using the SasTool program (Liu et al., 2012 ▶). UV spectroscopy data are processed in a batch mode by another in-house program AbsTool (Liu et al., 2012 ▶). The integrated data collection environment enables the user to concentrate on evaluating data quality and revising data collection strategy if necessary in a timely fashion, resulting in highly effective use of beam time. The sample changer hardware is compact and weighs just a couple of kilograms, facilitating frequent configuration changes with other X-ray scattering experiments including lipid and fibre diffraction.
Acknowledgments
The authors acknowledge K. Sartain and C. Weidner (SSRL) for hardware fabrication. AM thanks her collaborators and beamline users who made available their data for evaluation purposes and acknowledges useful discussion with Manfred Roessle (EMBL-Hamburg), Greg Hura and Michal Hammel (ALS), Petra Pernot (ESRF) and Javier Perez (SOLEIL). TMW thanks Kristin Schmidt (SSRL) for her assistance in preparing the graphics of Figs. 1 ▶ and 2 ▶. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the US Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program (5 P41 RR001209). The project described and its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
References
- David, G. & Pérez, J. (2009). J. Appl. Cryst. 42, 892–900.
- Hura, G. L., Menon, A. L., Hammel, M., Rambo, R. P., Poole, F. L., Tsutakawa, S. E., Jenney, F. E., Classen, S., Frankel, K. A., Hopkins, R. C., Yang, S. J., Scott, J. W., Dillard, B. D., Adams, M. W. & Tainer, J. A. (2009). Nat. Methods, 6, 606–612. [DOI] [PMC free article] [PubMed]
- Konarev, P. V., Petoukhov, M. V., Volkov, V. V. & Svergun, D. I. (2006). J. Appl. Cryst. 39, 277–286.
- Kuwamoto, S., Akiyama, S. & Fujisawa, T. (2004). J. Synchrotron Rad. 11, 462–468. [DOI] [PubMed]
- Liu, P., McPhillips, S. E., Song, J., Weiss, T. M. & Tsuruta, H. (2012). In preparation.
- McPhillips, T. M., McPhillips, S. E., Chiu, H.-J., Cohen, A. E., Deacon, A. M., Ellis, P. J., Garman, E., Gonzalez, A., Sauter, N. K., Phizackerley, R. P., Soltis, S. M. & Kuhn, P. (2002). J. Synchrotron Rad. 9, 401–406. [DOI] [PubMed]
- Mathew, E., Mirza, A. & Menhart, N. (2004). J. Synchrotron Rad. 11, 314–318. [DOI] [PubMed]
- Petoukhov, M. V., Konarev, P. V., Kikhney, A. G. & Svergun, D. I. (2007). J. Appl. Cryst. 40, s223–s228.
- Putnam, C. D., Hammel, M., Hura, G. L. & Tainer, J. A. (2007). Q. Rev. Biophys. 40, 191–285. [DOI] [PubMed]
- Round, A. R., Franke, D., Moritz, S., Huchler, R., Fritsche, M., Malthan, D., Klaering, R., Svergun, D. I. & Roessle, M. (2008). J. Appl. Cryst. 41, 913–917. [DOI] [PMC free article] [PubMed]
- Smolsky, I. L., Liu, P., Niebuhr, M., Ito, K., Weiss, T. M. & Tsuruta, H. (2007). J. Appl. Cryst. 40, s453–s458.
- Tsuruta, H. & Irving, T. C. (2008). Curr. Opin. Struct. Biol. 18, 601–608. [DOI] [PMC free article] [PubMed]