Abstract
Background:
Our genome-wide association study of schizophrenia found association signals at the Kalirin gene (KALRN) and EPH receptor B1 gene (EPHB1) in a Japanese population. The importance of these synaptogenic pathway genes in schizophrenia is gaining independent supports. Although there has been growing interest in rare (<1%) missense mutations as potential contributors to the unexplained heritability of schizophrenia, there are no population-based studies targeting rare (<1%) coding mutations with a larger effect size (eg, OR >1.5) in KALRN or EPHB1.
Methods and Results:
The present study design consisted of 3 phases. At the discovery phase, we conducted resequencing analyses for all exon regions of KALRN and EPHB1 using a DNA microarray–based method. Seventeen rare (<1%) missense mutations were discovered in the first sample set (320 schizophrenic patients). After the prioritization phase based on frequencies in the second sample set (729 cases and 562 controls), we performed association analyses for each selected mutation using the third sample set (1511 cases and 1517 controls), along with a combined association analysis across all selected mutations. In KALRN, we detected a significant association between schizophrenia and P2255T (OR = 2.09, corrected P = .048, 1 tailed); this was supported in the combined association analysis (OR = 2.07, corrected P = .006, 1 tailed). We found no evidence of association of EPHB1 with schizophrenia. In silico analysis indicated the functional relevance of these rare missense mutations.
Conclusion:
We provide evidence that multiple rare (<1%) missense mutations in KALRN may be genetic risk factors for schizophrenia.
Keywords: synaptogenic pathway, rare missense mutations, GWAS, Japanese population
Introduction
Schizophrenia is a genetically heterogeneous disorder with heritability estimated at up to 80%.1 According to a recent simulation based on genome-wide association study (GWAS) datasets, a highly polygenic model involving a number of common variants of very small effect may explain more than one-third of the total variation in risk of schizophrenia.2 On the other hand, interest has been growing in rare variants as potential contributors to the unexplained heritability of schizophrenia.3 This is partly triggered by recent studies establishing an important role for rare genomic copy number variants (CNVs) in the etiology of schizophrenia.4 Another potential genetic variation to explain the remaining heritability is rare missense mutations. Kryukov et al5 reported that ∼20% of new (de novo) missense mutations in humans result in a loss of function, whereas ∼53% have mildly deleterious effects and ∼27% are effectively neutral with respect to phenotype by a combined analysis of mutations causing human Mendelian diseases, mutations driving human-chimpanzee sequence divergence, and systematic data on human genetic variation. Their results were supported by an independent study.6 Because the pressure of purifying selection acting on the mildly deleterious mutations is weak, their cumulative high frequency in the human population is being maintained by “mutation-selection balance.” This provides support to a speculation that the accumulation of mildly deleterious missense mutations in individual human genomes can be a genetic basis for complex diseases.5 The importance of rare missense mutations in schizophrenia is demonstrated by a study of the ABCA13 gene in which multiple rare (<1%) coding variants were associated with schizophrenia.7
We recently performed a GWAS for schizophrenia in a Japanese population.8 Although single locus analysis did not reveal genome-wide support for any locus, a shared polygenic risk of schizophrenia between the Japanese and the Caucasian samples was confirmed. In our GWAS, association signals were detected at the regions of the Kalirin gene (KALRN) on 3q21.2 and the EPH receptor B1 gene (EPHB1) on 3q21-q23, both of which are in the same synaptogenic pathway9 (supplementary figure S1). Associations of each gene with schizophrenia have recently received support from independent GWASs in different populations.10,11 Furthermore, a rare de novo CNV overlapping with the EPHB1 gene locus was detected in a patient with schizophrenia.12
KALRN is a large neuronal dual Rho guanine nucleotide exchange factor (GEF) that activates small guanosine triphosphate–binding proteins of the Rho family, including Rac1.13 This activation enables KALRN to regulate neurite initiation, axonal growth, dendritic morphogenesis, and spine morphogenesis. Consistent with its biological function, KALRN is a key factor responsible for reduced densities of dendritic spines on pyramidal neurons in the dorsolateral prefrontal cortex (DLPFC)14 observed in postmortem brains from schizophrenic patients. The messenger RNA expression level of KALRN is significantly reduced in DLPFC of patients with schizophrenia and strongly correlated with spine density.15 In addition, KALRN-knockout mice not only exhibit spine loss and reduced glutamatergic transmission in the frontal cortex but also schizophrenia-like phenotypes including robust deficits in working memory, sociability, prepulse inhibition, and locomotor hyperactivity reversible by clozapine, an atypical antipsychotic.16 These synaptic and behavioral dysfunctions are apparent during young adulthood in mice (12 weeks old), which coincides with the onset of schizophrenia in patients. Notably, Disrupted-in-Schizophrenia 1, a prominent schizophrenia risk factor, was shown to be involved in the maintenance of spine morphology and function by regulating access of KALRN to Rac1.17 EPHB1 belongs to a receptor tyrosine kinase family and controls multiple aspects of neuronal development, including synapse formation and maturation, as well as synaptic structural and functional plasticity. In neurons, activation of EphB receptors by its ligand B-type ephrins induces the rapid formation and enlargement of dendritic spines, as well as rapid synapse maturation. One of the downstream effectors of ephrinB/EphB signaling is KALRN. In young hippocampal neurons, KALRN is reported to play an important role in the maturation of synapses induced by trans-synaptic ephrinB/EphB signaling.18
According to the above-mentioned study,5 most missense mutations with a frequency of <1% are mildly deleterious, indicating that a low frequency of missense mutation per se can serve as a strong predictor of a deleterious effect of variants. Therefore, the working hypothesis of the present study is that rare (<1%) missense or nonsense mutations with a larger effect size (eg, OR >1.5) in KALRN and EPHB1 may be genetic risk factors for schizophrenia. Recently, a DNA microarray–based resequencing method has been developed to enable accurate and rapid resequencing analysis of candidate genes.19 Using this system, we conducted resequencing analyses for all exon regions of KALRN and EPHB1 in 320 schizophrenic patients and found evidence that rare (<1%) missense mutations in KALRN are significantly associated with schizophrenia using the 3-phase study design.
Methods and Materials
Subjects
Three sample sets were used in this study. The first sample set, comprising 320 schizophrenic patients (mean age, 54.2 ± 14.1 years, 49.1% male), with long-term hospitalization for severe symptoms, was used to search for rare missense or nonsense mutations. We used the first sample set for mutation screenings because patients with extreme phenotypes (severe symptoms) can be expected to carry more deleterious mutations.20 The second sample set, including 729 cases (45.4 ± 15.1 years, 52.2% male) and 562 controls (44.0 ± 14.4 years, 49.8% male), was used to prioritize detected functional variants for subsequent association analyses. The third sample set, including 1511 cases (45.9 ± 14.0 years, 49.6% male) and 1517 controls (46.0 ± 14.6 years, 49.6% male), was used for association analyses. Age and gender were matched in the second and third sample sets, respectively. All patients were diagnosed according to Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, criteria, and controls were evaluated using unstructured interviews to exclude individuals with history of mental disorders. Detailed information regarding diagnostic procedures is available elsewhere.21 All subjects were ethnically Japanese and provided written informed consent. This study was approved by the ethics committees at each participating university.
Array Design for Resequencing Analyses
We used the Affymetrix GeneChip CustomSeq Resequencing Array (Affymetrix, Santa Clara, California) for exon sequencing in the first sample set. These arrays rely on allele-specific hybridization for determining DNA sequence.19 Each individual nucleotide of both the sense and the antisense DNA strands is interrogated with four 25-mer probes that differ only with respect to the central position (A, C, G, and T). According to Affymetrix’s Custom-Seq Array Design Guide, we designed arrays covering all exon regions of KALRN and EPHB1 (Ensembl release 52 [Human CCDS set]; Transcript: ENST00000360013, ENST00000240874, and ENST00000291478 for KALRN; ENST00000398015 for EPHB1). Because the principle of the resequencing arrays is based on hybridization, it is necessary to avoid cross-hybridization for accurate resequencing. For this purpose, we removed repetitive elements and highly homologous sequences from the array design.
Array-Based Resequencing
The experiments were conducted according to the manufacturer's instructions (supplementary figure S2). Genomic DNA was extracted from peripheral blood using standard methods. To generate enough target-enriched subject material for hybridization to the arrays, we generated 47 and 14 amplicons per sample for KALRN and EPHB1, respectively, using long-range polymerase chain reaction (PCR). The PCR conditions were as follows: 94°C for 2 minutes followed by 30 cycles consisting of 94°C for 15 seconds, 68°C for 3 minutes, followed by a final extension of 68°C for 8 minutes, using TaKaRa LA Taq™ (Takara Bio, Otsu, Shiga, Japan). Each PCR product was quantified using PicoGreen (Molecular Probes, Eugene, Oregon), pooled in an equimolar fashion. The PCR products were then purified, fragmented, labeled, and hybridized to the arrays, following the protocol. Finally, the arrays were washed and stained using the GeneChip Fluidics Station 450 (Affymetrix) and scanned using the GeneChip Scanner 3000 (Affymetrix). The data were analyzed using the GeneChip Operating Software (GCOS; Affymetrix), the GeneChip Sequence Analysis Software (GSEQ; Affymetrix), and SeqC (JSI Medical Systems, Kippenheim, Germany; http://www.jsi-medisys.de/html/products/SeqC/SeqC.htm) to automate the generation of sequence and genotype calls from the intensity data. In this study, around 17 kb was sequenced per sample, meaning that more than 5.4 Mb was sequenced in total. All missense mutations presented in this study were confirmed using both Sanger sequencing and Custom TaqMan SNP genotyping assays (Applied Biosystems, Foster City, California).
Association Analysis of Each Missense Mutation
Although the rare (<1%) missense mutations were originally discovered among 320 schizophrenic patients, it was possible that a portion of them might have neutral or protective effects.5 In addition, it was necessary to reduce the number of statistical tests for multiple comparison problems. To accomplish this, we prioritized rare (<1%) deleterious variants for subsequent association analyses based on the frequencies in the second case-control sample set because rare deleterious variants relevant to schizophrenia can be assumed to have higher frequency in cases than in controls. The criteria for prioritization were as follows: (1) frequencies of mutations were <1% in controls and (2) frequencies of mutations were higher in cases (ie, OR > 1). Mutations not detected in the second sample set were not followed up in this analysis. The frequencies of such mutations can be so low (<0.0005) that the results of association analyses are unlikely to be statistically significant in our sample size. For mutations meeting the above criteria, we conducted association analyses with schizophrenia using the third sample set. Genotyping was conducted by Custom TaqMan SNP genotyping assays (Applied Biosystems). For quality control, samples with missing call rates of 10% or higher were excluded from the analyses.
Combined Association Analyses
In general, it is difficult to establish an association of a rare mutation with a phenotype because statistical power is limited by low population frequency and because the number of rare variants requires a strict multiple test correction. Therefore, we conducted combined association analyses across rare mutations observed in each gene in the third sample set, comparing the number of mutations in cases with the number in controls. The criteria for mutations included in these analyses were same as the above criteria with 1 exception: Mutations not detected in the second sample set were included in the combined association analyses.
In Silico Analysis
The potential influence of missense mutations was evaluated using PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) and22 PMut (http://mmb2.pcb.ub.es:8080/PMut/)23 softwares. PolyPhen-2 uses 8 sequence-based and 3 structure-based predictive features and compares a property of the wild-type allele and the corresponding property of the mutant allele. PolyPhen-2 trained on HumDiv datasets is reported to achieve true positive prediction rates of 92% with a false-positive rate of 20%.22 A mutation is appraised qualitatively as benign, possibly damaging, or probably damaging based on naive Bayes posterior probability that a given mutation is damaging. PMut also allows the fast and accurate prediction (∼80% success rate in humans) of the pathological character of missense mutations based on the use of neural networks. The final output is a pathogenicity index ranging from 0 to 1 (indexes >0.5 signal pathological mutations).
We also examined evolutionary conservation of the mutated residues and surrounding amino acids. Multiple sequence alignment of human KALRN or EPHB1 with 6 orthologs was performed for this purpose.
Power Calculation
Power calculation was performed with a power calculator called CaTS (http://www.sph.umich.edu/csg/abecasis/CaTS/).24 Power was estimated under the following parameter assumptions with respect to association test statistics: genetic relative risk = 2, prevalence of disease = 0.01, risk allele frequency = the values frequency observed in controls, and α = .05; a multiplicative model was used.
Statistical Analysis
For the association analysis of each variant, Fisher exact test was used to examine whether rare deleterious variants were significantly overrepresented in the patient group rather than the control group.
A combined association test was performed following a previous study.7 In brief, to account for variable sample size, sample size was adjusted to , where Ni is the sample size at the ith variant, and n is the number of variants. The number of observed variants was adjusted as , where pi is the frequency of the ith variant. Fisher exact test was used in this test as well to examine an overrepresentation of rare deleterious missense mutations in the patient group rather than control group.
All statistical tests were 1 tailed, and a P value less than 0.05 was considered significant. Bonferroni correction was used for solving multiple testing problems.
Results
Discovery of Mutations
We detected 12 and 6 missense mutations with a frequency of <5% in KALRN and EPHB1, respectively, among 320 cases in the first sample set (table 1). All but 2 mutations (N2973S in KALRN and T981M in EPHB1) were novel. All mutations were validated by both Sanger sequencing and Custom TaqMan SNP genotyping assays. In the first sample set, 2 patients were compound heterozygotes for rare missense mutations in the 2 genes. One patient had R410H in KALRN and R905C in EPHB1. The other had A2382V in KALRN and D375N in EPHB1. There were no clinical characteristics shared between these patients. No nonsense mutations were identified in this study.
Table 1.
KALRN And EPHB1 Missense Mutations Identified in The First Sample Set And Their Frequencies in The Second Sample Set
| First Sample Set |
Second Sample Set |
||||||||||
| Genotype Counts |
Mutation Frequency |
||||||||||
| Gene | Genomic Position | Base Change | dbSNP Reference | AA Change | Homo | Hetero | SCZ | CONT | SCZ | CONT | OR >1 |
| KALRN | 125527659 | G → A | ss250607852 | R410H | 0 | 1 | 0/0/701 | 0/0/541 | 0 | 0 | |
| KALRN | 125531474 | T → A | ss250607853 | L452Q | 0 | 1 | 0/1/709 | 0/2/541 | 0.0007 | 0.0018 | |
| KALRN | 125600376 | C → A | ss250607854 | Q770K | 0 | 1 | 0/0/706 | 0/0/544 | 0 | 0 | |
| KALRN | 125656787 | C → T | ss250607855 | T1207M | 0 | 1 | 0/2/705 | 0/1/542 | 0.0014 | 0.0009 | + |
| KALRN | 125764534 | C → A | ss250607856 | P1695Q | 0 | 1 | 0/59/636 | 1/44/492 | 0.0425 | 0.0428 | |
| KALRN | 125764599 | A → T | ss250607857 | M1717L | 0 | 1 | 0/0/705 | 0/1/540 | 0 | 0.0009 | |
| KALRN | 125860927 | G → A | ss250607858 | R2049K | 0 | 1 | 0/1/696 | 0/1/540 | 0.0007 | 0.0009 | |
| KALRN | 125873259 | C → A | ss250607859 | P2255T | 0 | 7 | 1/14/684 | 0/7/536 | 0.0114 | 0.0064 | + |
| KALRN | 125873289 | C → T | ss250607860 | P2265S | 1 | 0 | 0/6/701 | 0/7/533 | 0.0042 | 0.0065 | |
| KALRN | 125873382 | G → T | ss250607861 | G2296C | 0 | 1 | 0/1/703 | 0/1/542 | 0.0007 | 0.0009 | |
| KALRN | 125876103 | C → T | ss250607862 | A2382V | 0 | 1 | 0/0/697 | 0/0/540 | 0 | 0 | |
| KALRN | 125920964 | A → G | rs16835896 | N2973S | 0 | 3 | 0/3/698 | 0/6/538 | 0.0021 | 0.0055 | |
| EPHB1 | 136153231 | T → C | ss252863894 | F151S | 0 | 1 | 0/0/710 | 0/0/543 | 0 | 0 | |
| EPHB1 | 136334407 | G → A | ss252863895 | D375N | 0 | 1 | 0/0/708 | 0/0/544 | 0 | 0 | |
| EPHB1 | 136368508 | G → A | ss252863896 | D577N | 0 | 1 | 0/0/707 | 0/0/544 | 0 | 0 | |
| EPHB1 | 136394134 | C → T | ss252863897 | R637C | 0 | 2 | 1/1/707 | 0/2/541 | 0.0021 | 0.0018 | + |
| EPHB1 | 136450890 | C → T | ss252863898 | R905C | 0 | 3 | 0/9/695 | 0/1/543 | 0.0064 | 0.0009 | + |
| EPHB1 | 136460639 | C → T | rs56186270 | T981M | 0 | 2 | 0/0/706 | 0/0/541 | 0 | 0 | |
Note: Genomic position based on NCBI build 36, chromosome 3. Amino acid changes based on NCBI Reference Sequence NP_001019831.2 (2986 aa) for KALRN and NP_004432.1 (984 aa) for EPHB1. All but N2973S (rs16835896) and T981M (rs56186270) are novel. AA change, amino acid change; dbSNP, Single Nucleotide Polymorphism Database; Homo, homozygote; Hetero, heterozygote; SCZ, schizophrenia; CONT, control; NCBI, National Center for Biotechnology Information.
Association Analysis of Each Missense Mutation
In the prioritization phase using the second sample set, T1207M and P2255T in KALRN and R637C and R905C in EPHB1 showed a higher frequency in cases than in controls (table 1). Seven missense mutations (R410H, Q770K, and A2382V in KALRN and F151S, D375N, D577N, and T981M in EPHB1) were not detected. The frequency of P1695Q was more than 4% both in cases and in controls. Based on our criteria, we selected 4 missense mutations (T1207M and P2255T in KALRN and R637C and R905C in EPHB1) for subsequent association analyses using the third sample set.
In the third phase, P2255T showed a nominally significant association with schizophrenia (OR = 2.09, P = .012) in the third sample set (table 2). This remained significant after correction for multiple testing of 4 variants (corrected P = .048). T1207M in KALRN and R637C and R905C in EPHB1 were also more frequent in cases, although differences were not significant.
Table 2.
Association Analyses of Each Missense Mutation in the Third Sample Set
| Third Sample Set |
|||||||
| Genotype Counts |
Mutation Frequency |
||||||
| AA Change | SCZ | CONT | SCZ | CONT | OR | P Value | |
| KALRN | T1207M | 0/7/1477 | 0/3/1482 | 0.0024 | 0.0010 | 2.34 | .171 |
| KALRN | P2255T | 0/31/1448 | 0/15/1473 | 0.0105 | 0.0050 | 2.09 | .012 |
| EPHB1 | R637C | 0/4/1477 | 0/4/1478 | 0.0014 | 0.0014 | 1.00 | .636 |
| EPHB1 | R905C | 0/15/1458 | 0/12/1466 | 0.0051 | 0.0041 | 1.26 | .347 |
Note: Abbreviations are explained in the first footnote to table 1. P values were calculated by Fisher exact test (1 tailed).
We excluded mutations not detected in the second sample set from this analysis. This was supported by a power analysis showing that the third sample set had only 10% power in analysis of very rare mutations.
Combined Association Analysis
In addition to 4 mutations (T1207M and P2255T in KALRN and R637C and R905C in EPHB1), 7 very rare mutations (R410H, Q770K, and A2382V in KALRN and F151S, D375N, D577N, and T981M in EPHB1), which were not detected in the second samples set, were included in the combined association analysis. A global comparison of the frequencies of 5 selected mutations in KALRN between cases and controls in the third sample set showed a significant increase in frequency in schizophrenic patients (OR = 2.07, P = .003) (table 3). This remained significant after correction for multiple testing (corrected P = .006). On the other hand, a global comparison of the frequencies of 6 selected mutations in EPHB1 did not show a significant difference (OR = 1.09, P = .438).
Table 3.
Combined Association Analysis in The Third Sample Set
| Third Sample Set |
Combined Analysis |
||||||
| Gene | Genotype Counts |
Mutation Frequency |
Gene Based |
||||
| AA Change | SCZ | CONT | SCZ | CONT | OR | P value | |
| KALRN | R410H | 0/0/1481 | 0/0/1484 | 0 | 0 | 2.07 | .003 |
| KALRN | Q770K | 0/0/1486 | 0/0/1490 | 0 | 0 | ||
| KALRN | T1207M | 0/7/1477 | 0/3/1482 | 0.0024 | 0.0010 | ||
| KALRN | P2255T | 0/31/1448 | 0/15/1473 | 0.0105 | 0.0050 | ||
| KALRN | A2382V | 0/7/1473 | 0/4/1480 | 0.0024 | 0.0013 | ||
| EPHB1 | F151S | 0/0/1478 | 0/0/1484 | 0 | 0 | 1.09 | .438 |
| EPHB1 | D375N | 0/0/1483 | 0/0/1490 | 0 | 0 | ||
| EPHB1 | D577N | 0/0/1486 | 0/2/1483 | 0 | 0.000673 | ||
| EPHB1 | R637C | 0/4/1477 | 0/4/1478 | 0.0014 | 0.0014 | ||
| EPHB1 | R905C | 0/15/1458 | 0/12/1466 | 0.0051 | 0.0041 | ||
| EPHB1 | T981M | 0/5/1481 | 0/4/1484 | 0.0017 | 0.0013 | ||
Note: Abbreviations are explained in the first footnote to table 1. P values were calculated by Fisher exact test (1 tailed).
In Silico Analysis
Results of in silico analysis are shown in table 4. All missense mutations but A2382V in KALRN were predicted to have functional relevance by PolyPhen-2 or PMut software.
Table 4.
Results of In Silico/Conservation Analysis
| KALRN | |||||||
| Analysis | R410H | Q770K | T1207M | P2255T | A2382V | ||
| PolyPhen-2 | Probably damaging | Probably damaging | Probably damaging | Benign | Benign | ||
| PMut | Pathological | Neutral | Pathological | Pathological | Neutral | ||
| Conservation analysis | Human (NP_001019831.2) | LDERSTI | IFLQLRI | IHATEIR | RSQPARL | SILAPLT | |
| Chimpanzee (XP_516703.2) | LDERSTI | IFLQLRI | IHATEIR | RSQPARL | SILAPLT | ||
| Dog (XP_535768.2) | LDERSTI | IFLQLRI | IHATEIR | RSQPSRV | SVLAPLT | ||
| Cattle (XP_001790302.1) | LDERSTI | IFLQLRI | IHATEIR | RSQPARV | SILTPLT | ||
| Mouse (XP_001481079.1) | LDERSTI | IFLQLRI | IHATEIR | RSQPPRV | SILAPLA | ||
| Rat (NP_114451.1) | LDERSTI | IFLQLRI | IHATEIR | RSQPPRV | SILAPLT | ||
| EPHB1 | |||||||
| Analysis | F151S | D375N | D577N | R637C | R905C | T981M | |
| PolyPhen-2 | Benign | Probably damaging | Possibly damaging | Probably damaging | Probably damaging | Probably damaging | |
| PMut | Pathological | Neutral | Neutral | Pathological | Pathological | Pathological | |
| Conservation analysis | Human (NP_004432.1) | QVDFGGR | RCDDNVE | VYSDKLQ | YKGRLKL | LLDRSIP | QSPTAMA |
| Chimpanzee (XP_001150963.1) | QVDFGGR | RCDDNVE | LLVEQWQ | YKGRLKL | LLDRSIP | QSPTAMA | |
| Dog (XP_542791.2) | QVDFGGR | RCDDNVE | VYSDKLQ | YKGRLKL | LLDRSIP | QSPTTMA | |
| Cattle (XP_614602.4) | QVDFGGR | RCDDNVE | VYSDKLQ | YKGRLKL | LLDRSIP | QSPTAMA | |
| Mouse (NP_775623.2) | QVDFGGR | RCDDNVE | AYSDKLQ | YKGRLKL | LLDRSIP | QSPSVMA | |
| Rat (XP_217250.1) | QVDFGGR | RCDDNVE | VYSDKLQ | YKGRLKL | LLDRSIP | QSPSVMA | |
Note: The bold are the mutated amino acids.
A multiple alignment of the region of KALRN or EPHB1 containing rare missense mutations with 6 orthologs is shown in table 4. Most of the rare missense mutations showed a high degree of amino acid conservation in different species.
Discussion
In this study, we conducted resequencing analyses for the 2 synaptogenic pathway genes (KALRN and EPHB1) in schizophrenia using a DNA microarray–based method. After resequencing more than 5.4 Mb, we discovered 17 rare (<1%) missense mutations in KALRN or EPHB1 and detected a significant association between schizophrenia and P2255T in KALRN, as well as in the combined association analysis for KALRN. These findings are consistent with an estimation that most rare (<1%) missense mutations are mildly deleterious and are associated with a heterozygous fitness loss.5
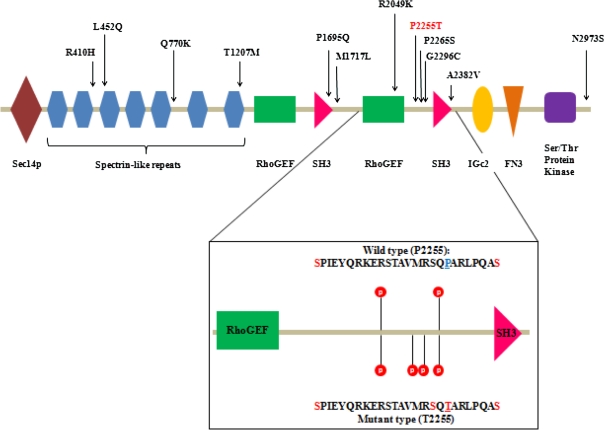
Schizophrenia is a genetically heterogeneous disorder, with both very rare variants with a high effect size (eg, CNVs in 1q21.1, 15q13.3) and common variants with a low effect size (eg, rs1344706 in ZNF804A) involved in its genetic architecture. In this frequency-effect size spectrum, P2255T (OR: ∼2, risk allele frequency in controls: ∼0.005) is located between the CNV in 1q21.1 (OR: ∼10, frequency in controls: ∼0.0001)25 and rs1344706[T] in ZNF804A (OR: ∼1.1, risk allele frequency in controls: ∼0.6),26 both of which have been recently associated with schizophrenia. The relatively modest effect size of P2255T compared with that of the above CNVs can be attributable to the difference in the effect of each variant on gene(s): Although CNVs strongly influence the expression of multiple genes, missense mutations in KALRN are presumed to have limited effects on KALRN function. P2255T is located in the evolutionally conserved proline-rich region between the C-terminal GEF and SH3 domains27 and is surrounded by 2 nearby phosphorylation sites (S2237 and S2262), according to Human Protein Reference Database (http://www.hprd.org/index_html)28 (figure 1). In silico analysis with PhosphoMotif Finder29 shows that T2255 itself can be recognized and phosphorylated by many kinases, suggesting functional implications of P2255T (figure 1). In addition, in silico analysis predicts that phosphorylation of T2255 will induce that of nearby S2253. Thus, P2255T may greatly change the phosphorylation status in a narrow region between the C-terminal GEF and SH3 domain. A protein with multiple phosphorylated sites like KALRN can be assumed to have an exponential number of phospho-forms, and individual phospho-forms may have distinct biological effects. The diffuse distribution of these phospho-forms at steady state enables the phosphoproteome to encode information and flexibly respond to varying demands.30 Thus, it is conceivable that P2255T may influence such plasticity in KALRN by changing the number of phosphorylated sites. Interestingly, detailed examination of clinical information from the first sample set, which was uniquely available to us, revealed that congenital or early-onset vascular disease was observed in 5 of 7 cases with P2255T (supplementary table S1). Because KALRN may represent a candidate gene for vascular diseases,31,32 it is tempting to speculate that P2255T may be a potential risk factor for vascular disease.
Fig. 1.
Rare Missense Mutations in KALRN and Change in Phosphorylation Status by P2255T.
In addition to P2255T, we detected multiple rare (<1%) missense mutations in KALRN or EPHB1. Such variants are not sufficiently frequent to be covered by GWAS nor do they have sufficiently large effect sizes to be detected by linkage analysis in family studies. For modest effect sizes, it is suggested that association testing may require composite tests of overall mutational load, comparing frequencies of mutations of potentially similar functional effect in cases and controls. Thus, we also performed combined association analyses for KALRN or EPHB1 and found evidence that multiple rare (<1%) missense mutations in KALRN as a whole are associated with schizophrenia. This finding is supported by in silico analyses showing that most of the mutations are predicted as being of functional relevance and that they are located in evolutionally conserved regions. In contrast, there were no significant differences in the cumulative frequencies of rare missense mutations in EPHB1. This might be due to a type II error. The cumulative frequency of rare mutations of EPHB1 in controls is almost same as the one of KALRN in controls (0.0075 vs 0.0073), indicating that cumulative effect size of rare missense mutations in EPHB1 may be smaller than the one in KALRN. In the mammalian genome, there are 5 different EphB receptors (EphB1, EphB2, EphB3, EphB4, and EphB6), with a high similarity at the amino acid level. Analysis of double and triple knockout mice lacking EphB1, EphB2, and EphB3 in different combinations revealed that EphBs have functional redundancy even though all these EphBs are responsible for spine morphogenesis and synapse formation to varying degrees.33 This is in contrast with the drastic phenotypes observed in KALRN-knockout mice.16 Therefore, biological effects of rare missense mutations in EPHB1 may be compensated for by other intact EPHBs. This might lower the ORs of rare missense mutations in EPHB1. Given that all the mutations detected in EPHB1 were predicted to have pathogenicity by PolyPhen-2 or PMut, a larger-scale case-control study with sufficient power may provide a significant result in a combined analysis for EPHB1.
One important aspect of the present study is that we found rare mutations associated with schizophrenia in the KALRN gene, in which GWASs detected association signals for schizophrenia. Several studies have recently reported the 1 gene may harbor both rare and common variants associated with the same diseases, including schizophrenia,34 type 2 diabetes,35 and hypertriglyceridemia.36 Given that the cost of whole-genome sequencing is still high to search for rare mutations, resequencing analyses for genes with support from GWAS might be a better strategy for detection of rare mutations with larger effect size.
There are several limitations to this study. First, we could not conduct segregation analyses for mutations due to limited access to family members. Furthermore, given the modest risk (OR ∼2), these mutations would show incomplete penetrance. In fact, it is reported that penetrance estimates of CNVs at 1q21.1 and 15q13.3, both of which show higher ORs, are 0.061 and 0.074, respectively.25 Therefore, a population-based study is a better choice to evaluate genetic associations for missense mutations with modest risk.37 The second limitation is population stratification. Although a Japanese population is considered relatively homogenous, small population stratifications may have influenced our findings.38 However, we believe that the recruitment of subjects in local regions minimized this concern. Third, we did not conduct functional analyses for detected missense mutations. The detailed effects of these mutations on the pathophysiology of schizophrenia need to be examined in a future study. Fourth, our resequencing analyses were not comprehensive in terms of the kind of variants and the number of genes. In other words, the present study did not cover indels or CNVs because of the methodological limitation of the DNA microarray–based method. Because these classes of variants could have a more profound effect on protein function, their genetic contribution to schizophrenia might be revealed in future studies. Also, as shown in EPHB1, it is assumed that a variety of molecules or pathways have a role in spine formation or synapse plasticity, which are impaired in patients with schizophrenia, to compensate for each other. A combined analysis of a large number of genes relevant for synaptic function might provide more robust evidence that rare missense mutations as a whole contribute to pathomechanisms of schizophrenia.
In conclusion, we provide the first evidence that multiple rare (<1%) missense mutations in KALRN may be genetic risk factors for schizophrenia. Further studies will be needed to examine the pathogenicity of these mutations from a biologic point of view.
Funding
This work was supported by research grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan; Ministry of Health, Labor and Welfare of Japan; Grant-in-Aid for Scientific Research on Pathomechanisms of Brain Disorders from the Ministry of Education, Culture, Sports, Science and Technology of Japan; Academic Frontier Project for Private Universities, Comparative Cognitive Science Institutes; Core Research for Evolutional Science and Technology.
Supplementary Material
Supplementary material is available at http://schizophreniabulletin.oxfordjournals.org.
Acknowledgments
We sincerely thank the patients and healthy volunteers for their participation in this study. We also thank Ryoko Ishihara and Junko Tsuda for technical assistance.
The authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 2.Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyko AR, Williamson SH, Indap AR, et al. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4:e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knight HM, Pickard BS, Maclean A, et al. A cytogenetic abnormality and rare coding variants identify ABCA13 as a candidate gene in schizophrenia, bipolar disorder, and depression. Am J Hum Genet. 2009;85:833–846. doi: 10.1016/j.ajhg.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikeda M, Aleksic B, Kinoshita Y, et al. Genome-wide association study of schizophrenia in a Japanese population [published online ahead of print September 10, 2010] Biol Psychiatry. doi: 10.1016/j.biopsych.2010.07.010. doi: 10.1016/j.biopsych.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 9.Penzes P, Beeser A, Chernoff J, et al. Rapid induction of dendritic spine morphogenesis by trans-synaptic ephrinB-EphB receptor activation of the Rho-GEF kalirin. Neuron. 2003;37:263–274. doi: 10.1016/s0896-6273(02)01168-6. [DOI] [PubMed] [Google Scholar]
- 10.St Jean PL. Genes associated with schizophrenia identified using a whole genome scan. 2008 http://www.freepatentsonline.com/y2008/0176240.html. Accessed August 8, 2010. [Google Scholar]
- 11.Sullivan PF, Lin D, Tzeng JY, et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu B, Roos JL, Levy S, et al. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 13.Rabiner CA, Mains RE, Eipper BA. Kalirin: a dual Rho guanine nucleotide exchange factor that is so much more than the sum of its many parts. Neuroscientist. 2005;11:148–160. doi: 10.1177/1073858404271250. [DOI] [PubMed] [Google Scholar]
- 14.Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57:65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- 15.Hill JJ, Hashimoto T, Lewis DA. Molecular mechanisms contributing to dendritic spine alterations in the prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2006;11:557–566. doi: 10.1038/sj.mp.4001792. [DOI] [PubMed] [Google Scholar]
- 16.Cahill ME, Xie Z, Day M, et al. Kalirin regulates cortical spine morphogenesis and disease-related behavioral phenotypes. Proc Natl Acad Sci U S A. 2009;106:13058–13063. doi: 10.1073/pnas.0904636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi-Takagi A, Takaki M, Graziane N, et al. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci. 2010;13:327–332. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penzes P, Jones KA. Dendritic spine dynamics—a key role for kalirin-7. Trends Neurosci. 2008;31:419–427. doi: 10.1016/j.tins.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kothiyal P, Cox S, Ebert J, et al. An overview of custom array sequencing. Curr Protoc Hum Genet. 2009;61:7.17.1–7.17.11. doi: 10.1002/0471142905.hg0717s61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda M, Aleksic B, Kirov G, et al. Copy number variation in schizophrenia in the Japanese population. Biol Psychiatry. 2010;67:283–286. doi: 10.1016/j.biopsych.2009.08.034. [DOI] [PubMed] [Google Scholar]
- 22.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrer-Costa C, Gelpi JL, Zamakola L, et al. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21:3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- 24.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 25.Vassos E, Collier DA, Holden S, et al. Penetrance for copy number variants associated with schizophrenia. Hum Mol Genet. 2010;19:3477–3481. doi: 10.1093/hmg/ddq259. [DOI] [PubMed] [Google Scholar]
- 26.O'Donovan MC, Craddock N, Norton N, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 27.Johnson RC, Penzes P, Eipper BA, Mains RE. Isoforms of kalirin, a neuronal Dbl family member, generated through use of different 5'- and 3'-ends along with an internal translational initiation site. J Biol Chem. 2000;275:19324–19333. doi: 10.1074/jbc.M000676200. [DOI] [PubMed] [Google Scholar]
- 28.Keshava Prasad TS, Goel R, Kandasamy K, et al. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amanchy R, Periaswamy B, Mathivanan S, et al. A curated compendium of phosphorylation motifs. Nat Biotechnol. 2007;25:285–286. doi: 10.1038/nbt0307-285. [DOI] [PubMed] [Google Scholar]
- 30.Thomson M, Gunawardena J. Unlimited multistability in multisite phosphorylation systems. Nature. 2009;460:274–277. doi: 10.1038/nature08102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Hauser ER, Shah SH, et al. Peakwide mapping on chromosome 3q13 identifies the kalirin gene as a novel candidate gene for coronary artery disease. Am J Hum Genet. 2007;80:650–663. doi: 10.1086/512981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krug T, Manso H, Gouveia L, et al. Kalirin: a novel genetic risk factor for ischemic stroke. Hum Genet. 2010;127:513–523. doi: 10.1007/s00439-010-0790-y. [DOI] [PubMed] [Google Scholar]
- 33.Henkemeyer M, Itkis OS, Ngo M, Hickmott PW, Ethell IM. Multiple EphB receptor tyrosine kinases shape dendritic spines in the hippocampus. J Cell Biol. 2003;163:1313–1326. doi: 10.1083/jcb.200306033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinberg S, Mors O, Borglum AD, et al. Expanding the range of ZNF804A variants conferring risk of psychosis. doi: 10.1038/mp.2009.149. [published online ahead of print January 05, 2010]. Mol Psychiatry. doi: 10.1038/mp.2009.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johansen CT, Wang J, Lanktree MB, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamaguchi-Kabata Y, Nakazono K, Takahashi A, et al. Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. Am J Hum Genet. 2008;83:445–456. doi: 10.1016/j.ajhg.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.