Abstract
Clostridium difficile, a causative agent of antibiotic-associated diarrhea and its potentially lethal form, pseudomembranous colitis, produces two large protein toxins that are responsible for the cellular damage associated with the disease. The level of toxin production appears to be critical for determining the severity of the disease, but the mechanism by which toxin synthesis is regulated is unknown. The product of a gene, txeR, that lies just upstream of the tox gene cluster was shown to be needed for tox gene expression in vivo and to activate promoter-specific transcription of the tox genes in vitro in conjunction with RNA polymerases from C. difficile, Bacillus subtilis, or Escherichia coli. TxeR was shown to function as an alternative sigma factor for RNA polymerase. Because homologs of TxeR regulate synthesis of toxins and a bacteriocin in other Clostridium species, TxeR appears to be a prototype for a novel mode of regulation of toxin genes.
Clostridium difficile, a Gram-positive, anaerobic, spore-forming bacterium, has been identified as one of the major causative agents of antibiotic-associated diarrhea and has been implicated in almost all cases of pseudomembranous colitis (PMC; ref. 1). Virulent strains of C. difficile generally produce two very large protein toxins (ToxA and ToxB) that have been identified as major virulence factors (2). Both ToxA and ToxB monoglucosylate mammalian Ras-related, small GTP-binding proteins involved in the regulation of F-actin cytoskeletal integrity leading to depolymerization of actin filaments and cytotoxicity (2–5).
Toxin production varies greatly among different toxigenic strains and appears to be highly influenced by environmental conditions (6, 7). Synthesis increases as cells enter stationary phase (6, 7) and is stimulated or inhibited by various amino acids present in the medium (8–11), stimulated by butyric acid but inhibited by butanol (12), repressed by rapidly metabolizable carbon sources (7), induced by addition of certain antibiotics (13), and stimulated by biotin limitation (14). However, the regulation of toxin synthesis is poorly understood at the molecular level, as is the mechanism that triggers an increase in the level of toxin synthesis leading to the transition from mild diarrhea to the potentially lethal PMC. The toxin genes, toxA and toxB (also known as tcdA and tcdB, respectively), are in close proximity on the chromosome and are among five ORFs found in a 19.6-kb DNA element called the “pathogenicity locus” that is typical of toxin-producing strains (15–17).
The tox promoters do not resemble the canonical σ70 promoters of bacteria (18) but do show strong similarity to the UV-inducible P1 promoter of the Clostridium perfringens bacteriocin locus (7). The expression of tox gene transcripts is coordinate, induced at the entry into stationary phase, and repressed by rapidly metabolizable carbon sources (6, 7), suggesting that the tox genes are likely to be under a common mode of regulation.
A potential breakthrough in the study of toxin regulation came from the analysis of txeR (also known as tcdD), the gene upstream of toxB. Moncrief and colleagues (19) demonstrated that, when expressed in trans in Escherichia coli, txeR activates toxA and toxB reporter fusions, suggesting that TxeR is a positive regulator of the tox genes. TxeR is an ≈22-kDa protein, contains a potential C-terminal helix–turn–helix DNA-binding motif, and shows sequence similarities to TetR, a positive regulator of the tetanus toxin gene in Clostridium tetani, BotR, a positive regulator of the botulism toxin genes in Clostridium botulinum and UviA, a putative positive regulator of the UV-inducible bacteriocin gene of C. perfringens (19–21). Here, we present evidence that TxeR is an alternative RNA polymerase sigma factor required for the activation of toxin gene expression in C. difficile.
Materials and Methods
Bacterial Strains and Growth Medium.
C. perfringens and C. difficile strains were grown in TY or TY medium supplemented with 1% glucose (TYG) as described previously (7). All routine plasmid constructions and cloning in E. coli were done by using standard techniques (22).
Construction of a GusA Reporter Fusion System in C. perfringens.
A transcriptional fusion vector for studying gene expression in C. perfringens was constructed by modifying the C. perfringens– E. coli shuttle vector pJIR750 (23). After deleting the lac promoter–operator sequences, we introduced the E. coli gusA gene engineered to contain the toxB ribosome-binding site upstream of the gusA start codon to create the transcriptional fusion vector pTUM177. Fragments corresponding to the toxA or toxB promoter regions (7) were cloned upstream of the gusA gene in pTUM177, creating plasmids pTUM181 and pTUM182, respectively.
Construction of a txeR-Expressing Plasmid for C. perfringens.
The txeR gene was amplified from the C. difficile chromosome by PCR and cloned in the E. coli– Bacillus subtilis shuttle vector pDG1664 (24). The txeR gene then was excised and ligated to the EcoRV-digested, broad host-range shuttle vector, pTRKL2 (25), to yield the plasmid pTUM307. To test the effect of TxeR on tox gene expression, plasmid pTUM181 or pTUM182 (see above) was introduced into electrocompetent C. perfringens strain SM101 (26) harboring either pTRKL2 or pTUM307. β-Glucuronidase activity of C. perfringens was assayed as described previously (7).
Overproduction and Partial Purification of TxeR.
To overproduce TxeR, the txeR gene was amplified from the C. difficile chromosome by PCR and cloned between the NcoI and XhoI sites of the expression vector pET28-b (Novagen). This construction created a translational fusion adding six carboxyl-terminal histidine residues to the txeR coding sequence and placed it under the control of the T7 promoter. The resulting plasmid, pCD54, was introduced into E. coli strain BL21λDE3 (pLysS), carrying the isopropyl β-d-thiogalactoside (IPTG)-inducible T7 RNA polymerase. To produce TxeR, a 2-liter culture of strain BL21λDE3 (pLysS, pCD54) was grown at 22°C in LB broth containing ampicillin and chloramphenicol. When the cells reached stationary phase (absorbance at 600 nm = 3), IPTG was added (1 mM) and incubation was continued for 3 h. The cells were harvested, resuspended in 20 ml of buffer I (50 mM sodium-phosphate, pH 8/300 mM NaCl/20 mM imidazole), and lysed by sonication, and the cell debris was removed by centrifugation. The supernatant was loaded on a 0.5-ml Ni+-NTA agarose column (Qiagen) and washed with buffer I, and TxeR was eluted with an imidazole gradient (30–500 mM). The protein was concentrated and exchanged into storage buffer (50 mM sodium-phosphate, pH 8/300 mM NaCl/50% glycerol). Protein purity was analyzed by Coomassie blue staining of an SDS-polyacrylamide gel (27), and protein concentration was measured by the method of Bradford (28).
Preparation of Antibodies to TxeR.
Rabbit serum was prepared against TxeR that was purified after overexpression in E. coli strain BL21λDE3 (pLysS) harboring pCD32, a derivative of pET16-b (Novagen) carrying the txeR gene under an IPTG-inducible T7 promoter. Induction of TxeR synthesis at 37°C resulted in formation of inclusion bodies, which were subjected to SDS/PAGE. TxeR was eluted from a gel slice, mixed with adjuvant, and injected s.c. into rabbits. Anti-TxeR antibodies were purified by absorption against crude extracts of E. coli strain BL21λDE3 (pLysS) harboring pET16-b.
Gel Retardation Experiments.
Fragments of 372, 347, and 353 bp containing the toxA, toxB, and gdh (C. difficile glutamate dehydrogenase gene) promoters, respectively, were amplified by PCR and labeled by using T4 polynucleotide kinase and [γ-32P]ATP (3,000 Ci/mmol; Amersham Pharmacia). E. coli RNA polymerase holoenzyme and core enzyme forms were purchased from Epicenter. B. subtilis RNA polymerase holoenzyme was prepared by F. W. Whipple (29). B. subtilis RNA polymerase core enzyme was a gift from J. Helmann (Cornell University, Ithaca, NY). Purification of C. difficile RNA polymerase will be described elsewhere (N.M., A. L. Sonenshein, and B.D., unpublished data). Labeled fragments (0.2 nM) were mixed with RNA polymerase holoenzyme or core enzyme (50–200 nM) in glutamate buffer (40 mM Hepes, pH 8/100 mM MgCl2/100 mM potassium glutamate/500 μg BSA per ml) and incubated for 60 min at room temperature in a total volume of 10 μl. Three microliters of a heparin–dye solution (150 μg heparin per ml/0.1% dye/50% sucrose) in glutamate buffer was added, and the mixture was loaded on a 4.5% polyacrylamide gel in TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) during electrophoresis. After electrophoresis, the gels were dried and analyzed by autoradiography. Holoenzyme was reconstituted by incubating one volume of 5 μM E. coli core enzyme with a 4-fold molar excess of TxeR for 30 min at 37°C. The concentration of TxeR was estimated based on its apparent abundance relative to that of other Coomassie blue-stained bands.
In Vitro Run-Off Transcription Reactions.
In vitro transcription reactions were carried out as described previously (30). The activities of all RNA polymerases were assayed by using poly(dA-dT) as template, and equal amounts of activity were used in any given experiment.
TxeR–Core Enzyme Interaction Experiments.
Samples (1 μg) of E. coli RNA polymerase core enzyme were immobilized on nitrocellulose filters (Amersham Pharmacia), and the membranes were blocked with 5% powdered milk in PBS at room temperature. Filters then were incubated for 2 h at 37°C with 200 μg of whole-cell extract of BL21λDE3 (pET28-b) (used as a negative control) or with 50–200 μg of whole-cell extract of BL21λDE3 (pCD54). The filters were washed and immunoblotted by using anti-TxeR serum and developed by using the enhanced chemiluminescence kit (Amersham Pharmacia).
Results
TxeR Stimulates toxA and toxB Expression in C. perfringens.
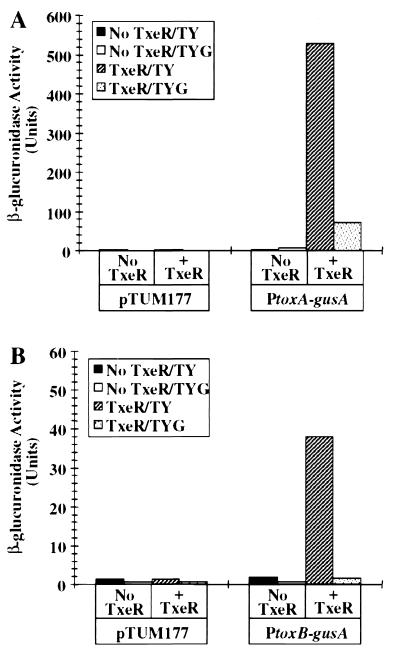
To study the mechanism of tox gene regulation, we created a transcriptional fusion vector, pTUM177 (see Materials and Methods). We chose C. perfringens because, unlike C. difficile, it can be manipulated genetically (31) and because C. perfringens is sufficiently close to C. difficile evolutionarily that the rules of promoter recognition and gene regulation should be conserved between the two species. When we introduced either the PtoxA-gusA fusion (Fig. 1A) or the PtoxB-gusA fusion (Fig. 1B) into C. perfringens carrying the compatible plasmid pTRKL2, we saw no detectable Gus activity in cells grown in either TY or TYG medium. However, when we introduced the PtoxA-gusA fusion or PtoxB-gusA fusion into a C. perfringens strain carrying txeR in trans (plasmid pTUM307), high levels of Gus activity were observed in cells grown in TY medium. This result is consistent with and confirms the observation of Moncrief et al. (19) that the txeR gene in trans stimulates tox gene expression in E. coli. We observed further that the TxeR-mediated activation of tox-gusA fusions was repressed in glucose-containing medium (TYG, Fig. 1), consistent with the behavior of the tox genes in their natural host (7).
Figure 1.
In vivo transcription activation of Ptox-gusA fusions by txeR in trans in C. perfringens. Fragments of DNA carrying either toxA (A) or toxB (B) gene promoters were cloned in the reporter fusion vector pTUM177 and introduced into C. perfringens with or without the txeR gene in trans. β-Glucuronidase activity of late stationary phase cells grown in TY or TYG medium was assayed as described previously (7). In control experiments, cells carrying the fusion vector alone were assayed. Solid bars, TY medium, no TxeR; open bars, TYG medium, no TxeR; striped bars, TY medium with TxeR; dotted bars, TYG medium, with TxeR.
Overexpression and Partial Purification of TxeR.
To understand the mechanism by which TxeR stimulates tox gene expression, we sought to purify TxeR for in vitro studies. Initial attempts were frustrated by the insolubility of TxeR overexpressed in E. coli. After trying many different approaches, we finally succeeded in overcoming this problem by cloning the txeR gene in pET28-b (as in pCD54), creating TxeR with six C-terminal histidine residues, by growing cells at low temperature (22°C) and by inducing expression of TxeR only when cells had reached stationary phase (see Materials and Methods).
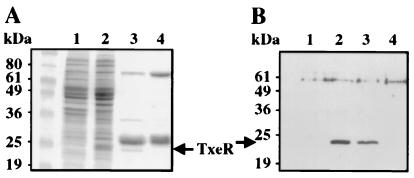
Analysis by SDS/PAGE of the soluble fraction of a crude extract of E. coli cells containing pCD54 showed an extra band with an apparent molecular mass of 24 kDa, which was absent in the control extract from cells carrying the pET28-b vector alone (Fig. 2A, lanes 1 and 2). His-tagged TxeR then was purified in a single step by using a nickel affinity column. Analysis by SDS/PAGE of partially purified TxeR protein, confirmed by Western blot analysis using anti-TxeR antibodies (Fig. 2B), showed some major contaminating proteins (Fig. 2A, lane 3). Unfortunately, we have never been able to improve the purity of TxeR. For this reason, we also purified the contaminating proteins in the same way from an E. coli strain carrying the vector pET28-b (Fig. 2A, lane 4) and used this preparation as a negative control for the in vitro experiments.
Figure 2.
Overproduction and purification of TxeR. (A) SDS/PAGE analysis of protein extracts from E. coli BL21λDE3 (pLysS) carrying either pCD54 or pET28-b. Lanes: 1, crude cell extracts from E. coli carrying the vector pET28-b; 2, crude cell extracts from E. coli carrying pCD54 (expressing TxeR); 3, TxeR-containing fraction from E. coli carrying pCD54 retained and eluted from a Ni+-NTA column; 4, an equivalent protein fraction from crude extract of E. coli carrying the vector pET28-b after Ni+-NTA chromatography. (B) Immunodetection of TxeR by using anti-TxeR antibodies. The protein samples in each lane correspond to those in A. The bands in lanes 2 and 3 corresponding to TxeR are indicated by an arrow.
RNA Polymerase Binds Specifically to tox Promoter Regions in the Presence of TxeR.
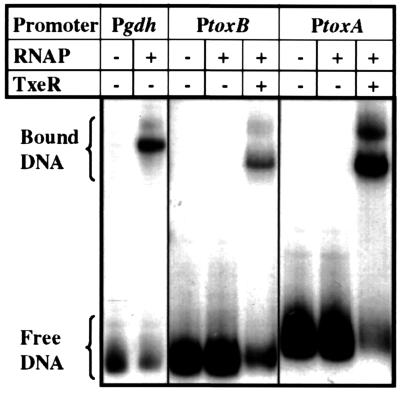
To determine whether TxeR activates toxA and toxB transcription by directly binding to the promoter regions of tox genes, we first used partially purified TxeR in gel mobility-shift assays with DNA fragments that contained the promoter regions of toxA and toxB. No shift in the mobility of the toxA or toxB promoter regions was seen (data not shown), even when we used large amounts of TxeR or when we modified the stringency of the binding reactions. This result suggests that TxeR may activate tox gene expression indirectly via other regulatory factors. One possibility is that TxeR interacts with RNA polymerase to activate tox gene transcription. To test this idea, RNA polymerase was purified from C. difficile (N.M. et al., unpublished data) and the ability of C. difficile RNA polymerase to bind to tox promoter regions in the presence and absence of TxeR was assayed. All binding reactions were challenged with the polyanion heparin, which is known to release RNA polymerase from weak, closed complexes with target DNA, without interfering with stable, open complexes (32). As shown in Fig. 3, C. difficile RNA polymerase holoenzyme was unable to bind to tox promoter-containing DNA fragments in the absence of added TxeR, even though it could bind well to the promoter region of the gdh gene, a gene expressed during the exponential growth phase (N.M. et al., unpublished data). Addition of partially purified TxeR greatly increased the ability of RNA polymerase to form heparin-resistant complexes at the tox promoters. Similar results were obtained with E. coli RNA polymerase (data not shown). On the other hand, addition of a fraction containing the proteins that contaminate our preparation of TxeR (Fig. 2A, lane 4) did not increase the affinity of RNA polymerase for the tox promoters (data not shown). These results suggest that TxeR is either a positive regulator that stabilizes binding of RNA polymerase to the tox promoters or a protein that alters the structure of RNA polymerase and thereby changes its promoter recognition properties.
Figure 3.
Gel mobility retardation of tox promoters with C. difficile RNA polymerase and TxeR; gel mobility retardation of the C. difficile glutamate dehydrogenase gene promoter (Pgdh), toxin B gene promoter (PtoxB), and toxin A gene promoter (PtoxA) by C. difficile RNA polymerase. For gdh promoter, left lane is Pgdh DNA alone and right lane is Pgdh DNA incubated with C. difficile RNA polymerase. For toxB and toxA promoters, left lane is tox promoter DNA alone, middle lane is tox promoter DNA with C. difficile RNA polymerase, and right lane is tox promoter DNA with C. difficile RNA polymerase and TxeR, respectively.
TxeR Activates Transcription from tox Promoters in Vitro with RNA Polymerase Holoenzyme.
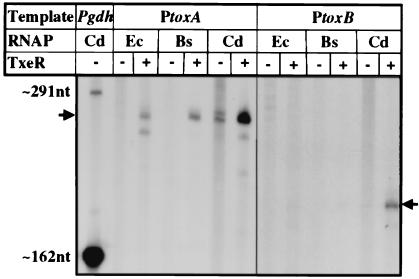
To see whether TxeR activates transcription specifically from the tox promoters used in vivo, the ability of TxeR to stimulate transcription was tested by in vitro run-off transcription assays. As shown in Fig. 4, synthesis of a toxA-specific transcript (255 nt) and a toxB-specific transcript (175 nt) was greatly stimulated when TxeR was added to the reaction when any one of the three RNA polymerases was tested. In the absence of TxeR, no detectable tox transcripts were observed with either E. coli RNA polymerase or B. subtilis RNA polymerase. The C. difficile RNA polymerase showed low levels of tox transcript without added TxeR, perhaps because of the presence of small amounts of TxeR in this RNA polymerase preparation. Nevertheless, it was very clear that the addition of TxeR greatly elevated tox transcription in all cases. Addition of the contaminating proteins (Fig. 2, lane 4) had no effect on the ability of RNA polymerase holoenzymes to transcribe from the tox promoters (data not shown). These results confirm that TxeR interacts with a variety of RNA polymerases and show that TxeR activates transcription from the bona fide tox promoters.
Figure 4.
In vitro transcription activation from tox promoters by RNA polymerase and TxeR. In vitro run-off transcription reactions were performed by using RNA polymerases from E. coli (Ec), B. subtilis (Bs), or C. difficile (Cd) and DNA fragments containing either the toxA promoter (PtoxA) or the toxB promoter (PtoxB) incubated in the absence or presence of partially purified TxeR. A control transcription reaction performed on the gdh promoter DNA with C. difficile RNA polymerase is shown in the leftmost lane. The expected sizes of run-off transcripts (indicated by arrows) are: Pgdh, 162 nt; PtoxA, 255 nt; PtoxB, 175 nt. An additional read-through transcript of 291 nt is made from the gdh promoter.
TxeR Allows Core RNA Polymerase to Bind to the tox Promoters.
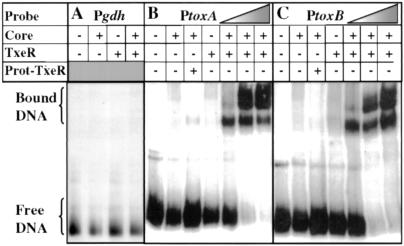
A blast search of TxeR against the GenBank and SwissProt databases revealed that TxeR is distantly related to certain sigma factors of bacterial RNA polymerase, including members of the extracytoplasmic function (ECF) family of sigma factors (33, 34). To test the ability of TxeR to act as a sigma factor, we performed gel mobility-shift assays with the tox promoter DNA fragments and E. coli core RNA polymerase with or without addition of TxeR and challenged the complexes with heparin. Neither core enzyme alone nor core enzyme mixed with TxeR was able to shift the mobility of the gdh promoter-containing fragment (Fig. 5A), indicating that the core enzyme was devoid of the major vegetative sigma factor and that any effect of TxeR would be specific to the tox promoters. When we mixed TxeR with the core enzyme, binding to the tox promoters was observed in a dose-dependent manner (Fig. 5 B and C). A mixture of core enzyme and the proteins equivalent to those that contaminate our preparation of TxeR (Fig. 2) was unable to bind to the promoter regions of toxA (Fig. 5B) and toxB (Fig. 5C). We obtained very similar results when we used the core RNA polymerase of B. subtilis (data not shown).
Figure 5.
Gel mobility retardation of tox promoters with E. coli RNA polymerase core enzyme and TxeR. Gel mobility retardation of the C. difficile gdh gene promoter (Pgdh) (A), the toxA gene promoter (PtoxA) (B), and toxB gene promoter (PtoxB) (C) by E. coli RNA polymerase core enzyme alone or in the presence of either TxeR or contaminating proteins (Prot-TxeR) from E. coli not expressing TxeR. The triangles in B and C indicate the use of increasing amounts of TxeR (50, 100, and 200 nM).
For both toxA and toxB, the addition of an excess of unlabeled heterologous DNA [1 μg of poly(dI-dC)] did not prevent DNA binding, whereas the addition of an excess of unlabeled homologous DNA effectively prevented DNA binding (data not shown). Thus, it is clear that neither TxeR nor the RNA polymerase core enzyme independently associates with the tox promoters, but the simultaneous presence of both of these components is required for specific, open complex formation at the toxA and toxB promoters.
TxeR Directs Core Enzyme to Initiate Transcription from tox Promoters in Vitro.
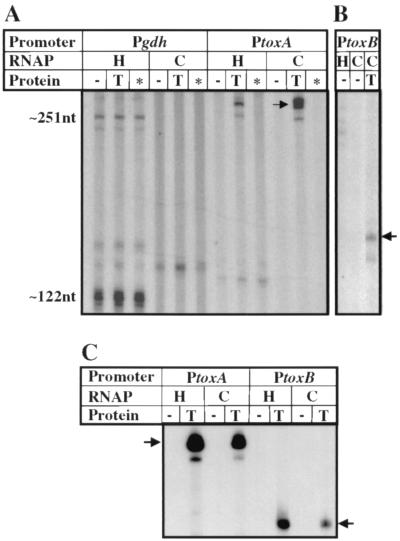
To test directly the hypothesis that TxeR is an RNA polymerase sigma factor, we carried out in vitro run-off transcription reactions with core enzyme of E. coli RNA polymerase in the presence and absence of TxeR. As shown in Fig. 6 A and B, E. coli RNA polymerase core enzyme was unable to initiate transcription from either the toxA or toxB promoters. However, addition of TxeR to E. coli core RNA polymerase activated transcription from both tox promoters (Fig. 6 A and B). Similar results were obtained with B. subtilis RNA polymerase core enzyme (Fig. 6C). These findings suggest strongly that TxeR is an alternative sigma factor required for the expression of toxin genes. No effect on transcription by core RNA polymerase was observed with either the toxA (Fig. 6A) or the toxB (data not shown) promoter upon addition of the contaminating proteins.
Figure 6.
In vitro transcription activation from tox promoters by RNA polymerase core enzyme and TxeR. In vitro transcription reactions were carried out in the presence of E. coli RNA polymerase holoenzyme (H) or core enzyme (C) (A and B) or B. subtilis RNA polymerase holoenzyme (H) or core enzyme (C) (C) with or without TxeR by using as templates DNA fragments containing the promoters of the gdh, toxA, or toxB genes. In lanes marked with an asterisk, contaminating proteins from E. coli not expressing TxeR were added. The arrows indicate the positions of the toxA and toxB transcripts. The sizes of expected gdh transcripts are 122 and 251 nt.
TxeR Interacts with the E. coli Core RNA Polymerase.
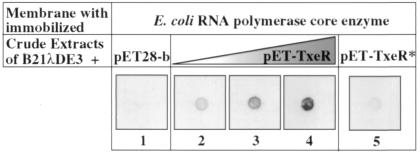
To verify that TxeR stimulates transcription by interacting with core RNA polymerase rather than by binding independently to DNA, we immobilized E. coli core RNA polymerase on membranes and, then, after saturating the membranes with blocking proteins, incubated the membranes with whole-cell extracts of E. coli strain BL21λDE3 either containing or lacking TxeR. Retention of TxeR was detected by using anti-TxeR antibodies. As shown in Fig. 7, a signal appeared in a dose-dependent manner only when the core enzyme was incubated with a crude extract of TxeR-expressing cells (filters 2–4) and not with a crude extract of cells that do not express TxeR (filter 1). We applied the same procedure to unrelated proteins (BSA, restriction enzyme NsiI) immobilized on filters to check the specificity of TxeR–core enzyme interaction (data not shown), and there was no indication of nonspecific binding. Moreover, no specific binding of anti-TxeR antibodies appeared with immobilized core enzyme alone (data not shown), and the signal of the TxeR–core enzyme interaction was decreased strongly when we incubated the immobilized core enzyme with the heat-inactivated crude extract of TxeR-expressing cells (filter 5). These results show that TxeR is able to bind directly to RNA polymerase core enzyme, forming a TxeR–holoenzyme complex that is recognized by anti-TxeR antibodies.
Figure 7.
Interaction of purified TxeR with E. coli RNA polymerase core enzyme. Core RNA polymerase was spotted onto nitrocellulose membranes and immunoblotted by using anti-TxeR antibodies after incubation with crude extract of E. coli (200 μg) carrying the vector pET28-b (filter 1), crude extract of E. coli carrying pCD54 (expressing TxeR) (filters 2–4), or heat-inactivated crude extract of TxeR-expressing cells (filter 5). The triangle indicates the use of increasing amounts of crude extract of E. coli carrying pCD54 (50, 100, and 200 μg).
Discussion
Our results provide genetic and biochemical evidence that TxeR is required for specific transcription from the toxA and toxB promoters and that TxeR functions as an alternative sigma factor of RNA polymerase. The putative promoter elements of the toxA and toxB genes show striking similarities with each other but do not resemble the canonical σ70 consensus promoters of prokaryotes (6, 7, 18), consistent with the notion that the tox genes are likely to be under a common mode of regulation that depends on a novel form of RNA polymerase.
TxeR shows similarities, on the one hand, to transcriptional activators from different Clostridium species (19, 20), and, on the other, to RNA polymerase sigma factors (data not shown). Our conclusion that TxeR is a sigma factor rather than a positive regulator of RNA polymerase holoenzyme is based on the following results: (i) TxeR is unable to bind to tox promoters by itself; (ii) TxeR interacts directly with RNA polymerase core enzyme in the absence of promoter DNA; and (iii) TxeR confers upon RNA polymerase core enzyme the ability to recognize the tox promoters. We cannot exclude the possibility, however, that TxeR acts in conjunction with another protein present in E. coli extracts. The relatedness of TxeR to BotR of C. botulinum (19, 21), TetR of C. tetani (20), and UviA, a putative activator of the bcn gene in C. perfringens (35), leads us to speculate that these other regulators are also sigma factors. In fact, the −35 regions of the target genes of these regulators [the C. botulinum ntnH bonT operon, the C. tetani tetX gene, and the C. perfringens bcn gene, respectively (35, 36)] are nearly identical to those of the toxA and toxB genes (7). Thus, a common molecular mechanism may control production of toxins and bacteriocins in several Clostridium species. In support of this idea, we have found that a complex of TxeR and E. coli core enzyme is able to bind to all of these promoters (data not shown). Interestingly, the botR, tetR, and uviA genes, as txeR, lie just upstream of the genes they regulate (35, 36). Thus, TxeR, BotR, TetR, and UviA seem to define a new subgroup of the σ-70 family. These proteins have some similarity with the extracytoplasmic function (ECF) family (33, 34, 37–42) but are sufficiently different in structure and function to leave open the question of their phylogenetic relationship. TxeR may be the first example described of an alternate sigma factor that directly affects the transcription of toxin genes.
Previous studies showed that toxin production and toxA and toxB mRNA levels in C. difficile vary in response to environmental signals, including the amino acid composition of the medium and the nature of the carbon source (7, 11, 14). Dupuy and Sonenshein (7) and Hundsberger et al. (6) also observed that the tox transcripts are induced as cells approach stationary phase in rich medium. Consistent with these data, C. difficile TxeR-mediated activation of tox-gusA fusions in C. perfringens also is repressed by the presence of glucose. Interestingly, TxeR not only activates transcription of tox genes but also activates its own transcription in C. perfringens, and TxeR synthesis in C. difficile is greatly decreased by the presence of glucose (N.M. et al., unpublished data).
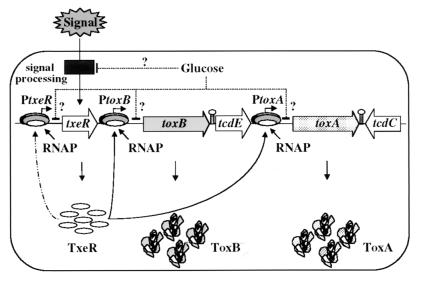
On the basis of our results and those of others, we offer a model for tox gene expression and regulation (Fig. 8). According to this model, as C. difficile cells reach stationary phase (in the absence of a rapidly metabolizable carbon source), an extracellular signal (nutrient depletion, stress, etc.) results in increased synthesis of TxeR. TxeR, an alternative sigma factor, activates transcription of the toxA and toxB genes by directing RNA polymerase core enzyme to bind to the tox promoters. Our preliminary data indicating that synthesis of TxeR is positively autoregulated (N.M. et al., unpublished data) suggest that a small initial increase in TxeR accumulation will be amplified rapidly. The inhibitory effects of glucose and other nutritional sources on tox gene transcription may be exerted directly through TxeR or through other regulatory proteins that act at either the txeR or tox promoters. However, we cannot exclude the possibility that the tox promoters may be independently subject to environmental regulation. A detailed understanding of the key players involved in regulating toxin synthesis is likely to provide useful insights into the mechanism by which C. difficile-induced, antibiotic-associated diarrhea evolves into the more debilitating and life-threatening disease, pseudomembranous colitis.
Figure 8.
A model for toxin regulation in C. difficile. See text for details.
Acknowledgments
We thank A. L. Sonenshein for advice, encouragement, and assistance with the manuscript, T. Msadek and A. Chastonet for their valuable help in protein purification, A. Kolb and G. Orsini for helpful advice on gel-retardation experiments, J. Helmann for the gift of B. subtilis core enzyme, L. Barroso and T. Wilkins for making the gdh gene sequence available before publication, and G. Reysset, S. T. Cole, A. Wright, and B. Belitsky for critical reading of the manuscript. This work was supported by funds from the Institut Pasteur and by a research grant (GM42219) from the U.S. Public Health Service to A. L. Sonenshein. N.M. was a fellow of the Charles A. King Trust. Preliminary studies were aided by pilot project funds from a U.S. Public Health Service grant (DK34928) to the Gastroenterology Research and Secretory Proteins Center.
References
- 1.Kelly C P, LaMont J T. Annu Rev Med. 1998;49:375–390. doi: 10.1146/annurev.med.49.1.375. [DOI] [PubMed] [Google Scholar]
- 2.Von Eichel-Streiber C, Boquet P, Sauerborn M, Thelestam M. Trends Microbiol. 1996;4:375–382. doi: 10.1016/0966-842X(96)10061-5. [DOI] [PubMed] [Google Scholar]
- 3.Dillon S T, Rubin E J, Yakubovich M, Pothoulakis C, LaMont J T, Feig L A, Gilbert R J. Infect Immunol. 1995;63:1421–1426. doi: 10.1128/iai.63.4.1421-1426.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Just I, Wilm M, Selzer J, Rex G, Von Eichel-Streiber C, Mann M, Aktories K. J Biol Chem. 1995;270:13932–13936. doi: 10.1074/jbc.270.23.13932. [DOI] [PubMed] [Google Scholar]
- 5.Wilkins T D, Lyerly D M. Trends Microbiol. 1996;4:49–51. doi: 10.1016/0966-842X(96)81508-3. [DOI] [PubMed] [Google Scholar]
- 6.Hundsberger T, Braun V, Weidmann M, Leukel P, Sauerborn M, Von Eichel-Streiber C. Eur J Biochem. 1997;244:735–742. doi: 10.1111/j.1432-1033.1997.t01-1-00735.x. [DOI] [PubMed] [Google Scholar]
- 7.Dupuy B, Sonenshein A L. Mol Microbiol. 1998;27:107–120. doi: 10.1046/j.1365-2958.1998.00663.x. [DOI] [PubMed] [Google Scholar]
- 8.Yamakawa K, Kamiya S, Meng X Q, Karasawa T, Nakamura S. J Med Microbiol. 1994;41:319–323. doi: 10.1099/00222615-41-5-319. [DOI] [PubMed] [Google Scholar]
- 9.Karasawa T, Maegawa T, Nojiri T, Yamakawa K, Nakamura S. Microbiol Immunol. 1997;41:581–585. doi: 10.1111/j.1348-0421.1997.tb01895.x. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda D, Karasawa T, Yamakawa K, Tanaka R, Namiki M, Nakamura S. Zentralbl Bakteriol. 1998;287:375–386. doi: 10.1016/s0934-8840(98)80174-6. [DOI] [PubMed] [Google Scholar]
- 11.Karlsson S, Burman L G, Akerlund T. Microbiology. 1999;145:1683–1693. doi: 10.1099/13500872-145-7-1683. [DOI] [PubMed] [Google Scholar]
- 12.Karlsson S, Lindberg A, Norin E, Burman L G, Akerlund T. Infect Immunol. 2000;68:5881–5888. doi: 10.1128/iai.68.10.5881-5888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura S, Mikawa M, Tanabe N, Yamakawa K, Nishida S. Microbiol Immunol. 1982;26:985–992. doi: 10.1111/j.1348-0421.1982.tb00248.x. [DOI] [PubMed] [Google Scholar]
- 14.Yamakawa K, Karasawa T, Ohta T, Hayashi H, Nakamura S. J Med Microbiol. 1998;47:767–771. doi: 10.1099/00222615-47-9-767. [DOI] [PubMed] [Google Scholar]
- 15.Hammond G A, Johnson J L. Microb Pathog. 1995;19:203–213. doi: 10.1016/s0882-4010(95)90263-5. [DOI] [PubMed] [Google Scholar]
- 16.Braun V, Hundsberger T, Leukel P, Sauerborn M, von Eichel-Streiber C. Gene. 1996;181:29–38. doi: 10.1016/s0378-1119(96)00398-8. [DOI] [PubMed] [Google Scholar]
- 17.Cohen S H, Tang Y J, Silva J., Jr J Infect Dis. 2000;181:659–663. doi: 10.1086/315248. [DOI] [PubMed] [Google Scholar]
- 18.Hawley D K, McClure W R. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moncrief J S, Barroso L A, Wilkins T D. Infect Immunol. 1997;65:1105–1108. doi: 10.1128/iai.65.3.1105-1108.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marvaud J C, Eisel U, Binz T, Niemann H, Popoff M R. Infect Immunol. 1998;66:5698–5702. doi: 10.1128/iai.66.12.5698-5702.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marvaud J C, Gibert M, Inoue K, Fujinaga Y, Oguma K, Popoff M R. Mol Microbiol. 1998;29:1009–1018. doi: 10.1046/j.1365-2958.1998.00985.x. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 23.Bannam T L, Rood J I. Plasmid. 1993;29:233–235. doi: 10.1006/plas.1993.1025. [DOI] [PubMed] [Google Scholar]
- 24.Guérout-Fleury A M, Frandsen N, Stragier P. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- 25.O'Sullivan D J, Klaenhammer T R. Gene. 1993;137:227–231. doi: 10.1016/0378-1119(93)90011-q. [DOI] [PubMed] [Google Scholar]
- 26.Zhao Y, Melville S. J Bacteriol. 1998;180:136–142. doi: 10.1128/jb.180.1.136-142.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 29.Whipple F W, Sonenshein A L. J Mol Biol. 1992;223:399–414. doi: 10.1016/0022-2836(92)90660-c. [DOI] [PubMed] [Google Scholar]
- 30.Jourlin-Castelli C, Mani N, Nakano M M, Sonenshein A L. J Mol Biol. 2000;295:865–878. doi: 10.1006/jmbi.1999.3420. [DOI] [PubMed] [Google Scholar]
- 31.Rood J I, Cole S T. Microbiol Rev. 1991;55:621–648. doi: 10.1128/mr.55.4.621-648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnston D E, McClure W R. In: RNA Polymerase. Losick R, Chamberlin M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1976. pp. 413–428. [Google Scholar]
- 33.Lonetto M A, Brown K L, Rudd K E, Buttner M J. Proc Natl Acad Sci USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Missiakas D, Raina S. Mol Microbiol. 1998;28:1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x. [DOI] [PubMed] [Google Scholar]
- 35.Garnier T, Cole S T. Mol Microbiol. 1988;2:607–614. doi: 10.1111/j.1365-2958.1988.tb00069.x. [DOI] [PubMed] [Google Scholar]
- 36.Popoff M R, Marvaud J C. In: The Comprehensive Sourcebook of Bacterial Protein Toxins. Freer J H, Alouf J E, editors. London: Academic; 1999. pp. 174–201. [Google Scholar]
- 37.Walker S L, Hiremath L S, Wozniak D J, Galloway D R. Gene. 1994;150:87–92. doi: 10.1016/0378-1119(94)90863-x. [DOI] [PubMed] [Google Scholar]
- 38.Sexton R, Gill P R, Jr, Callanan M J, O'Sullivan D J, Dowling D N, O'Gara F. Mol Microbiol. 1995;15:297–306. doi: 10.1111/j.1365-2958.1995.tb02244.x. [DOI] [PubMed] [Google Scholar]
- 39.Hershberger C D, Ye R W, Parsek M R, Xie Z D, Chakrabarty A M. Proc Natl Acad Sci USA. 1995;92:7941–7945. doi: 10.1073/pnas.92.17.7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ochsner U A, Johnson Z, Lamont I L, Cunliffe H E, Vasil M L. Mol Microbiol. 1996;21:1019–1028. doi: 10.1046/j.1365-2958.1996.481425.x. [DOI] [PubMed] [Google Scholar]
- 41.Stintzi A, Johnson Z, Stonehouse M, Ochsner U A, Meyer J M, Vasil M L, Poole K. J Bacteriol. 1999;181:399–413. doi: 10.1128/jb.181.13.4118-4124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasil M L, Ochsner U A. Mol Microbiol. 1999;34:399–413. doi: 10.1046/j.1365-2958.1999.01586.x. [DOI] [PubMed] [Google Scholar]