Abstract
Recent studies have shown that the genes of the gibberellin (GA) biosynthesis pathway in the fungus Gibberella fujikuroi are organized in a cluster of at least seven genes. P450–1 is one of four cytochrome P450 monooxygenase genes in this cluster. Disruption of the P450–1 gene in the GA-producing wild-type strain IMI 58289 led to total loss of GA production. Analysis of the P450–1-disrupted mutants indicated that GA biosynthesis was blocked immediately after ent-kaurenoic acid. The function of the P450–1 gene product was investigated further by inserting the gene into mutants of G. fujikuroi that lack the entire GA gene cluster; the gene was highly expressed under GA production conditions in the absence of the other GA-biosynthesis genes. Cultures of transformants containing P450–1 converted ent-[14C]kaurenoic acid efficiently into [14C]GA14, indicating that P450–1 catalyzes four sequential steps in the GA-biosynthetic pathway: 7β-hydroxylation, contraction of ring B by oxidation at C-6, 3β-hydroxylation, and oxidation at C-7. The GA precursors ent-7α-hydroxy[14C]kaurenoic acid, [14C]GA12-aldehyde, and [14C]GA12 were also converted to [14C]GA14. In addition, there is an indication that P450–1 may also be involved in the formation of the kaurenolides and fujenoic acids, which are by-products of GA biosynthesis in G. fujikuroi. Thus, P450–1 displays remarkable multifunctionality and may be responsible for the formation of 12 products.
Keywords: cytochrome P450, ent-kaurenoid metabolism, GA14 synthase
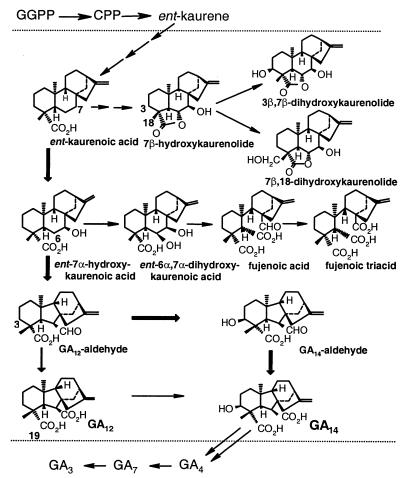
Gibberellins (GAs) constitute a large family of tetracyclic diterpenoid carboxylic acids, some members of which function as growth hormones in higher plants, promoting processes such as seed germination, stem elongation, flower development, and fruit growth (1). GAs were first identified as secondary products of the fungus Gibberella fujikuroi (mating population C) and are also produced by other fungal species (2–4). The early steps of the GA-biosynthetic pathway from transgeranylgeranyl diphosphate to GA12-aldehyde (Fig. 1) are identical for plants and fungi, but the pathways diverge thereafter. The major end product of the pathway in G. fujikuroi is gibberellic acid (GA3), which is a minor GA component in most plant species.
Figure 1.
Biosynthetic pathways to GAs and ent-kaurenoids in G. fujikuroi, indicating the reactions we propose to be catalyzed by P450–1 (between dotted lines). Thick arrows depict proposed major pathway from ent-kaurenoic acid to GA14.
Considerable progress has been made in isolating and characterizing the genes of GA biosynthesis in plants (5). These include genes encoding the membrane-associated cytochrome P450-monooxygenases and soluble dioxygenases that catalyze, respectively, intermediate and late steps of the pathway. Recently, the genes involved in GA biosynthesis in G. fujikuroi were found to be organized in a cluster containing at least seven genes (ref. 6 and unpublished work). The identification of these genes has provided the opportunity to study the fungal enzymes and metabolic steps they catalyze, which had previously proved difficult at the protein level (2). It has been possible to demonstrate enzyme activities in vitro for only the early part of the fungal GA pathway; these activities include the formation (7) and metabolism (8) of the intermediate, ent-kaurene, the 7β-hydroxylation of ent-kaurenoic acid (9), and the conversion of GA12-aldehyde to GA14 (10). Each of these reactions after ent-kaurene is catalyzed by microsomal monooxygenases with the properties expected for cytochrome P450s.
Approximately 18 steps are required for the formation of GA3 from transgeranylgeranyl diphosphate (GGPP), yet the cluster apparently contains only seven genes, one of which is GGPP synthase (6). The limited number of genes suggests that either not all of the genes have been identified or some genes encode multifunctional enzymes. On the basis of amino acid sequence similarities (ref. 6 and unpublished work), four of the seven genes in the GA-biosynthesis cluster in G. fujikuroi appear to encode cytochrome P450 monooxygenases. These genes are designated P450–1, P450–2, P450–3, and P450–4; the P450–1 gene is closely linked to P450–4 in the gene cluster, sharing the same promoter sequence but being transcribed in the opposite direction (Fig. 2). P450–1 is classified with P450–2 in the family CYP68 (6). In the present study, we have used two methods to characterize its function in the GA-biosynthetic pathway. We have shown by disruption of P450–1 that the gene is involved in the conversion of ent-kaurenoic acid. Furthermore, by expressing P450–1 in a G. fujikuroi mutant lacking the entire gene cluster, we have demonstrated that the gene product catalyzes four steps in the metabolism of ent-kaurenoic acid to GA14. Our results also indicate that P450–1 is responsible for the formation of ent-kaurenoid-derived by-products of the GA-biosynthetic pathway.
Figure 2.
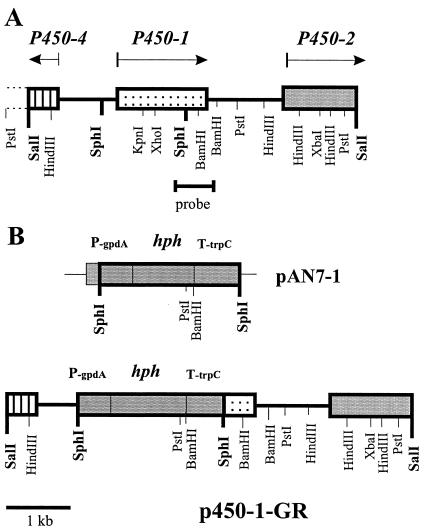
The P450–1 locus in the GA gene cluster in G. fujikuroi. (A) Physical map of the P450–1 ORF and surrounding genomic region. (B) Construction of the gene replacement vector pP450–1-GR.
Materials and Methods
Fungal Strains and Culture Conditions.
The wild-type strain G. fujikuroi IMI 58289 and the GA-defective mutant strain SG139 (11) were kindly provided by E. Cerda-Olmedo and J. Avalos (University of Sevilla, Sevilla, Spain).
Bacterial Strains and Plasmids.
For gene replacement experiments, a 6.5-kb SalI fragment carrying the P450–1 gene was cloned into pUC19 with a deleted SphI site (pUC19ΔSphI). An internal 1,519-bp fragment of this genomic SalI fragment was excised with SphI and replaced by a 2.5-kb SphI fragment from the vector pAN7–1 (12), carrying the hygromycin B resistance cassette. For gene complementation, a 4-Kb HindIII fragment carrying the whole P450–1 gene was cloned into pAN7–1 to give p450–1-GC.
Media and Culture Conditions.
For DNA isolation, fungal strains were grown in complete liquid medium optimized for Fusarium spp (100 ml) (13) for 3 days at 28°C on a rotary shaker set at 200 rpm. The mycelium was harvested by filtration through a sterile glass filter (G2, Schott Jena, Jena, Germany), washed with sterile distilled water, frozen in liquid nitrogen, and lyophilized for 24 h. The lyophilized mycelial tissue was ground into a fine powder. For RNA isolation, the fungal strains were grown in 20% ICI medium (14) for 48 h on a rotary shaker at 28°C. Cultures for metabolism studies were established in 100% ICI medium (100 ml) for 4 days at 25°C on a rotary shaker, then subcultured into 40% ICI medium (100 ml) and, after 5 days, into 10% ICI medium containing 1 mM AMO-1618 (Calbiochem) (100 ml).
Southern and Northern Blot Analysis.
Genomic DNA was isolated from lyophilized mycelium according to ref. 15. Plasmid DNA was extracted by using Genomed columns following the manufacturer's protocol (Genomed, Bad Oeynhausen, Germany). After endonuclease digestion and electrophoresis, genomic DNA was transferred onto Hybond N+ filters (Amersham Pharmacia). The 32P-labeled probes were prepared by using the random oligomer-primer method. Filters were hybridized at 65°C in 5 × Denhardt's solution (0.02% polyvinylpyrrolidone/0.02% Ficoll/0.02% BSA containing 5% dextran sulfate) (16). Filters were washed at the hybridization temperature in 2 × 0.18 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA (SSPE)/0.1% SDS, and 1 × SSPE/0.1% SDS.
RNA was isolated by using the RNAgents Total RNA Isolation Kit (Promega). Northern blot hybridization was accomplished by the method of Church and Gilbert (17). The conserved Botrytis cinerea actin gene (J. van Kan, personal communication) was used as a control for RNA transfer.
Transformation of G. fujikuroi.
Preparation of protoplasts and the transformation procedure were carried out as described (18). For gene replacement, 107 protoplasts (100 μl) of strain IMI 58289 were transformed with 10 μg of the SacI-digested gene replacement vector P450–1-GR. For complementation of the mutant strain SG139 with the intact P450–1 gene, protoplasts of SG139 were transformed with 10 μg of the circular complementation vector P450–1-GC. For complementation of the restriction enzyme-meditated integration-deletion mutant Gib1 (18) carrying an additional nia-D mutation, a cotransformation was performed by using 10 μg each of the vector P450–1-GK and the vector pJ1, containing the intact G. fujikuroi niaD gene (19).
Transformed protoplasts were regenerated at 28°C in a complete regeneration agar [0.7 M sucrose/0.05% yeast extract/0.1% (NH4)2SO4] containing 120 μg/ml of hygromycin B (Calbiochem) for 6–7 days or, in the case of cotransformation, in a stabilized (0.8 M KCl) Czapex–Dox agar (Merck). Single conidial cultures were established from hygromycin B-resistant transformants and used for DNA isolation and Southern blot analysis.
PCRs.
G. fujikuroi genomic DNA (50 ng) was used as template for amplification of the P450–1 gene in the wild type and transformants of strain SG139 with the P450–1 gene copy. The specific primers (50 ng), which were synthesized by MWG–Biotech (Munich), had the following sequences: forward (P450–1-F), 5′-TAT ACA CCC TCG CCT GTG AGG G-3′; reverse (P450–1-R), 5′-GTG AGG AGA CTT GCC GTC ATA GG-3′. DNA amplification was performed in 50-μl mixtures by using 2 units of Taq DNA polymerase (Red-Taq, Sigma–Aldrich). PCR was carried out for 36 cycles, each comprising 1 min denaturation at 94°C, 0.5 min annealing at 56°C, and 1.5 min synthesis at 72°C.
GA Analysis.
For analysis of GA formation, strain IMI 58289 and P450–1-disrupted mutants were cultivated in 100-ml Erlenmeyer flasks containing 20 ml of 20% ICI medium. The cultures were incubated for 7 days on a rotary shaker (200 rpm) at 28°C. GA production was determined by HPLC (20) by using a Merck HPLC system with a Lichrispher 100 RP18 column (5 μm; 250 × 4; Merck) or by TLC (18). Extracts of culture filtrates and mycelia were analyzed by combined GC/MS, as described previously (18).
Incubations with Radiolabeled Substrates.
ent-[14C4]kaurenoic acid (specific radioactivity, 7.51 TBq⋅mol−1), ent-7α-hydroxy[14C4]kaurenoic acid (7.91 TBq⋅mol−1), ent-6α,7α-dihydroxy[14C4]kaurenoic acid (4.64 TBq⋅mol−1), [14C4]GA12 (4.40 TBq⋅mol−1), and [14C4]GA12-aldehyde (6.81 TBq⋅mol−1) were prepared from R-[2-14C]mevalonic acid (Amersham Pharmacia) by using a cell-free system from pumpkin endosperm, as described (21). Fungal cultures, grown in 40% ICI medium for 2 days at 28°C, were harvested and the mycelia washed with 0% ICI medium (15). For experiments to characterize SG139 transformants, mycelia were resuspended in 0% ICI and 1-ml aliquots transferred to 25-ml sterile flasks containing 5 ml of the same medium. The radiolabeled substrates (500 Bq per flask) or ent-[17-2H]kaurenoic acid (20 μg to 4 mg per flask) were added to the cultures in methanol (10–40 μl). After incubation on an orbital shaker at 28°C for 2 days, cultures were filtered, and filtrates were adjusted to pH 3.0 with 2 M HCl and partitioned against ethyl acetate (3 × equal volume). The combined organic layers were evaporated under vacuum and the residue dissolved in 20% methanol/H2O, pH 3.0. This solution was applied to a C18 cartridge (SepPak, Waters) and the products eluted with methanol and further analyzed by HPLC on a C18 column (Symmetry, Waters) in a Waters 600 instrument by using a linear gradient from 60 to 100% methanol/H2O, pH 3.0, over 15 min, followed by 100% methanol for 15 min. The flow rate was 1 ml/min. Fractions (1 ml) were collected and the radioactivity measured by liquid scintillation counting.
For incubations with P450–1-disrupted mutants, radiolabeled substrates (2–4 kBq) were added in methanol (20–50 μl) to 5-day-old cultures in 10% ICI medium (5 ml) in sterile 50 ml Falcon tubes. After incubation on an orbital shaker at 25°C for 3 days, the cultures were filtered and the filtrates extracted, as described above. The mycelia were extracted with 10 ml of methanol overnight, the extracts passed although silica gel (500 mg), which was eluted with a further 10 ml of methanol, and the combined eluates taken to dryness in vacuo. The extracts were analyzed by reverse-phase HPLC on a C18 Hypersil column (25 × 0.46 cm) with on-line radioactivity monitoring (22); culture extracts were eluted with a linear gradient of 20–100% solvent B over 24 min at 1 ml⋅min−1, after which the column was eluted with solvent B for a further 5 min (solvent A, 10% methanol-water; solvent B, methanol, each containing 50 μl⋅liter−1 acetic acid) and mycelial extracts, by using a gradient of 75–100% B over 30 min followed by 30 min with B.
Microsomes were prepared from the mycelia of the SG139-T7 transformant as described previously (10) and incubated with [14C4]GA12-aldehyde or [14C4]GA12 (10,000 dpm) in the presence of 1 mM NADPH and 5 μM FAD in a final volume of 100 μl. Products were extracted and analyzed by HPLC, as described (10).
Results
Disruption of P450–1.
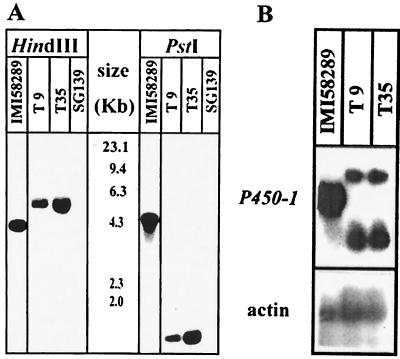
The P450–1 gene was mapped between P450–4 and P450–2 in the GA biosynthesis gene cluster (Fig. 2A). It was shown in a previous experiment (6) that replacement of a central part of genes P450–1 and P450–2 led to the loss of GA biosynthesis. To identify the biosynthetic steps in which the P450–1 gene product is involved, the P450–1 locus was specifically inactivated in the wild-type strain IMI 58289 by transformation with the linearized replacement vector, P450–1-GR, containing the hygromycin resistance cassette as selection marker (Fig. 2B). Among the 35 transformants generated, two, ΔP450–1-T9 and -T35, failed to produce GAs when cultured in a GA production medium. Southern blot analysis of genomic DNA from these two transformants and the wild-type after digestion with HindIII and PstI and probing with the 460-bp P450–1 cDNA fragment (see Fig. 2) indicated clear differences (Fig. 3A). The probe hybridized to a 4.0-kb HindIII and a 4.3-kb PstI fragment from the wild-type strain and to 5.0-kb HindIII and 1.8-kb PstI fragments (because of an additional PstI-site in the hygromycin cassette) from the transformants. These restriction patterns were consistent with a double-crossover event between the disruption vector and the P450–1 locus. Northern blot analysis revealed two transcripts in ΔP450–1-T9 and -T35 of altered lengths and lower abundance compared with the 1.6-kb transcript from the wild type (Fig. 3B). The larger transcript may be produced from reading through the terminator of the hygromycin B resistance cassette, but the origin of the smaller transcript is unclear.
Figure 3.
Molecular analysis of P450–1 replacement mutants (A) Southern blot analysis of G. fujikuroi strains IMI 58289 (wild type), SG139 (mutant lacking GA-biosynthesis gene cluster), and gene-disruption mutants ΔP450–1-T9 and ΔP450–1-T35. Genomic DNA from each strain was digested with either HindIII (lanes 1–3) or PstI (lanes 4–6), and the blots were probed with P450–1 cDNA. The size standards are indicated in kilobases. (B) Northern blot analysis of the wild-type strain IMI 58289 and the replacement mutants T9 and T35. The blot was probed with the P450–1 cDNA and the Botrytis cinerea actin gene (loading control)
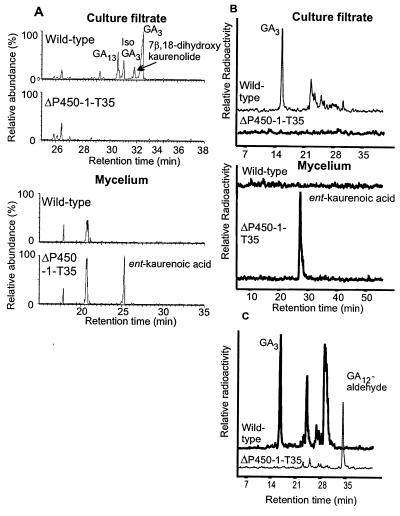
The wild-type strain and T35 were grown under GA-inducing conditions for 5 days, after which the culture filtrate and mycelia were analyzed by full-scan GC/MS (Fig. 4A). The culture filtrate from the wild type contained mainly GA3, with lower amounts of GA13, and 7β,18-dihydroxykaurenolide. The isolactone of GA3, which was also present, may be an artifact of the chromatography. Strain ΔP450–1-T35 accumulated ent-kaurenoic acid in the mycelium but not later intermediates in the culture filtrate (Fig. 4A), so that the block appeared to be in the conversion of ent-kaurenoic acid to ent-7α-hydroxykaurenoic acid. To confirm this, we incubated T35 and the wild-type strain with 14C-labeled ent-kaurenoic acid and GA12-aldehyde. The mutant strain T35 metabolized neither ent-kaurenoic acid, which accumulated in the mycelia (Fig. 4B), nor GA12-aldehyde, which was extracted from the culture filtrate (Fig. 4C). The wild-type strain converted both intermediates efficiently to GA3. These results suggest that the P450–1 enzyme catalyzes not only the conversion of ent-kaurenoic acid but also at least one subsequent oxidative step in the pathway.
Figure 4.
Characterization of gene disruption mutant ΔP450–1-T35. (A) Total ion currents from GC/MS analyses of extracts of culture filtrates and mycelia from G. fujikuroi strains IMI 58289 (wild type) and ΔP450–1-T35. (B and C) HPLC radiochromatograms of extracts from the culture filtrates and mycelia after incubation of ent-[14C4]kaurenoic acid (B) and [14C4]GA12-aldehyde (culture filtrate only) (C) with IMI58289 and ΔP450–1-T35.
Functional Analysis of P450–1.
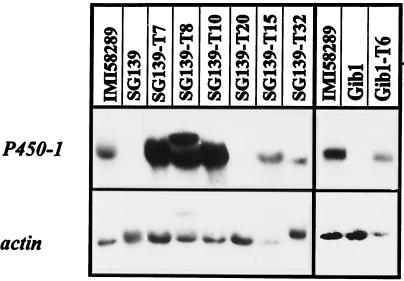
To define precisely the metabolic steps catalyzed by the P450–1 enzyme, we transformed the GA-deficient mutant strain SG139 with the gene complementation vector, pP450–1-GC, carrying the whole wild-type P450–1 gene. Mutant SG139, which was produced by UV treatment (11), lacks the entire GA gene cluster, as demonstrated by Southern blot (see Fig. 3A) and PCR analysis (data not shown). In an additional experiment, the GA-deficient mutant Gib1, which also lacks the GA gene cluster as part of an ≈300-Kb fragment that was lost from chromosome IV after restriction enzyme-meditated integration (18), was also transformed with pP450–1-GC. Of 45 transformants analyzed, 38 contained the expected 2,310-bp PCR fragment between primers P450–1-F and P450–1-R. To confirm that the gene is expressed in the transformants, Northern blot analysis was performed on five PCR-positive lines derived from SG139 (SG139-T7, -T8, -T10, -T15, and -T32), one transformant that did not reveal a PCR fragment of the expected size (SG139-T20), and one PCR-positive line (Gib1-T6) derived from Gib1. IMI58289, SG139, and Gib1 were also analyzed as controls. As shown in Fig. 5, P450–1 is expressed in all PCR-positive transformants tested and in the wild-type strain. No transcript for P450–1 was present in the deletion mutants SG139 and Gib1 or in SG139-T20.
Figure 5.
Northern blot analysis of the G. fujikuroi deletion mutants SG139 and Gib1, and strains transformed with the vector P450–1-GC carrying the complete P450–1 gene copy. The blot was probed with the P450–1 cDNA and the Botrytis cinerea actin gene (loading control)
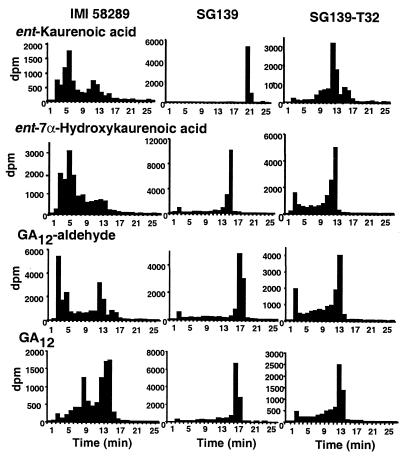
Three SG139 transformants (SG139-T7, SG139-T10, and SG139-T32) and one transformant of Gib1 (Gib1-T6) were incubated for 2 days with ent-[14C]kaurenoic acid, ent-7α-hydroxy[14C]kaurenoic acid, [14C]GA12-aldehyde, and [14C]GA12 (not with Gib1-T6) under GA-inducing conditions, and the products were analyzed by HPLC (shown for SG139-T32 in Fig. 6). Each transformant metabolized all four substrates, the major product in each case having a retention time (Rt) of 13–14 min. The main radiolabeled compound in this fraction was identified by GC/MS as [14C]GA14 (Table 1). After shorter (2- to 6-h) incubations of SG139-T32 with ent-[14C]kaurenoic acid, small amounts of ent-7α-hydroxy[14C]kaurenoic acid (Rt, 16 min) and 7β,18-dihydroxy[14C]kaurenolide (Rt, 13 min) were identified, whereas no other product besides [14C]GA14 could be detected by GC/MS after incubations with ent-7α-hydroxy[14C]kaurenoic acid or [14C]GA12-aldehyde. Neither GA12-aldehyde nor GA14-aldehyde, which has been demonstrated to be an intermediate on the main pathway to GA3 in G. fujikuroi (23), accumulated in any of the incubations. The wild-type IMI 58289 strain, from which SG139 was obtained, converted ent-[14C]kaurenoic acid, ent-7α-hydroxy[14C]kaurenoic acid, and [14C]GA12-aldehyde to products that are more polar than GA14, including GA3 (Rt, 5–6 min), whereas the major products from [14C]GA12 eluted with a Rt of 13–15 min. None of the substrates were metabolized by the deletion mutants SG139 (Fig. 6) and Gib1 (not shown).
Figure 6.
HPLC-radioactivity profiles of total extracts from incubations of ent-[14C4]kaurenoic acid, ent-7α-hydroxy[14C4]kaurenoic acid, [14C4]GA12-aldehyde, or [14C4]GA12 with the G. fujikuroi wild-type strain IMI 58289, deletion mutant SG139, and the P450–1 transformant SG139-T32. Retention times for ent-kaurenoic acid, ent-7α-hydroxykaurenoic acid, GA12-aldehyde, and GA12 are 22, 16, 18, and 17 min, respectively. HPLC and incubation conditions were as described in Materials and Methods, except for ent-kaurenoic acid with SG139-T32, for which the incubation time was 4 h.
Table 1.
Products identified by GC/MS from incubations of isotopically labeled intermediates with mutant strain SG139 transformed with P450-1
| Substrate | Product |
|---|---|
| ent-[17-2H2]kaurenoic acid* | ent-7α-hydroxy[2H2]kaurenoic acid |
| [2H2]GA12 | |
| [2H2]GA14 | |
| 7β-hydroxy[2H2]kaurenolide | |
| 7β,18-dihydroxy[2H2]kaurenolide | |
| 3β,7β-dihydroxy[2H2]kaurenolide | |
| [2H2]fujenoic acid | |
| ent-7α-hydroxy-[14C4]kaurenoic acid | [14C4]GA14 |
| ent-6α, 7α-dihydroxy-[14C4]kaurenoic acid* | [14C4]fujenoic acid |
| [14C4]fujenoic triacid | |
| [14C4]GA12-aldehyde | [14C4]GA14 |
| [14C4]GA12 | [14C4]GA14 |
The mass spectrometric data are provided as Table 2, which is published as supplemental data on the PNAS web site, www.pnas.org.
Total extracts of culture filtrates were analyzed by GC/MS. Otherwise, extracts were separated by HPLC, and fractions containing radioactivity were analyzed.
When milligram amounts of ent-[2H]kaurenoic acid were incubated with the SG139-T7 transformant, 41% of substrate remained, and [2H]GA14 was produced as the main product (45%). The intermediates ent-7α-hydroxy[2H]kaurenoic acid and [2H]GA12 were detected by GC/MS as minor products, together with 2H-labeled 7β-hydroxykaurenolide, 7β,18-dihydroxykaurenolide, 3β,7β-dihydroxykaurenolide, and fujenoic acid (Table 1). The P450–1 transformants were also incubated with ent-6α,7α-dihydroxy[14C]kaurenoic acid and the products separated by TLC. In each case, there was complete conversion to give a radioactive band at Rf 0.25–0.38, which was shown by GC/MS for SG139-T7 to contain 14C-labeled fujenoic acid and fujenoic triacid (Table 1). After incubation with the deletion mutants, a single band of radioactivity was obtained at the same Rf (0.62–0.75) as the substrate, indicating there had been no conversion.
The metabolism of [14C]GA12-aldehyde and [14C]GA12 was investigated also with isolated microsomes from SG139-T7 mycelia. In the presence of 1 mM NADPH and 5 μM FAD, [14C]GA12-aldehyde was efficiently converted to [14C]GA14 (60%) plus smaller amounts of [14C]GA12 (15%), whereas [14C]GA12 remained unchanged under these conditions.
Discussion
We have used two methods to determine the function of the cytochrome P450 monooxygenase encoded by P450–1, one of a cluster of seven genes involved in GA biosynthesis in G. fujikuroi. First, specific disruption of this gene in a GA-producing strain of the fungus indicated that the encoded enzyme catalyzed the 7β-hydroxylation of ent-kaurenoic acid. However, we showed that at least one later step in the pathway was also affected by disrupting P450–1. Second, we examined the function of P450–1 in the absence of the other GA-biosynthetic enzymes by expressing the gene in G. fujikuroi mutants that lacked the GA-biosynthesis gene cluster. By this means, we could demonstrate that the enzyme converts ent-kaurenoic acid to GA14 and thus catalyzes four steps in the GA-biosynthetic pathway: hydroxylation at C-7, oxidation at C-6 resulting in contraction of ring B (24, 25), 3β-hydroxylation, and finally oxidation at C-7 (Fig. 1). Intermediates in this sequence, ent-7α-hydroxykaurenoic acid and GA12-aldehyde, as well as GA12, which is not thought to be a normal precursor of 3β-hydroxylated GAs (26), were converted efficiently by the enzyme to GA14. Even when ent-kaurenoic acid was applied at very high concentrations, GA14 was the predominant product, and only low amounts of ent-7α-hydroxykaurenoic acid and GA12 were found in the culture medium. This result suggests that the first step in the reaction sequence may limit the overall reaction rate, although uptake of the substrates into the cell may be the limiting factor.
Microsomes from G. fujikuroi have been shown to catalyze the reaction sequence from ent-kaurenoic acid to GA12 and GA14 (9, 10, 27). However, microsomal preparations failed to convert GA12 to GA14 by 3β-hydroxylation, regardless of whether they were prepared from a wild-type strain (10) or from the P450–1 transformants (this work). This apparent inconsistency between the experiments by using intact cultures of the P450–1 transformants and those with microsomal preparations could be explained if GA12 is a relatively poor substrate for 3β-hydroxylation. If GA12 is converted to GA14 at a slow rate, this reaction could occur only in cultures of the P450–1 transformant, where the substrate is in contact with high levels of enzyme for long periods of time in the absence of competing enzyme activities. Thus, the major pathway to GA14 is presumably via GA14-aldehyde (23), which was not detected as an intermediate in the present study. In our experiments, the wild-type strain converted GA12 to nonpolar products, consistent with the results of Bearder et al. (28), who found that GA12 is metabolized mainly to the non-3β-hydroxylated GA, GA9. The small amount of GA3 formed from GA12 may be the result of the low rate of 3β-hydroxylation.
We provide evidence to suggest that P450–1, as well as catalyzing reactions of the GA-biosynthetic pathway, may be involved in two branch pathways, those from ent-kaurenoic acid to the kaurenolides and those from ent-7α-hydroxykaurenoic acid to seco-ring B products (3) (Fig. 1). We detected 7β-hydroxy-, 3β,7β-dihydroxy-, and 7β,18-dihydroxykaurenolides together with fujenoic acid as products after incubations of ent-kaurenoic acid with cultures of the P450–1 transformants, whereas the deletion mutants lacking the P450–1 gene failed to metabolize ent-kaurenoic acid or ent-7α-hydroxykaurenoic acid. Furthermore, the transformants formed fujenoic acid and fujenoic triacid from ent-6α,7α-dihydroxykaurenoic acid, whereas the deletion mutants did not metabolize this intermediate. ent-6α,7α-dihydroxykaurenoic acid, which was postulated as the precursor of the seco-ring B products (29), has been shown to be formed together with GA12-aldehyde in cell-free preparations from pumpkin endosperm by oxidation of ent-7α-hydroxykaurenoic acid on C-6 (25). Full confirmation for the involvement of P450–1 in the branch pathways will require demonstration of each metabolic step in the P450–1 transformants and its absence in the disruption or deletion mutants. However, the seven reactions required for the branch pathways (Fig. 1), involving oxidation on C-3, C-6, C-7, and C-18, are consistent with the demonstrated activities of P450–1 for the conversion of ent-kaurenoic acid to GA14.
Sequential oxidations at a single carbon atom that are mediated by cytochrome P450s have been described as, for example, in the loss of C-14 in steroid biosynthesis (30) and the three-step oxidation of ent-kaurene to ent-kaurenoic acid in G. fujikuroi (8). However, the reactions catalyzed by P450–1 involve oxidations at four different carbon atoms, C-6, C-7, C-3, and C-18, most of which are nonadjacent. Oxidation at multiple sites by a single P450 enzyme has been reported for monooxygenases involved in mammalian steroid biosynthesis as, for example, in the 11β- and 18-hydroxylation of 11-deoxycorticosterone by porcine adrenocortical cytochrome P450 (11β) (31) and 11β-, 18-, and 19-hydroxylations of the same steroid by rat cytochrome P450 (11β) (32). Furthermore, in the fungus Curvularia lunata, 11-deoxycortisol is hydroxylated at positions 11β and 14α by the same P450 monooxygenase (33). P450–1 contains a single heme-binding motif so that all reactions catalyzed, which include oxidations at seemingly remote carbon centers, must occur at the same active site. However, depending on conformation, the sites oxidized by P450–1, for example C-3β and C-7 in GA12-aldehyde, are sufficiently close that little change in substrate-binding orientation may be required for either reaction to take place. Differences in the requirement for FAD for GA12-aldehyde 7-oxidase and 3β-hydroxylase activities (10) might suggest interaction of the P450–1 heme with different electron transport proteins for these two reactions. A small change in the binding orientation of 7β-hydroxykaurenolide relative to that of ent-7α-hydroxykaurenoic acid would allow the former substrate, but not the latter, to be hydroxylated at C-18 or C-3β.
Analysis of its secondary structure indicates that P450–1, in common with most eukaryotic cytochrome P450s, contains a single membrane-spanning domain at its N terminus. However, the enzyme is apparently loosely bound to the microsomal fraction, as it can be easily solubilized in the presence of high salt concentrations (9).
We have recently become aware of parallel work showing that barley and Arabidopsis contain cytochrome P450 monoxygenases with catalytic activity similar to that of P450–1 (34). The plant enzymes, which belong to the CYP88A subfamily, convert ent-kaurenoic acid to GA12. There must, however, be important differences between the fungal and plant enzymes because in plants, 3β-hydroxylation is catalyzed by 2-oxoglutarate-dependent dioxygenases and, in most cases, occurs late in the pathway, after C-20 has been removed (see ref. 5). The plant enzymes responsible for GA12 synthesis are not apparently capable of 3β-hydroxylation.
The involvement of dioxygenases in GA biosynthesis in plants has been shown for oxidation at C-20, C-2β, C-3β, and, in pumpkin, C-7. Oxidation at C-20, resulting in the loss of this C atom, occurs also in G. fujikuroi. However, as no dioxygenase genes have been identified within the GA-biosynthesis gene cluster of the fungus, it seems likely that the 20-oxidase is a different type of enzyme in G. fujikuroi, most probably a cytochrome P450. The conversion of GA12-aldehyde to GA12 (7-oxidation) in pumpkin seed can be catalyzed by either monooxygenase or dioxygenase activities (35, 36), but it is not known whether the dioxygenase is of more general significance. In Arabidopsis, there are no candidate dioxygenases GA 7-oxidase genes (37), so it appears likely that this activity is caused solely by a monooxygenase.
Research of GA biosynthesis has reached an exciting stage in plants and in G. fujikuroi. As more genes and enzymes are characterized, it is becoming clear that very significant differences exist in the genetics and biochemistry of GA biosynthesis in plants and fungus. For example, they differ in the organization of the genes, in the nature and substrate specificities of the enzymes, and in the mechanisms of regulation. For G. fujikuroi, most of the genes are now available, allowing new opportunities for detailed studies of their regulation. Furthermore, mutants lacking the GA-biosynthesis gene cluster provide a highly efficient and convenient expression system for the preparation of enzymes for studies of function and structure.
Supplementary Material
Acknowledgments
We thank Jessica Schulte and Katja Hölter (Münster) and Reju D'Cunha and James Melichar (Long Ashton) for technical assistance. This project was supported by the Deutsche Forschungsgemeinschaft (Tu 101/7) and the Deutscher Akademischer Austauschdienst/Consejo Nacional de Ciencia y Technología Cooperation Program. The Institute of Arable Crops Research–Long Ashton Research Station receives grant-aided support from the Biotechnology and Biological Sciences Research Council of the United Kingdom.
Abbreviation
- GA
gibberellin
References
- 1.Hooley R. Plant Mol Biol. 1994;26:1529–1555. doi: 10.1007/BF00016489. [DOI] [PubMed] [Google Scholar]
- 2.Rademacher W. Plant Growth Regul. 1994;15:303–314. [Google Scholar]
- 3.MacMillan J. Nat Prod Res. 1997;14:221–243. [Google Scholar]
- 4.Tudzynski B. In: The Mycota V, Plant Relationship, Part A. Esser K, Lemke P A, editors. Berlin: Springer; 1997. pp. 167–184. [Google Scholar]
- 5.Hedden P, Phillips A L. Trends Plant Sci. 2000;5:523–530. doi: 10.1016/s1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- 6.Tudzynski B, Hölter K. Fungal Genet Biol. 1998;25:157–170. doi: 10.1006/fgbi.1998.1095. [DOI] [PubMed] [Google Scholar]
- 7.Fall R R, West C A. J Biol Chem. 1971;246:6913–6928. [PubMed] [Google Scholar]
- 8.Ashman P J, Mackenzie A, Bramley P M. Biochim Biophys Acta. 1990;1036:151–157. doi: 10.1016/0304-4165(90)90027-t. [DOI] [PubMed] [Google Scholar]
- 9.Jennings J C, Coolbaugh R C, Nakata D A, West C A. Plant Physiol. 1993;101:925–930. doi: 10.1104/pp.101.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urrutia O, Hedden P, Rojas M C. Phytochemistry. 2001;56:505–511. doi: 10.1016/s0031-9422(00)00381-2. [DOI] [PubMed] [Google Scholar]
- 11.Fernández-Martín R, Reyes F, Domenech C E, Cabrera E, Bramley P M, Barrero A F, Avalos J, Credá-Olmedo E. J Biol Chem. 1995;270:14970–14974. doi: 10.1074/jbc.270.25.14970. [DOI] [PubMed] [Google Scholar]
- 12.Punt P J, Oliver R P, Dingemanse M A, Pouwels P H, van den Hondel C A. Gene. 1987;56:117–124. doi: 10.1016/0378-1119(87)90164-8. [DOI] [PubMed] [Google Scholar]
- 13.Pontecorvo G V, Poper J A, Hemmonns L M, MacDonal K D, Buften A W J. Adv Genet. 1953;141:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 14.Geissman T A, Verbiscar A J, Phinney B O, Cragg G. Phytochemistry. 1966;5:933–947. [Google Scholar]
- 15.Doyle J J, Doyle J L. Focus. 1990;12:13–15. [Google Scholar]
- 16.Sambrook J, Fritisch W F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 17.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linnemannstöns P, Voss T, Hedden P, Gaskin P, Tudzynski B. Appl Environ Microbiol. 1999;65:2558–2564. doi: 10.1128/aem.65.6.2558-2564.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tudzynski B, Mende K, Weltring K M, Kinghorn J R, Unkles S E. Microbiology. 1996;142:533–539. doi: 10.1099/13500872-142-3-533. [DOI] [PubMed] [Google Scholar]
- 20.Barendse G W M, van de Werken P H, Takahashi N. J Chromatogr. 1980;198:449–455. [Google Scholar]
- 21.Graebe J E, Hedden P, Gaskin P, MacMillan J. Phytochemistry. 1974;13:1413–1440. [Google Scholar]
- 22.MacMillan J, Ward D A, Phillips A L, Sanchez-Beltran M J, Gaskin P, Lange T, Hedden P. Plant Physiol. 1997;113:1369–1377. doi: 10.1104/pp.113.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hedden P, MacMillan J, Phinney B O. J. Chem. Soc. Perkin Trans. I. 1974. , 587–592. [Google Scholar]
- 24.Evans R, Hanson J R, White A F. J. Chem. Soc. C. 1970. , 2601–2603. [Google Scholar]
- 25.Graebe J E, Hedden P, MacMillan J. J. Chem. Soc. Chem. Commun. 1975. , 161–162. [Google Scholar]
- 26.Bearder J R, MacMillan J, Phinney B O. Phytochemistry. 1973;12:2173–2179. [Google Scholar]
- 27.West C A. In: Biosynthesis and Its Control in Plants. Milborrow B V, editor. London: Academic; 1973. pp. 143–169. [Google Scholar]
- 28.Bearder J R, MacMillan J, Phinney B O. J. Chem. Soc. Perkin Trans. I. 1975. , 721–726. [Google Scholar]
- 29.Cross B E, Stewart J C, Stoddart J L. Phytochemistry. 1970;9:1065–1071. [Google Scholar]
- 30.Lamb D C, Kelly D E, Manning N J, Kaderbhai M A, Kelly S L. FEBS Lett. 1999;462:283–288. doi: 10.1016/s0014-5793(99)01548-3. [DOI] [PubMed] [Google Scholar]
- 31.Sun T, Zhao Y, Nonaka Y, Okamoto M. J Steroid Biochem Biol. 1995;52:227–232. doi: 10.1016/0960-0760(94)00167-k. [DOI] [PubMed] [Google Scholar]
- 32.Nonaka Y, Okamoto M. Eur J Biochem. 1991;202:897–902. doi: 10.1111/j.1432-1033.1991.tb16449.x. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki K, Sanga K-I, Chikaoka Y, Itagaki E. Biochim Biophys Acta. 1993;1203:215–223. doi: 10.1016/0167-4838(93)90086-7. [DOI] [PubMed] [Google Scholar]
- 34.Helliwell C A, Chandler P M, Poole A, Dennis E S, Peacock W J. Proc Natl Acad Sci USA. 2001;98:2065–2070. doi: 10.1073/pnas.041588998. . (First Published February 6, 2001; 10.1073/pnas.041588998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hedden P, Graebe J E, Beale M H, Gaskin P, MacMillan J. Phytochemistry. 1984;23:569–574. [Google Scholar]
- 36.Lange T. Proc Natl Acad Sci USA. 1997;94:6553–6558. doi: 10.1073/pnas.94.12.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.The Arabidopsis Genome Initiative. Nature (London) 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.