Abstract
We report altogether 3-μs molecular dynamics (MD) simulations of the antimicrobial peptide CM15 to systematically investigate its interaction with two model lipid bilayers, pure POPC and mixed POPG:POPC (1:2). Starting with either an α-helical or a random-coil conformation, CM15 is found to insert into both bilayers. Peptide-lipid interaction is stronger with the anionic POPG:POPC than the zwitterionic POPC, which is largely attributed to the electrostatic attraction between CM15 and the negatively charged POPG. Simulations initiated with CM15 as a random coil allowed us to study peptide folding at the lipid-water interface. Interestingly, CM15 folding appears to be faster in POPC than POPG:POPC, which may be explained by a lower activation energy barrier of structural rearrangement in the former system. Our data also suggest that compared with the random-coil conformation, CM15 in a pre-folded α-helix has significantly reduced interactions with the lipids, indicating that peptide initial structures may bias the simulation results considerably on the 100-ns timescale. The implications of this result should be considered when preparing and interpreting future AMP simulations.
1. Introduction
Many living organisms can produce small, cationic and amphipathic peptides that exhibit a wide spectrum of antimicrobial activity against Gram-positive and Gram-negative bacteria, fungi, viruses and parasites [1, 2, 3, 4, 5]. These antimicrobial peptides (AMPs) are part of the innate immune system in a large number of species, where they form the first line of defense against pathogenic invasion. While AMPs acting on intracellular targets have received increasing attention, the majority of them target the cellular membranes through disrupting the physical integrity of the bilayers. Many AMPs in the latter category self-organize to form pores once the peptide/lipid ratio is beyond a certain threshold [6, 7, 8, 9, 10]. These AMP-induced pores destroy the transmembrane electrochemical gradient, thereby, causing osmolysis, cell swelling, and eventually, cell death.
Apart from their antimicrobial activity, some AMPs also cause lysis of human red blood cells (hemolytic), making them unsuitable for therapeutic use. For instance, melittin, an AMP with 26 amino acids and a +7 net charge, is a hemolytic peptide first isolated from the venom of European honeybee [11]. In comparison, a well-studied non-hemolytic peptide is cecropin A from the silk moth [12], which contains 35 amino acids and a +6 net charge. Combining residues 1–7 of cecropin A and residues 2–9 of melittin, a synthetic AMP, CM15 (KWKLFKKIGAVLKVL-NH2), has been found to retain the potency of cecropin A without the hemolytic activity of melittin [13].
Understanding the selectivity of different AMPs for mammalian and bacterial membranes is of obvious interest in the development of these peptides as novel antibiotic agents. Molecular dynamics (MD) simulations, which provide an atomic-resolution model of the system under investigation, have been used extensively to investigate AMP-lipid interactions [14, 15, 16, 17, 18, 19, 20]. In particular, simulations have been successfully applied to explore pore formation induced by AMPs [21, 22]. However, relatively few MD studies focused on the comparison of AMP interactions with different types of lipid bilayers([16] and references therein). Furthermore, although many AMPs adopt a random-coil conformation in water and only fold into α-helices upon membrane insertion [23, 7], most AMP simulations in the literature use a pre-folded α-helix as the initial structure. To further reduce computational cost, the peptide is often placed in a parallel orientation close to the membrane (≤10 Å). These conditions inevitably affect the simulation results, especially the early stage of peptide binding and insertion.
In this study, we report over 3 μs simulations on CM15 to systematically investigate its interaction with two model lipid bilayers, POPC and mixed POPG:POPC (1:2), which mimic the mammalian and bacterial membranes, respectively. The peptide is placed ~30 Å away from the bilayer in either a random-coil or an α-helical initial conformation, in order to analyze the impact of initial conditions on simulation results. Altogether ten sets of 100-ns simulations were performed for each peptide-lipid system, with two of the simulations extended to 180 ns. Analysis of these simulations clearly indicates that CM15 binds and inserts into both the anionic POPG:POPC and the zwitterionic POPC. This result is similar with our experimental measurements, which will be described in a separate report. Simulations with initially unfolded CM15 allowed us to study peptide folding induced by membrane insertion. Interestingly, compared with POPG:POPC, CM15 folding appears to be faster in the POPC bilayer. This difference is discussed in the context of peptide-lipid electrostatic interactions in the two systems. Our analysis also shows that compared with the random-coil initial conformation, CM15 with an α-helical structure has a significantly reduced number of interactions with the bilayers. Therefore, even with multiple 100-ns trajectories, simulation results may still be biased by peptide initial conformations, suggesting that the starting conditions should be chosen carefully for AMP simulations on this timescale.
2. Methods
Lipid bilayers
A POPC bilayer with 39 lipid molecules in each monolayer was taken from a 70-ns equilibration performed previously [24]. The system, consisting of 78 POPC and 6169 water molecules, was used to build a POPG:POPC bilayer with a 1:2 mixing ratio. Specifically, 13 out of the 39 POPC molecules in each monolayer were chosen randomly and “mutated” into POPG molecules. Similar to Zhao, et al. [25], we created a racemic mixture of POPG by generating an equal number of molecules in the L- and D- configurations. Using the autoionize plugin of VMD [26], 26 Na+ ions were added to neutralize the charge of the POPG:POPC system. Both bilayers were minimized for 1000 steps and equilibrated under constant temperature and pressure (NPT) conditions for 100 ps with phosphorus atoms of the lipids restrained (spring constant k=10 kcal/mol/Å 2). A 500-ps NPT simulation was then performed for the POPC bilayer with all atoms free to move. The area per lipid is 68.2 Å 2 at the end of this simulation, which is in the range of experimental values (68.3±1.5Å 2) [27]. For the mixed POPG:POPC, a 5-ns NPT simulation was performed to further equilibrate this system. The end structures of these simulations were used to build the peptide-lipid systems described below.
Peptide-lipid systems
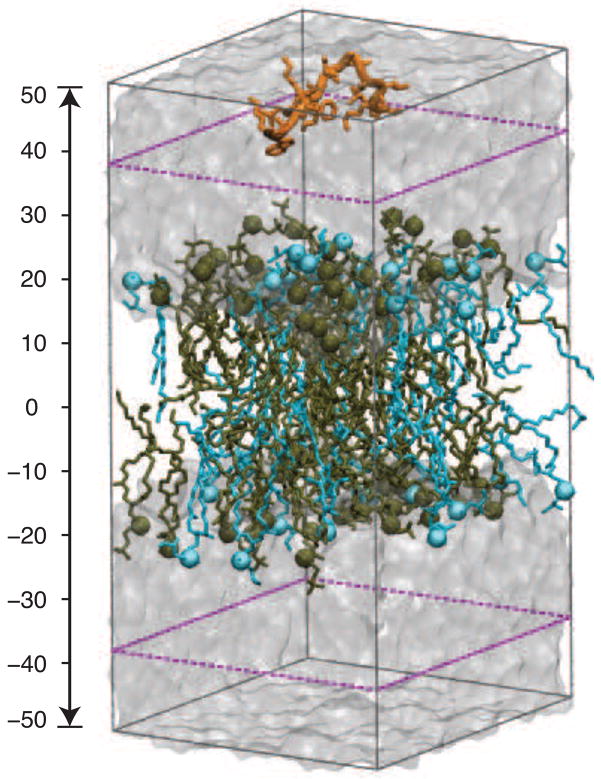
In order to study the effect of initial structure on simulation results, we constructed the CM15-lipid systems with the peptide in either a random-coil or an α-helical conformation. To generate the random-coil structure, the peptide was simulated in a water box with a fully extended initial conformation. Three independent, 1-ns NPT simulations were performed. The 0.5–1 ns of these simulations were combined and clustering analysis was performed using the program GROMACS [28] to select ten representative structures of CM15. These structures were used to build CM15-lipid systems with the equilibrated POPC and POPG:POPC described above. Based on the peptide conformation and lipid species, we will refer to these systems as CM15r-POPC and CM15r-POPG:POPC, respectively. Additionally, a CM15H-POPC system was constructed using the α-helical conformation of the peptide (pdb code 2JMY [29]). Consistent with our experimental conditions, the C-terminus of CM15 is amidated in all peptide-lipid systems. To neutralize the systems, we added 7 Na+ and 13 Cl− to CM15r-POPC and CM15H-POPC, and removed 6 Na+ ions from CM15r-POPG:POPC (leaving 20 Na+ ions). In all systems, CM15 was initially placed on one side of the bilayer, approximately 30 Å above the phosphorus atoms (Fig 1).
Fig 1.
The CM15r-POPG:POPC simulation system. POPG and POPC are colored in blue and tan, respectively, and water molecules are shown as a transparent box. The ‘buffer zone’ is highlighted by purple dashed lines (see Methods section).
MD simulations
Ten sets of 100-ns simulations were performed for each of the peptide-lipid systems described above, two of which were continued to 180 ns to further study peptide insertion (Table 1). All simulations were performed under constant temperature and pressure conditions, with only the dimension along the membrane normal allowed to fluctuate (NPzT). The choice of NPzT over NPT conditions was made based on a 50-ns test simulation of the pure POPC bilayer. This NPT simulation was initiated from the equilibrated POPC bilayer used to construct the CM15-POPC system. Starting from an area per lipid (68.2 Å 2) similar to the experimental value (68.3±1.5Å 2), an 8.4% decrease in area per lipid is observed in this simulation (Fig S1). Based on this result, we chose to perform the peptide binding and insertion simulations under the NPzT conditions. Similar to a previous study [30], the NPzT conditions are found suitable for studying the initial association of a single CM15 with the lipid bilayers.
Table 1.
List of all peptide-lipid simulations.
| System | No. simulations | Length (ns) |
|---|---|---|
| CM15r-POPG:POPC | 8 | 100 |
| 2 | 180 | |
| CM15r-POPC | 8 | 100 |
| 2 | 180 | |
| CM15H-POPC | 8 | 100 |
| 2 | 180 | |
To improve sampling efficiency, a “soft boundary” condition was used to keep CM15 within a 20 Å “buffer zone” of the bilayer in all peptide-lipid simulations. Since the peptide was initially placed 30 Å away from the membrane, it can diffuse freely in water before reaching the boundary. As shown in Fig 1, once CM15 enters the buffer zone from either side, a weak restraining potential (spring constant 3 kcal/mol/Å 2) will be applied whenever its center-of-mass leaves the buffer zone. No external force is applied when the center-of-mass of CM15 is inside the buffer zone. Therefore, the soft boundary condition allows us to improve the sampling efficiency without interferering with the natural dynamics of the peptide.
Force field and simulation protocols
The CHARMM27 force field for proteins [31, 32] and the latest update for lipids (CHARMM36) [33] were used to prepare the aforementioned systems. All simulations were performed with the program NAMD (release 2.7 and 2.8) [34] using a timestep of 2 fs, with bonds involving hydrogens in the peptide constrained using RATTLE [35] and water geometries maintained using SETTLE [36]. The multiple-time-stepping algorithm was used, with short-range forces calculated every step and long-range electrostatics calculated every 2 steps. The cutoff for short-range non-bonded interactions was set to 12 Å, with a switching distance of 10 Å. Assuming periodic boundary conditions, the Particle Mesh Ewald method [37] with a grid density of at least 1/Å 3 was employed for computation of long-range electrostatic forces. The temperature was maintained at 303.15K for all simulations using Langevin dynamics, while the pressure was kept constant at 1 bar using a Nośe-Hoover-Langevin piston [38]. The soft boundary condition in peptide-lipid simulations was enforced through the Tcl force interface of NAMD [34].
Data analysis
Throughout this work, we define the bilayer center as the center of phosphorus atoms from both monolayers and place it at z = 0. To facilitate data analysis, the monolayer in contact with CM15 at the end of a simulation is designated as the ‘upper’ monolayer, regardless of its initial position. The center-of-mass (c.o.m.) distribution of CM15 along the membrane normal (z) is represented by a histogram with a bin width of 1 Å. The lipid contact of each CM15 residue is measured as the average number of non-hydrogen peptide atoms within 3 Å of the lipids. The result is then normalized by the total number of atoms in that residue. To characterize peptide-lipid interactions, the electrostatic (Eelec) and vdW (EvdW ) interaction energy between CM15 and the lipids are calculated using the NAMDEnergy plugin [34]. We also calculated the secondary structure content of CM15 using the programs DSSP [39] and GRO-MACS [40]. Both the interaction energy and the secondary structure content are plotted a function of the c.o.m. location of CM15 along the membrane normal.
3. Results
We performed altogether 30 sets of simulations (Table 1), each at least 100 ns long, to investigate the interaction of CM15 with two model lipid bilayers, POPC and POPG:POPC. The peptide was initially placed ~30 Å away from the bilayers, in either a random-coil (CM15r) or an α-helical (CM15H) conformation. The interaction of CM15 with the bilayers, the conformational change of the peptide, and the effect of initial structure on simulation results are examined through quantitative analysis of CM15 and lipid properties. Below we present these results in detail, and then discuss their implications in the following section.
CM15 inserts into both POPG:POPC and POPC
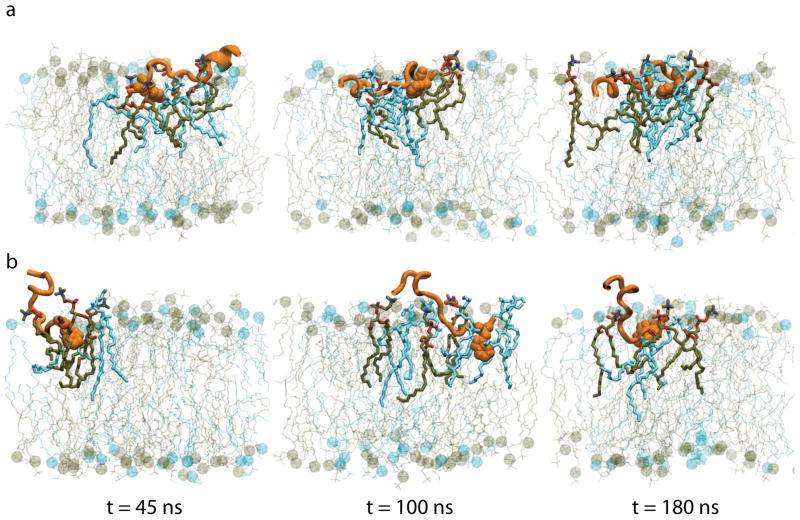
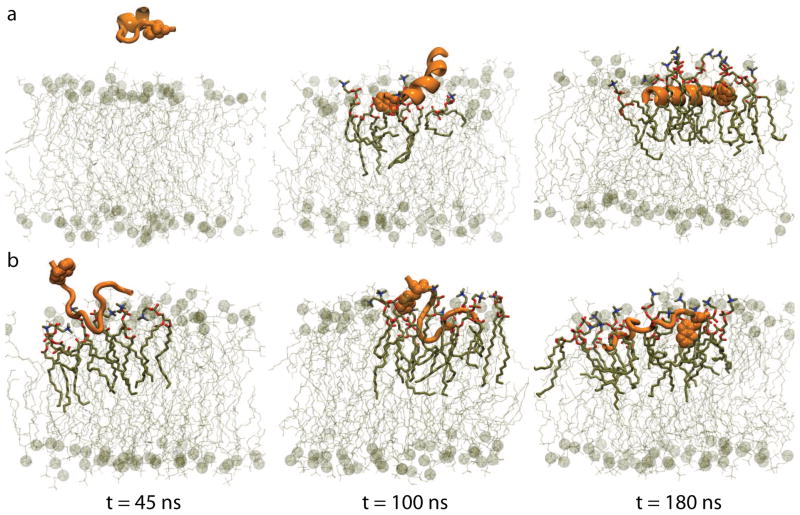
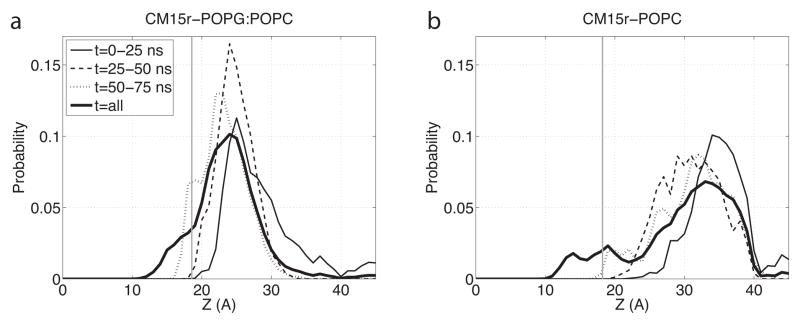
Representative snapshots from the 180-ns simulations of CM15r-POPG:POPC and CM15r-POPC are shown in Fig 2 and Fig 3, respectively. As revealed by these snapshots, CM15 binds and inserts into both POPG:POPC and POPC. Combining data from all ten sets of simulations for each system, we constructed the c.o.m. distribution of CM15 along the membrane normal (Fig 4). We define the peptide as ‘inserted’ if its c.o.m. lies below the phosphorus atoms from the upper monolayer. Based on the analysis of all simulation data, CM15 is inserted into both lipid bilayers in a significant portion of the trajectories: 12.2% for POPG:POPC and 10.6% for POPC (Table 2).
Fig 2.
Snapshots of two 180-ns CM15r-POPG simulations. The CM15 residue Trp2 is shown in vdW representation. Lipid molecules within 3 Å of the peptide are highlighted, with POPG and POPC colored in blue and tan, respectively.
Fig 3.
Snapshots of two 180-ns CM15r-POPC simulations. POPC molecules are colored in tan and lipids within 3 Å of the peptide are highlighted.
Fig 4.
Center-of-mass distribution of CM15 in the POPG:POPC and POPC bilayers. Calculations are performed using all ten sets of simulations for CM15r-POPG:POPC (a) and CM15r-POPC (b) with a 1-Å resolution along the mem- brane normal (z). Results obtained from the first 25, 50, and 75-ns simulations are shown in thin solid, dashed, and dotted lines, respectively, while results obtained from the entire 100-ns or 180-ns trajectories (t=all) are shown in thick solid lines. Lipid bilayers are centered at z = 0 and the average location of phosphorus atoms in the upper monolayer is shown in grey lines.
Table 2.
Interaction of CM15 with POPG:POPC and POPC. The percentage of inserted CM15, the average time of the first stable contact, the average number of hydrogen bonds and salt bridges between CM15 and the lipids are listed. The latter two calculations are performed using simulation snapshots with CM15 inserted into the bilayers.
| System | Inserted (%) | t1-st contact (ns) | H-bond | Salt bridge |
|---|---|---|---|---|
| CM15r-POPG:POPC | 12.2 | 6.1 | 0.9 | 5.9 |
| CM15r-POPC | 10.6 | 13.4 | 0.5 | 3.7 |
| CM15H-POPC | 4.0 | 21.1 | 0.6 | 4.0 |
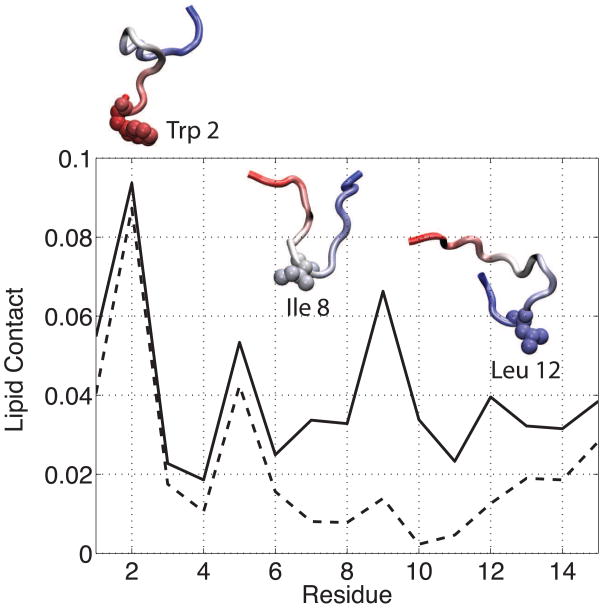
As described in the Methods section, each of the CM15r simulations begins with the peptide placed ~30 A away from the bilayer. During the simulations, CM15 can diffuse freely in water and adopt any orientation when it first reaches the bilayer, thereby, allowing unbiased analysis of the initial peptide-lipid contact. Our analysis suggests that CM15 interacts with the bilayers more frequently through its N-terminus than C-terminus. On average, the hydrophobic tryptophan residue (Trp 2) has the largest number of atoms in contact with the lipids. The average lipid contact of each CM15 residue, normalized by the size of that residue, is shown in Fig 5 for CM15-POPC simulations. Similar result is observed in the CM15r-POPG:POPC runs (Fig S2). These calculations suggest that Trp 2 plays the key role in mediating peptide-lipid interactions. Tryptophan residues are frequently found in AMPs [41, 42, 43, 16], where they drive the partition of the peptides between lipids and water. Experimentally, mutagenesis and omission studies have shown that the tryptophan residues in cecropin A and melittin are indispensable for the activity of the two AMPs [44, 45, 46]. Although no experimental data is available for CM15, our simulation results suggest that, similar to its two parent peptides, Trp 2 is likely crucial to the membrane binding and insertion of CM15.
Fig 5.
The number of peptide atoms in contact with lipid molecules in the CM15r-POPC (solid) and CM15H-POPC (dashed) simulations. The lipid contact is measured as the number of non-hydrogen atoms within 3 Å of POPC. Results are averaged over all simulations and normalized by the total number of atoms in a residue. Three representative conformations at the lipid-water interface are shown for CM15r-POPC. The peptide is colored by its primary sequence (red: N-terminus, blue: C-terminus) and the residue closest to the lipid bilayer in each structure is shown in vdW representation.
It is worth noting that although the simulations presented here are longer than many previous studies, they have not reached the equilibrium state of CM15 in the two bilayers yet. This is partly reflected in the continuously varying c.o.m. distributions constructed from different periods of the trajectories (Fig 4). Given longer simulation time, the results presented above should converge to their equilibrium values, and more peptides will likely insert into the bilayers. Nevertheless, calculations based on our current data clearly reveal the partition of CM15 into both POPG:POPC and POPC. Additionally, since the trajectories are of equal length, their comparison allows us to characterize the different interactions between CM15 and the two lipid membranes on the ~100-ns timescale.
Compared with the zwitterionic POPC, CM15 binds to the negatively charged POPG:POPC much faster. The c.o.m. distributions constructed using the first 25 ns of simulations have their peaks at z = 25 Å (POPG:POPC) and z = 34 Å (POPC), respectively, reflecting the strong electrostatic attraction between the peptide and the anionic POPG molecules. The faster association of CM15 with POPG:POPC is also characterized by the first-stable-contact-time between the peptide and the lipids: On average, CM15 first establishes a stable contact in 6.1 and 13.4 ns with POPG:POPC and POPC, respectively (Table 2). Furthermore, the average duration of such stable contacts is approximately four times longer in POPG:POPC than POPC, indicating that peptide binding to the former bilayer is not only faster, but also stronger. Here, a contact is considered stable if any non-hydrogen atom of CM15 is within 3 Å of the lipids for at least 1 ns. While the results clearly depend on the cutoff parameters, a similar relation between the two systems is observed by varying the cutoff values (data not shown).
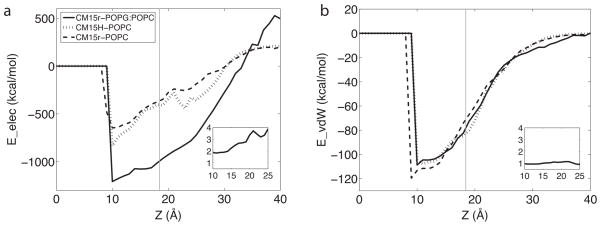
The preference of CM15 for the POPG:POPC bilayer can be explained by a highly favorable electrostatic interaction between the cationic peptide and the anionic POPG molecules. Through post-processing the trajectories, we calculated the electrostatic interaction energy (Eelec) between CM15 and the lipids. As shown in Fig 6, Eelec in the CM15r-POPG:POPC system is ~2–4 times stronger than CM15r-POPC. In line with this result, the peptide-lipid salt bridge and hydrogen bond interactions are both increased in the former system (Table 2). Interestingly, while the electrostatic interaction is highly favorable between CM15 and POPG:POPC, the vdW interaction energy profiles (EvdW ) are remarkably similar in the two peptide-lipid systems (Fig 6 b). As the latter interaction depends closely on atom contact, these results indicate that CM15 forms similar ‘amount’ of contact with the two bilayers. Indeed, distributions of the total number of lipid contacts are comparable in POPG:POPC and POPC (Fig S3). It is worth noting that Fig 6 only characterizes the enthalpic contribution from peptide-lipid interactions, while CM15 binding is also driven by an entropic contribution and an enthalpic contribution from peptide-solvent interactions, the latter of which may counterbalance the favorable peptide-lipid interactions. Nevertheless, Fig 6 suggests that the difference in peptide binding and insertion between CM15-POPG:POPC and CM15-POPC can be largely attributed to the different electrostatics in the two systems.
Fig 6.
The average electrostatic (a) and vdW (b) interaction energy between CM15 and lipid molecules in the CM15r-POPG:POPC (solid), CM15r-POPC (dashed) and CM15H-POPC (dotted) simulations. The calculation is performed with the program NAMD [34] using all ten sets of simulations for each system. Lipid bilayers are centered at z = 0 and the average location of phosphorus atoms in the upper monolayer is shown in grey lines. The inset figures show the ratio of CM15r-POPG:POPC and CM15r-POPC results for z = 10 to 25 Å.
Folding may occur at different speeds in POPG:POPC and POPC
Many AMPs, including CM15, adopt a random-coil structure in water and fold into an α-helix in the presence of lipids [23, 7]. However, as peptide folding is often time-consuming, the majority of AMP studies in the literature use a pre-folded, α-helical conformation as the initial structure of the peptide. In this work, we chose a unique, random-coil conformation for CM15 in each of the ten simulations with POPG:POPC or POPC (see Methods section). These conditions allow us to systematically study the early events of peptide folding at the lipid-water interface.
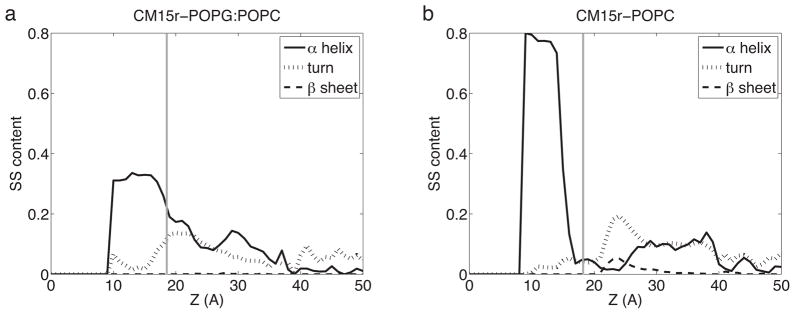
Overall, CM15 folding is observed in both POPG:POPC and POPC simulations. Using the programs DSSP [39] and GROMACS [40], we calculated the secondary structure content of the peptide and its distribution along the membrane normal (Fig 7). The average α-helical content of CM15 in water (z ≤ 30 Å) is found to be 3.8% and 6.2% in POPG:POPC and POPC, respectively, consistent with experimental findings that CM15 primarily adopts a random-coil conformation in water. However, as discussed earlier, we cannot exclude the possibility that peptide folding may occur in water given longer simulations. Despite this potential caveat, our data clearly reveal a stronger tendency of CM15 to form an α-helix inside the bilayer. The average α-helical content of inserted CM15 is found to be 28.3% in POPG:POPC and 52.1% in POPC. Note that a substantial portion of inserted CM15 remains as random-coils (67.7% in POPG:POPC and 20.0% in POPC), suggesting that helicity is not required for peptide-lipid interaction and that folding may continue after CM15 insertion.
Fig 7.
Secondary structure content distribution of CM15 in the POPG:POPC (a) and POPC (b) bilayers. Calculations are performed using all ten sets of simulations with a 1-Å resolution along the membrane normal (z). Secondary structure is calculated using GROMACS [40] and the program DSSP [39]. Lipid bilayers are centered at z = 0 and the average location of phosphorus atoms in the upper monolayer is shown in grey lines.
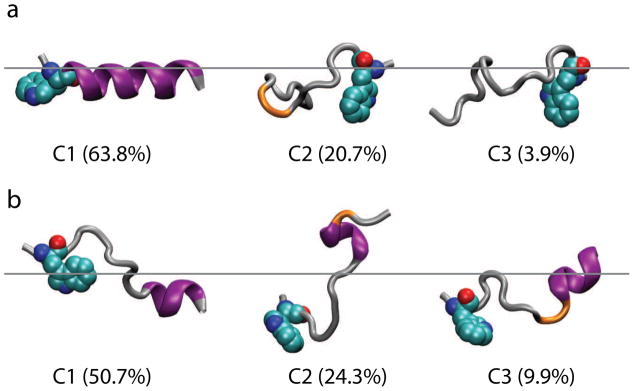
Compared with the anionic POPG:POPC, CM15 folding appears to be faster in the zwitterionic POPC. In one of the 180-ns CM15r-POPC simulations, the peptide folds into a nearly ideal α-helix (Fig 3), while only partially-folded α-helices are observed in the 180-ns POPG:POPC simulations (Fig 2). Using the program GROMACS [40], we performed clustering analysis to extract representative conformations of inserted CM15. Based on the parameter scan shown in Fig S4, a cutoff of 3.2 Å and 1.5 Å were chosen for the POPG:POPC and POPC clustering analysis, respectively. Centroid structures from the first three clusters, which represents 84.9% (POPG:POPC) and 88.3% (POPC) of the total populations, are shown in Fig 8. While the majority of CM15 residues participate in the α-helix formation in POPC, only partial folding is observed in POPG:POPC. Combining data from all the trajectories, a higher α-helical content is observed in POPC (52.1%) than POPG:POPC (28.3%). These results suggest that folding is faster in the former bilayer on the ~100-ns timescale.
Fig 8.
Secondary structure of inserted CM15. Clustering analysis was performed using GROMACS [40] for CM15r-POPC (a) and CM15r-POPG:POPC (b). The first three clusters and their populations are shown. CM15 is colored by its secondary structure calculated using the DSSP program [39] (purple: α-helix, orange: turn, silver: random coil). The residue Trp 2 is shown in vdW representation. The approximate locations of phosphorus atoms in the upper monolayers are shown in grey lines.
The different folding speeds in POPG:POPC and POPC may be explained by the strong electrostatic interaction between CM15 and POPG (Fig 6). While this interaction is highly favorable for peptide binding, it appears to increase the activation energy barrier of structural rearrangement: The strong hydrogen bond and salt bridge interactions between the peptide and lipids contribute to slow down helix formation in the negatively charged POPG:POPC (Table 2). Note, however, that more ‘turn’ conformations are adopted by the inserted CM15 in POPG:POPC than POPC (Fig 7). Since turns may serve as precursors of α-helices, more folding events are likely to occur in the former bilayer as the simulations continue. We have recently completed a 1-μs simulation with multiple copies of CM15 in the POPG:POPC bilayer, which confirms that folding continues after CM15 insertion and that the majority of peptides eventually adopt an α-helical conformation (data not shown).
Initial conformation affects CM15 binding and insertion
As mentioned earlier, many AMP studies use a pre-folded, α-helical conformation as the initial structure, and place the peptide in a parallel orientation close to the bilayer (≤ 10 Å). While these measures significantly reduce the computational cost, they inevitably introduce certain artifacts into the simulations, especially the early stage of peptide binding and insertion. To investigate the impact of initial conditions on AMP simulations, we performed ten CM15H-POPC simulations in addition to the CM15r runs described above, in which the peptide starts as an α-helix and is placed ~30 Å away from the bilayer. Comparison of these simulations suggests that the initial conformation plays an important role in the association of CM15 with lipid bilayers.
As shown in Fig S5, similar to the CM15r-POPC runs, the peptide binds and inserts into the POPC bilayer in the CM15H-POPC simulations. However, unlike in the former system, CM15 only inserted into the bilayer in one of the two 180-ns simulations, reflecting reduced peptide-lipid interactions. Furthermore, the first-stable-contact-time between CM15 and POPC increases from 13.4 to 21.1 ns, and the peak of c.o.m. distribution is shifted from z = 34 Å to z = 38 Å during the first 25-ns trajectories (Fig S6). The percentage of inserted CM15, calculated using all simulation data, drops from 10.6% to 4.0% (Table 2). These results clearly indicate that peptide binding and insertion is reduced when CM15 adopts a pre-folded, α-helical initial structure.
The difference in the CM15r and CM15H simulations cannot be explained by enthalpic contributions from peptide-lipid interactions: As shown in Fig 6, the two systems have similar electrostatic and vdW interaction energy profiles, suggesting that the reduced peptide binding and insertion in the latter system is not due to a less favorable interaction with the bilayer. Further analysis reveals that this difference is caused by the different flexibility of CM15 in the two initial conformations, reflecting an entropic contribution. As shown in Fig 5, while the tryptophan serves as the main anchoring residue in both CM15r and CM15H simulations, the remaining residues have significantly different lipid contact profiles. The peptide starting from a random-coil conformation frequently interacts with the bilayer through residues Lys 7 to Leu 12; in contrast, CM15 in an α-helical initial structure has much less interaction with the lipids at these sites (Fig 5). The flexibility of a random coil clearly increases the chance of CM15 to form contacts with the bilayers.
To further illustrate the above point, we performed clustering analysis for peptides at the lipid-water interface. Only translation or rotation in the xy plane is used to align the structures, in order to maintain the peptide orientation with reference to the membrane normal. Three representative centroid structures of CM15r-POPC are shown in Fig 5. As revealed by these structures, although Trp 2 remains the key residue mediating peptide-lipid interactions, contact with the bilayer can be initiated by other residues when the peptide adopts a random-coil initial conformation. Upon insertion, CM15 primarily adopts an α-helix in both CM15r and CM15H simulations (Fig 3 and Fig S5), where the peptide forms similar hydrogen bond and salt bridge interactions with the lipids (Table 2). However, due to the limited flexibility, a pre-folded initial structure leads to a significantly reduced number of peptide-lipid contact in the CM15H simulations.
While the physiological relevance of the above result remains to be examined, its implications should be considered when setting up AMP simulations and interpreting their results. Our analysis suggests that on the ~100-ns timescale, CM15 binding and insertion is significantly reduced with a pre-folded initial structure. As the peptide cannot fully unfold on this timescale, conclusions based on these simulations should take into account the effect of peptide initial conformations, which may be even stronger for longer AMPs due to the larger entropic difference in the random-coil and α-helical conformations. In this work, if we only performed ten sets of CM15H-POPC simulations and limited their length to 100 ns, we might have reached the conclusion that CM15 does not insert into the POPC bilayer, since very little peptide-lipid interaction is observed during these simulations. Therefore, even with multiple copies of trajectories on the 100-ns timescale, peptide initial conformation may still affect the simulation results considerably.
4. Discussion
Our simulations of CM15 in POPG:POPC and POPC clearly indicate that the peptide inserts into both lipid bilayers. This result is somewhat unexpected given the non-hemolytic nature of CM15 [13]. Based on the simulations alone, we cannot rule out the possibility that CM15 may still achieve its selectivity for bacterial membranes by only inducing pore formation in POPG:POPC. However, our experiments revealed significant fluorescence leakage in both POPG:POPC and POPC vesicles at a peptide/lipid ratio comparable to that used in the simulations (to be described in a separate report). Previous studies also demonstrated a similar leakage behavior of another non-hemolytic cecropin A-melittin hybrid AMP [47], suggesting that these peptides can indeed disrupt certain zwitterionic lipid bilayers. These results reflect the complex mechanism of AMP selectivity and might be explained by the different compositions of model bilayers and mammalian membranes. The former systems lack the complex components of the latter, such as cholesterol and various membrane proteins. Experimental and simulation studies of the POPC:cholesterol mixture will be the next step to further examine the activity of CM15 in different lipid environments.
Compared with the zwitterionic POPC, the binding and insertion of CM15 is found to be much faster in the anionic POPG:POPC. This result is largely attributed to the strong electrostatic interaction between the peptide and the lipids in the former system. However, as shown in Fig 6, the difference between electrostatic interactions in the two systems gradually decreases as CM15 inserts deeper into the bilayer: A four-fold difference in Eelec at z = 25 Å is reduced to approximately two-fold at z = 10 Å. This trend suggests that electrostatics may contribute more significantly to initial peptide binding than the subsequent insertion. Similar observations have also been made in previous studies, which suggest that electrostatic interactions between AMPs and anionic lipids only provide the long-range attraction to bring the peptides close to the bilayer [21].
The random-coil initial conformation used in our simulations allowed systematic study of peptide folding at the lipid-water interface. Although CM15 folding is observed in both POPG:POPC or POPC, helicity is not required for peptide binding and insertion in either bilayer. In fact, compared with a pre-folded α-helix, CM15 in a random-coil conformation has more flexibility, and thus, a higher probability to establish contact with the lipids. Once inserted into the bilayer, CM15 primarily adopts an α-helical conformation, which is similar to the NMR structure of the peptide in DPC micelles [29]. Interestingly, the strong electrostatic attraction between CM15 and POPG, which is highly favorable for peptide binding and insertion, appears to slow down CM15 folding in the negatively charged bilayer. It will be of interest to examine whether such a result also applies in other cationic AMPs and anionic lipids.
Several caveats should be noted for our simulations: First of all, with 39 lipid molecules in each leaflet, our systems are relatively small. While they allow us to study the initial binding and insertion of a single CM15, a larger lipid patch will be required to further examine the membrane disruption induced by the peptide. In particular, multiple copies of CM15 are likely needed to induce pore formation, as demonstrated by previous studies of magainin and melittin [21, 22]. Secondly, while an NPzT ensemble is sufficient for studying initial peptide binding and insertion in a single-CM15 system, the NPT conditions may be required if pore formation is expected. Studies of larger lipid bilayers with multiple CM15 are currently underway in the McCammon lab. Lastly, as with many AMP simulations, the limited timescale remains a bottleneck of our study. Apart from obtaining longer simulations, coarse-grained force fields [48, 49] and enhanced sampling methods [50, 51] may provide alternative directions for future studies.
5. Conclusions
Using altogether 3 μs of simulations, we investigated the interaction of CM15 with two model lipid bilayers. Our results indicate that CM15 binds and inserts into both POPC and mixed POPG:POPC (1:2), which mimic the mammalian and bacterial membranes, respectively. Compared with the former bilayer, the stronger electrostatic interaction between CM15 and the anionic POPG results in faster binding and insertion in the latter system. The peptide shows a strong tendency to form α-helices inside both membranes, although helicity is not required for binding or insertion. Our analysis also demonstrates that peptide initial conformation has a significant impact on simulation results. Compared with a pre-folded, α-helical conformation, the random-coil initial structure allows CM15 to form contacts with the lipids more frequently. The implications of this result should be considered when preparing and interpreting future AMP simulations.
Supplementary Material
Highlights.
We report over -μs MD simulations of CM15 with POPC and POPG:POPC.
CM15 binds and inserts into both bilayers despite its non-hemolytic nature.
CM15 folding is faster in POPC than POPG:POPC.
Peptide initial conformation affects simulation results on 100-ns timescale.
Acknowledgments
We thank Dr. Peter Tieleman for helpful discussions. Work at the Mc-Cammon group has been supported in part by the National Science Foundation (NSF), the National Institutes of Health (NIH), Howard Hughes Medical Institute, Center for Theoretical Biological Physics, the National Biomedical Computation Resource, and the NSF supercomputer centers. Work at the Kim group is supported by an NSF CAREER award to J.E.K. and a UCSD MBTG NIH training grant (GM08326) to D.E.S. The authors appreciate the computational resources provided by the Texas Advanced Computing Center (TG-MCA93S013 and TG-MCB100141). This research was also supported in part by the NSF through TeraGrid Supercomputer resources provided by a directors discretionary grant from the National Institute for Computational Science (TG-CHE100128) and the San Diego Supercomputer Center (TG-MCB090110) to Dr. Ross Walker.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zasloff M. Antimicrobial peptides of multicellular organism. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 2.Brogden KA. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat Rev Mol Cell Biol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 3.Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 4.Matsuzaki K. Control of cell selectivity of antimicrobial peptides. Biochim Biophys Acta. 2009;1788:1687–1692. doi: 10.1016/j.bbamem.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen L, Haney E, Vogel H. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotech. 2011;29:464–472. doi: 10.1016/j.tibtech.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Huang HW. Molecular mechanism of antimicrobial peptides: the origin of cooperativity. Biochim Biophys Acta. 2006;1758:1292–1302. doi: 10.1016/j.bbamem.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Sato H, Feix J. Peptide-membrane interactions and mechanisms of membrane destruction by amphipathic alpha-helical antimicrobial peptides. Biochim Biophys Acta Biomembr. 2006;1758:1245–1256. doi: 10.1016/j.bbamem.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 8.Lee MT, Huang WC, Chen FY, Huang HW. Mechanism and kinetics of pore formation in membranes by water-soluble amphipathic peptides. Proc Natl Acad Sci USA. 2007;105:5087–5092. doi: 10.1073/pnas.0710625105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian S, Wang LYW, Huang HW. Structure of transmembrane pore induced by bax-derived peptide: evidence for lipic pores. Proc Natl Acad Sci USA. 2008;105:17379–17383. doi: 10.1073/pnas.0807764105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almeida PF, Pokorny A. Mechanisms of antimicrobial, cytolytic, and cell-penetrating peptides: From kinetics to thermodynamics. Biochemistry. 2009;48:8083–8093. doi: 10.1021/bi900914g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raghuraman H, Chattopadhyay A. Melittin: a membrane-active peptide with diverse functions. Biosci Rep. 2007;27:189–223. doi: 10.1007/s10540-006-9030-z. [DOI] [PubMed] [Google Scholar]
- 12.Tamang DG, Saier MH. The cecropin superfamily of toxic peptides. J Mol Microbiol Biotech. 2006;11:94–103. doi: 10.1159/000092821. [DOI] [PubMed] [Google Scholar]
- 13.Andreu D, Ubach J, Boman A, Wåhlin B, Wade D, Merrifield R, Boman H. Shortened cecropin A-melittin hybrids. Significant size reduction retains potent antibiotic activity. FEBS Lett. 1992;296:190–194. doi: 10.1016/0014-5793(92)80377-s. [DOI] [PubMed] [Google Scholar]
- 14.Tieleman D, Sansom M. Molecular dynamics simulations of antimicrobial peptides: from membrane binding to transmembrane channels. Int J Quantum Chem. 2001;83:166–179. [Google Scholar]
- 15.Kandasamy SK, Larson RG. Effect of salt on the interactions of antimicrobial peptides with zwitterionic lipid bilayers. Biochim Biophys Acta Biomembr. 2006;1758(9):1274–1284. doi: 10.1016/j.bbamem.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 16.Mátyus E, Kandt C, Tieleman D. Computer simulation of antimicrobial peptides. Curr Med Chem. 2007;14:2789–2798. doi: 10.2174/092986707782360105. [DOI] [PubMed] [Google Scholar]
- 17.Hsu J, Yip C. Molecular dynamics simulations of indolicidin association with model lipid bilayers. Biophys J. 2007;92:L100–L102. doi: 10.1529/biophysj.107.108050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khandelia H, Ipsen J, Mouritsen O. Impact of peptides on lipid membranes. Biochim Biophys Acta Biomembr. 2008;1778:1528–1536. doi: 10.1016/j.bbamem.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Pan J, Tieleman D, Nagle J, Kucerka N, Tristram-Nagle S. Alamethicin in lipid bilayers: combined use of x-ray scattering and md simulations. Biochim Biophys Acta Biomembr. 2009;1788:1387–1397. doi: 10.1016/j.bbamem.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunkin C, Pokorny A, Almeida P, Lee H. Molecular dynamics studies of transportan 10 (tp10) interacting with a popc lipid bilayer. J Phys Chem B. 2011;115:1188–1198. doi: 10.1021/jp107763b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leontiadou H, Mark AE, Marrink SJ. Antimicrobial peptides in action. J Am Chem Soc. 2006;128:12156–12161. doi: 10.1021/ja062927q. [DOI] [PubMed] [Google Scholar]
- 22.Sengupta D, Leontiadou H, Mark A, Marrink S. Toroidal pores formed by antimicrobial peptides show significant disorder. Biochim Biophys Acta. 2008;1778:2308–2317. doi: 10.1016/j.bbamem.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Bhargava K, Feix JB. Membrane binding, structure, and localization of cecropin-mellitin hybrid peptides: A site-directed spin-labeling study. Biophys J. 2004;86:329–336. doi: 10.1016/S0006-3495(04)74108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Markwick PRL, de Oliveira CAF, McCammon JA. Enhanced lipid diffusion and mixing in accelerated molecular dynamics. J Chem Theor Comp. 2011;7:3199–3207. doi: 10.1021/ct200430c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao W, Róg T, Gurtovenko AA, Vattulainen I, Karttunen M. Atomic-scale structure and electrostatics of anionic palmitoyloleoylphosphatidyl-glycerol lipid bilayers with Na+ counterions. Biophys J. 2007;92(4):1114–1124. doi: 10.1529/biophysj.106.086272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Humphrey W, Dalke A, Schulten K. VMD – Visual Molecular Dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 27.Kucerka N, Tristram-Nagle S, Nagle JF. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J Membr Biol. 2005;208:193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 28.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theor Comp. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 29.Respondek M, Madl T, Göbl C, Golser R, Zangger K. Mapping the orientation of helices in micelle-bound peptides by paramagnetic relaxation waves. J Am Chem Soc. 2007;129:5228–5234. doi: 10.1021/ja069004f. [DOI] [PubMed] [Google Scholar]
- 30.Khandelia H, Kaznessis YN. Structure of the antimicrobial beta-hairpin peptide protegrin-1 in a DLPC lipid bilayer investigated by molecular dynamics simulation. Biochim Biophys Acta Biomembr. 2007;1768:509–520. doi: 10.1016/j.bbamem.2006.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher IWE, Roux B, Schlenkrich M, Smith J, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 32.MacKerell AD, Jr, Feig M, Brooks CL., III Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comp Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 33.Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Jr, Pastor RW. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys Chem. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comp Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersen HC. Rattle: A “velocity” version of the shake algorithm for molecular dynamics calculations. J Chem Phys. 1983;52:24–34. [Google Scholar]
- 36.Miyamoto S, Kollman PA. Settle: An analytical version of the shake and rattle algorithm for rigid water models. J Comp Chem. 1993;13:952–962. [Google Scholar]
- 37.Darden T, York D, Pedersen L. Particle mesh Ewald. An N·log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 38.Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation — the Langevin piston method. J Chem Phys. 1995;103(11):4613–4621. [Google Scholar]
- 39.Joosten R, Te Beek T, Krieger E, Hekkelman M, Hooft R, Schneider R, Sander C, Vriend G. A series of PDB related databases for everyday needs. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. Gromacs: Fast, flexible, and free. J Comp Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 41.Wimley W, White S. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 42.Shepherd CM, Vogel HJ, Tieleman DP. Interactions of the designed antimicrobial peptide MB21 and truncated dermaseptin S3 with lipid bi-layers: molecular-dynamics simulations. Biochem J. 2003;370:233–243. doi: 10.1042/BJ20021255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aliste M, Tieleman D. Computer simulation of partitioning of ten pen-tapeptides Ace-WLXLL at the cyclohexane/water and phospholipid/water interfaces. BMC Biochem. 2005;6:30. doi: 10.1186/1471-2091-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blondelle SE, Houghten RA. Hemolytic and antimicrobial activities of the twenty-four individual omission analogs of melittin. Biochemistry. 1991;30:4671–4678. doi: 10.1021/bi00233a006. [DOI] [PubMed] [Google Scholar]
- 45.Blondelle SE, Houghten RA. Probing the relationships between the structure and hemolytic activity of melittin with a complete set of leucine substitution analogs. Pept Res. 1991;4:12–18. [PubMed] [Google Scholar]
- 46.Andreu D, Merrifield RB, Steiner H, Boman HG. N-terminal analogs of cecropin A: synthesis, antibacterial activity, and conformational properties. Biochemistry. 1985;24:1683–1688. doi: 10.1021/bi00328a017. [DOI] [PubMed] [Google Scholar]
- 47.Mancheño J, Oñaderra M, Martínez del Pozo A, Díaz-Achirica P, Andreu D, Rivas L, Gavilanes J. Release of lipid vesicle contents by an antibacterial cecropin a-melittin hybrid peptide. Biochemistry. 1996;35:9892–9899. doi: 10.1021/bi953058c. [DOI] [PubMed] [Google Scholar]
- 48.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The martini forcefield: coarse grained model for biomolecular simulations. J Phys Chem B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 49.Monticelli L, Kandasamy S, Periole X, Marrink RLDTS. The martini coarse grained forcefield: extension to proteins. J Chem Theor Comp. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 50.Hamelberg D, Mongan J, McCammon J. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J Chem Phys. 2004;120(24):11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y, Harrison CB, Schulten K, McCammon JA. Implementation of accelerated molecular dynamics in NAMD. Comput Sci Discov. 4:015002. doi: 10.1088/1749-4699/4/1/015002. (11 pages) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.