Abstract
Alterations in DNA methylation frequently occur in hepatocellular cancer (HCC). We have previously demonstrated that hypermethylation in candidate genes can be detected in plasma DNA prior to HCC diagnosis. To identify with a genome-wide approach additional genes hypermethylated in HCC that could be used for more accurate analysis of plasma DNA for early diagnosis, we analyzed tumor and adjacent non-tumor tissues from 62 Taiwanese HCC cases using Illumina methylation arrays that screen 26,486 autosomal CpG sites. After Bonferroni adjustment, a total of 2,324 CpG sites significantly differed in methylation level, with 684 CpG sites significantly hypermethylated and 1,640 hypomethylated in tumor compared to non-tumor tissues. Array data were validated with pyrosequencing in a subset of 5 of these genes; correlation coefficients ranged from 0.92 to 0.97. Analysis of plasma DNA from 38 cases demonstrated that 37% to 63% of cases had detectable hypermethylated DNA (≥5% methylation) for these 5 genes individually. At least one of these genes was hypermethylated in 87% of cases, suggesting that measurement of DNA methylation in plasma samples is feasible. The panel of methylated genes indentified in the current study will be further tested in large cohort of prospectively collected samples to determine their utility as early biomarkers of hepatocellular carcinoma.
Keywords: Genome-wide, DNA mehtylation, Hepatocellular Carcinoma
Introduction
HCC is a complex disease and likely the result of the accumulation of both genetic and epigenetic aberrations. A number of mutations have been observed in HCC, most frequently in p53.1 Gene expression studies have found profiles associated with survival, recurrence and metastasis.2 These changes in gene expression may be related to gene-specific DNA hyper- or hypomethylation (reviewed in 3).
Most previous methylation studies looked at one or a few genes at a time (e.g.,4–11) although 105 genes were analyzed in one study.12 While reasonably consistent results have been observed across studies, the exact frequencies of hypermethylation in tumor tissues differ. CDKN2A/INK4 (p16) is methylated in 30–70% of HCCs.13–16 RASSF1A is methylated in up to 85% of HCCs,15,17 GSTP1 in 50–90%18–20 and MGMT in 40%21 Our studies also observed that frequent methylation of particular genes correlated with AFB1-DNA adduct levels in the liver tissues.15,16,18,21 We found correlations between gene-specific hypermethylation in tumor tissue and plasma DNA using blood collected at the time of diagnosis.16 Using samples from a prospective ~25,000 subject cohort, we found that methylation of three genes (RASSF1A, CDKN2A and INK4B (p15)) in plasma DNA was predictive of later HCC development.22 These prior studies used a candidate gene approach.
To identify additional differentially methylated genes with a genome-wide approach, we used Illumina Infinium Human Methylation 27K arrays to analyze 27,578 CpG sites covering 14,495 genes in paired HCC tumor and adjacent non-tumor tissues. The aims of the current study were first to identify DNA methylation markers that significantly differentiate tumor tissue from adjacent non-tumor tissue, and then to test the feasibility of detecting the hypermethylated markers in plasma samples and their correlations with relevant liver tissues. Because plasma DNAs are mostly derived from necrotic or apoptotic cells with little released from white blood cells, it is appropriate to use plasma to study circulating tumor DNA.23 Recently, three other studies have reported DNA methylation profiles in HCC tumor/adjacent tissues using Illumina arrays. Two used Illumina 1,500 Golden Gate arrays on 5 paired samples from Korea and 30 from France and the third used Illumina Human Methylation27K arrays on 12 samples from Germany.24–26 A fourth earlier study used methylated CpG island amplification microarrays to study 6,458 CpG islands in 10 paired samples from Japan.27 These prior studies differed in sample size, technology used and the major etiologic cause (HBV, HCV, alcohol). The current study is the largest to date and is comprised of Taiwanese cases who are predominantly HBV positive.
Materials and Methods
Patients and Biopsy Specimens
This study was approved by Institutional Review Boards of Columbia University and National Taiwan University (NTU). Written informed consent was obtained. Sixty-six frozen liver tissues collected in the Department of Surgery, NTU Hospital were assayed. Demographic data and clinicopathologic characteristics were obtained from hospital charts; HBV (HBsAg) and HCV (anti-HCV) status were determined by immunoassay. For 39 subjects missing HCV status, liver tissues were stained with monoclonal antibody NS3 (Novocastra™, Newcastle, UK). Specimens were kept at −70°C until shipment to Columbia University where pathologic analysis confirmed HCC status and indicated that adjacent tissues were primarily cirrhotic. Bloods were collected at the time of diagnosis for 30 patients and plasma frozen. Plasma from 8 additional cases from the same hospital were included in the analysis.
DNA Preparation and Illumina Infinium Human Methylation Platform
DNA was extracted by standard proteinase K/RNase treatment and phenol/chloroform extraction. Plasma DNAs were extracted using DNeasy Blood & Tissue Kits (Qiagen, Valencia, CA). Bisulfite modification of 1 μg DNA was conducted using an EZ DNA Methylation Kit (Zymo Research, Irvine, CA).
The Human Methylation 27 DNA Analysis BeadChips (Illumina, San Diego, CA) were used to interrogate 27,578 highly informative CpG sites covering 14,495 genes following their standard protocol. Paired samples (HCC tumor and adjacent non-tumor tissues) were processed on the same chip to avoid chip-to-chip variation; 4 pairs of tissues were repeat assayed as a quality control (QC). Information on location of CpG sites in promoter regions was provided by Dr. Kim.28
Pyrosequencing for Candidate Gene Methylation
Pyrosequencing was carried out with primers designed with the Pyromark Assay Design Software 2.0 (Qiagen). The region selected for interrogation included the CpG sites identified to be differentially methylated based on the array data, as well as surrounding sites. PCR was performed in a 25uL reaction mix containing 50ng bisulfite-converted DNA, 1X Pyromark PCR Master Mix (Qiagen), 1X Coral Load Concentrate (Qiagen), and 0.3uM forward and 5′ biotinylated reverse primers, using the cycling conditions outlined in Supporting Table 1. Each set of amplifications included bisulfite-converted CpGenome universal methylated (Millipore, Billerica, MA), unmethylated (whole genome amplified DNA) and non-template controls.
The sequencing reaction and quantitation of methylation was conducted using a PyroMark Q24 instrument and software (Qiagen). Percent methylation was calculated by averaging across all CpG sites interrogated. A plasma DNA sample was considered positive if % methylation was ≥5% since lower values are not reliable.29
Statistical Methods
β values were generated using the Illumina BeadStudio software.30 Sites on the sex chromosomes were removed from the analysis leaving 26,486 autosomal sites. For QC, methylation measures with a detection p-value >0.05 and samples with CpG coverage <95% were removed. This eliminated 4 pairs with a final sample size of 62 paired tissues. For these samples, the control panel in the BeadStudio analytical software showed excellent intensity for staining (>15,000), clear clustering for the hybridization probes, good target removal intensity (<400) and satisfactory bisulfite conversion.31 Demographic data for the 62 patients are presented in Table 1.
Table 1.
Clinical and Pathological Characteristic of 62 HCC Patients with Tissue DNA Methylation Array Data and 8 Additional Cases with only Plasma DNA Data
| Cases with Array Data | Cases with Only Plasma Data | |
|---|---|---|
| Variables | Mean ± SD | Mean ± SD |
| Age at diagnosis | 52.2 yr ± 14.2* | 53.0±11.5 † |
| N (%) | N (%) | |
| Gender | ||
| Male | 54 (87) | 7 (100) † |
| Female | 8 (13) | 0 |
| Viral status | ||
| HBV (−) and HCV (−) | 7 (11) | 0 |
| HBV (+) and HCV (−) | 36 (58) | 7 (88) |
| HBV (−) and HCV (+) | 6 (10) | 0 |
| HBV (+) and HCV (+) | 13 (21) | 1 (12) |
| Cigarette smoking | ||
| No | 25 (40) | 2 (25) |
| Yes | 24 (39) | 4 (50) |
| Missing | 13 (21) | 2 (25) |
| Alcohol drinking | ||
| No | 41 (66) | 8 (100) |
| Yes | 8 (13) | 0 |
| Missing | 13 (21) | 0 |
| AFB1-DNA adducts in tumor tissues | ||
| High to Medium | 37 (60) | |
| Low | 25 (40) | |
| AFB1-DNA adducts in non-tumor tissues | ||
| High to Medium | 15 (24) | |
| Low | 15 (24) | |
| Missing | 32 (52) | |
Age missing for five subjects
Data missing for one subject
Paired t-tests with Bonferroni correction for multiple testing were used to identify CpG sites that were differentially methylated between tumor and adjacent non-tumor tissues. A significant difference was defined as sites with a Bonferroni-corrected p-value ≤ 0.05. A volcano plot displayed mean DNA methylation differences for all 26,486 CpG sites. A Manhattan plot displayed the significance (-log10(adjusted-pvalue)) of the associations by chromosomes.
To select genes for validation of the methylation array data, we focused on hypermethylation since our long term goal is to detect hypermethylated plasma DNA for early diagnosis of HCC. Candidate CpG sites were selected for confirmatory analysis with two methods. In method A, we required: (1) the mean difference in methylation levels between tumor and adjacent tissues is ≥20%; (2) ≥70% of the tumor tissues had methylation levels greater than two SDs above the mean methylation level of all 62 adjacent tissues; and (3) the mean methylation level for adjacent tissues is ≤25%. In method B, we conducted three-fold cross-validation where we randomly chose 40 pairs out of 62 pairs to form a training set and the remaining 22 pairs as a testing set. We then repeated the paired t-test using the training set and selected the top 100 most significant CpG sites with the following loosened three criteria to ensure selection of enough candidate CpG sites at each cross-validation: (1) the mean difference in methylation levels between tumor and adjacent tissues is ≥20%; (2) ≥60% of the tumor tissues had methylation levels greater than two SDs above the mean methylation level of the 40 adjacent tissues; and (3) the mean methylation level for adjacent tissues is ≤40%. We repeated the three-fold cross-validation 1,000 times and selected the top most frequently selected CpG sites with the same number as in the list using method A. We then applied three different prediction methods, diagonal linear discriminant analysis, support vector machines, and k-nearest neighbor to determine the prediction accuracy of the selected panel (method B) using data on the remaining 22 pairs.32
The hierarchical clustering of the methylation data was performed with the top 1,000 most significantly differentially methylated sites and with the two selected panels of CpG sites using methods A and B. Gene ontology analysis was performed by the PANTHER classification system (htpp://www.pantherdb.org) to compare the significant methylated gene lists with the reference (NCBI, human genome build 36).33 The binomial test was used to identify significantly enriched pathways, biological processes, molecular functions, cellular components and protein class terms after Bonferroni correction for multiple comparisons with a cutoff of p≤0.05.
To investigate if methylation levels are affected by HCC risk factors such as HBsAg status, HCV status, cigarette smoking (ever/never), alcohol consumption (ever/never), AFB1-DNA adduct level, and gender within tumor and adjacent non-tumor tissues separately, we used a two-sample t-test with Bonferroni correction for multiple testing.
In the second stage confirmatory analysis, Pearson’s correlations between methylation levels using Illumina arrays and pyrosequencing on selected sites were calculated.
All analyses were conducted using the R language (http://www.r-project.org/).
Results
Clinical and Pathological Characteristics of the HCC Cases
Clinical and pathological characteristics are described in Table 1. Almost 90% of cases are male and 79% are HBsAg positive. About 31% of subjects are positive for HCV. Seven subjects are negative for both HBV and HCV, 36 subjects are positive for HBsAg and negative for HCV; 13 subjects are both HBV and HCV positive; and the remaining 6 subjects are negative for HBsAg and positive for HCV. Thus, viral infection, primarily HBV, is the major risk factor in this population. The average age at HCC diagnosis is 52.2±14.2 years. About 40% of cases smoke and 13% consume alcohol but data are missing for about 20% of subjects. Aflatoxin B1-DNA adducts, measured previously in all tumor tissues and in about half of adjacent tissues,34,35 are also summarized in Table 1.
Reproducibility of Methylation Array Data
The reproducibility of the Illumina platform was evaluated using replicates of four paired samples on a different day. High concordance was observed for all eight replicates with coefficients of determination (R2) ranging from 0.96 to 0.98. A representative example of the concordance between two replicates for an adjacent tissue sample is given in Supporting Fig. 1 and is consistent with previous studies.24,26 The site-by-site comparisons across the 26,486 sites between the 4 pairs of replicates gave an absolute mean differences in methylation level ranging from 0.003 to 0.04.
Methylation Profiles Differentiate HCC Tumor from Non-tumor Tissues
The methylation levels of the individual 26,486 autosomal CpG sites as well as the overall means were compared between the 62 pairs of tissues. There were 2,324 CpG sites that significantly differed in methylation level between tumor and non-tumor tissues after Bonferroni adjustment (for a complete list see Supporting Tables 2 and 3). Among all significant CpG sites, 684 were significantly hypermethylated (covering 548 genes) and 1,640 were significantly hypomethylated (covering 1,290 genes) in tumor compared to non-tumor tissues. Fig. 1 displays mean DNA methylation differences between the 62 paired tumor/adjacent tissues at all 26,486 CpG sites using a volcano plot. Both hyper- and hypomethylation alterations are common events in HCC tumor tissues. The top 20 hyper- or hypomethylated sites ranked by statistical significance are given in Table 2. Regardless of whether they were hypo- or hypermethylated, all significant CpG sites have similar mean methylation levels in tumor tissues (42.2% vs. 42.9%), while the mean methylation levels in non-tumor tissues were dramatically different (26.0% for hypermethylated vs. 58.4% for hypomethylated sites. Fig. 2 shows the heatmap of the top 1,000 CpG sites (based on statistical significance) distinguishing tumor from adjacent tissues. In general good separation of tumor and adjacent tissues was observed with a small amount of misclassification.
Fig. 1.
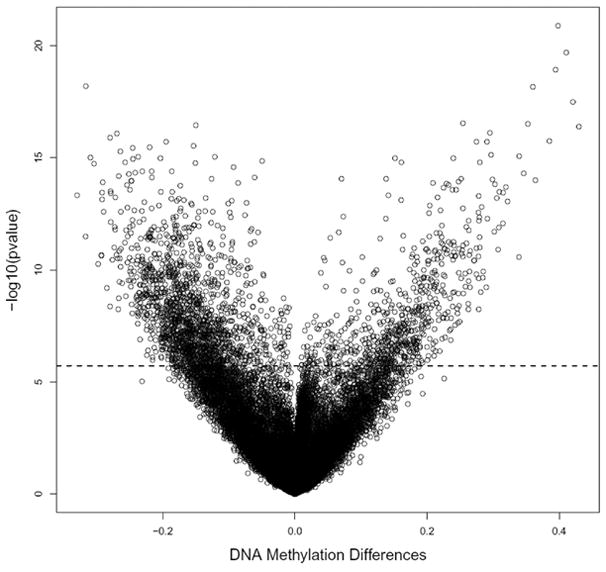
Volcano plot for differential DNA methylation analysis of 62 paired HCC tumor and adjacent non-tumor tissues.
The x-axis shows the mean DNA methylation difference, while the y-axis shows the – log10 of the adjusted p value for each CpG site, representing the strength of association. Above the dashed line indicates statistically significant (p≤0.05) after Bonferroni correction. Blue represents adjacent non-tumor tissue, and red represents tumor tissue.
Table 2.
Top 20 Ranked Hypermethylated or Hypomethylated genes *
| Hypermethylated in Tumor | Hypomethylated in Tumor | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | Target ID | Mean in tumor | Mean in non-tumor | Mean Difference | Adjusted P Value † | Genes | Target ID | Means in tumor | Mean in non-tumor | Mean Difference | Adjusted P Value † |
| DAB2IP | cg05684891 | 0.61 | 0.21 | 0.40 | 3.58E-17 | CCL20 | cg21643045 | 0.46 | 0.77 | −0.32 | 1.70E-14 |
| BMP4 | cg14310034 | 0.55 | 0.14 | 0.41 | 5.36E-16 | AKT3 | cg11314684 | 0.14 | 0.29 | −0.15 | 9.34E-13 |
| ZFP41 | cg12680609 | 0.51 | 0.12 | 0.39 | 3.25E-15 | SCGB1D1 | cg01772980 | 0.40 | 0.67 | −0.27 | 2.28E-12 |
| SPDY1 | cg04786857 | 0.55 | 0.19 | 0.36 | 1.79E-14 | WFDC6 | cg24765446 | 0.32 | 0.60 | −0.28 | 3.42E-12 |
| CDKN2A | cg09099744 | 0.50 | 0.08 | 0.42 | 8.69E-14 | PAX4 | cg08886154 | 0.30 | 0.50 | −0.19 | 5.19E-12 |
| TSPYL5 | cg15747595 | 0.71 | 0.45 | 0.25 | 7.56E-13 | GCET2 | cg25462303 | 0.15 | 0.30 | −0.15 | 7.92E-12 |
| CDKL2 | cg24432073 | 0.46 | 0.11 | 0.35 | 8.03E-13 | CD300E | cg04995095 | 0.26 | 0.48 | −0.22 | 8.87E-12 |
| ZNF154 | cg21790626 | 0.51 | 0.08 | 0.43 | 1.14E-12 | CD1B | cg04574507 | 0.36 | 0.60 | −0.25 | 9.89E-12 |
| ZNF540 | cg03975694 | 0.53 | 0.24 | 0.30 | 2.03E-12 | FLJ00060 | cg03602500 | 0.43 | 0.70 | −0.26 | 1.37E-11 |
| CCDC37 | cg00891278 | 0.60 | 0.31 | 0.29 | 5.07E-12 | MNDA | cg25119415 | 0.29 | 0.53 | −0.24 | 2.42E-11 |
| PKDREJ | cg11377136 | 0.74 | 0.47 | 0.27 | 5.18E-12 | CD1E | cg12200412 | 0.19 | 0.31 | −0.12 | 2.42E-11 |
| NKX6-2 | cg08441806 | 0.56 | 0.32 | 0.24 | 2.77E-11 | CYP11B1 | cg09120035 | 0.43 | 0.74 | −0.31 | 2.59E-11 |
| FOXD2 | cg15868302 | 0.70 | 0.55 | 0.15 | 2.88E-11 | KRTAP13-1 | cg02764897 | 0.26 | 0.46 | −0.20 | 2.84E-11 |
| RBAK | cg06914598 | 0.48 | 0.32 | 0.16 | 4.22E-11 | KLK9 | cg01144251 | 0.37 | 0.62 | −0.25 | 2.95E-11 |
| CFTR | cg25509184 | 0.59 | 0.31 | 0.28 | 4.23E-11 | KPNA1 | cg25564800 | 0.05 | 0.09 | −0.05 | 3.80E-11 |
| HIST1H3E | cg07922606 | 0.91 | 0.84 | 0.07 | 2.38E-10 | SPRR1B | cg18780284 | 0.32 | 0.58 | −0.26 | 4.44E-11 |
| LYPD3 | cg25340403 | 0.67 | 0.54 | 0.14 | 2.40E-10 | SPRR1A | cg04505023 | 0.35 | 0.66 | −0.30 | 4.82E-11 |
| DNM3 | cg23391785 | 0.55 | 0.18 | 0.36 | 2.75E-10 | CCR6 | cg13615963 | 0.14 | 0.24 | −0.09 | 7.20E-11 |
| RASSF1 | cg21554552 | 0.57 | 0.37 | 0.21 | 3.45E-10 | FLJ44674 | cg13897627 | 0.42 | 0.64 | −0.22 | 1.05E-10 |
| KCNC1 | cg27409364 | 0.65 | 0.42 | 0.23 | 3.70E-10 | OR51B4 | cg06353345 | 0.26 | 0.49 | −0.23 | 1.06E-10 |
Top 20 hyper- or hypomethylated genes in HCC tumor tissues compared to adjacent non-tumor tissues ranked by statistical significance
After Bonferroni adjustment
Fig. 2.
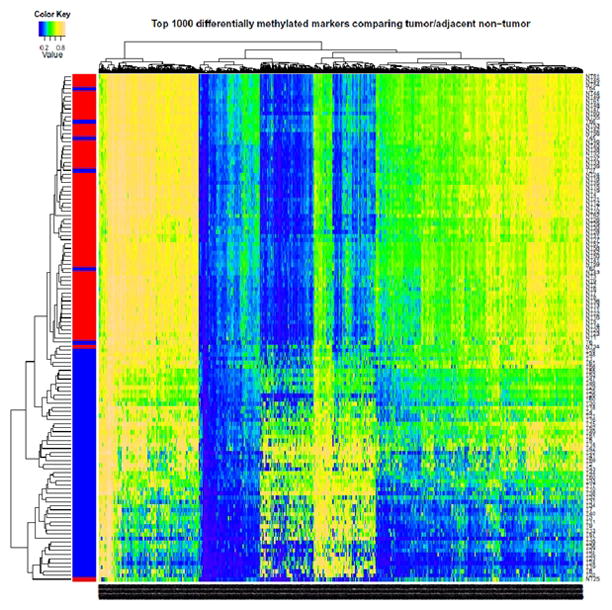
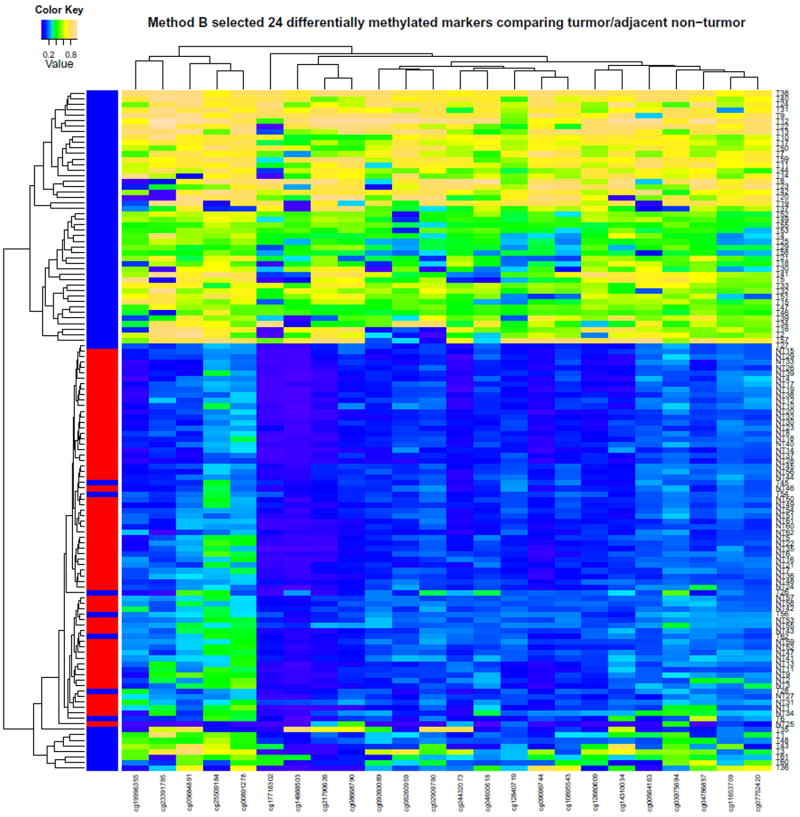
Hierachical cluster analysis of top 1,000 significantly differentially methylated CpG sites between 62 tumor and adjacent tissues
Characteristics of Significant CpG Sites in HCC Tissues
A Manhattan plot was used to display the log10(adjusted-pvalue) for the differences in methylation by chromosome (Supporting Fig. 2) and indicates that aberrant methylation is spread across all chromosomes. Among the 2,324 significantly differentially methylated CpG sites, >80% (82.3% and 85.8% for hyper- and hypomethylated sites, respectively) had a >10% absolute tumor/non-tumor difference in %methylation, and >50% had a >15% difference (Supporting Table 4). These data indicate that the methylation changes occurring during HCC development are robust and may provide useful biomarkers.
The majority of the significantly differentially methylated CpG sites are located within the proximal promoter regions. Among the 2,324 significant CpG sites, the distances to the transcription start site (TSS) ranged from 0 to 1,498bp with an average of 407bp and a standard deviation (SD) of 362bp. Hypermethylated CpG sites are more common within a short distance of TSS (50.7% within 250bp and 26.9% between 250 and 500bp) compared to hypomethylated sites (41.6% and 23.3%, respectively) (Supporting Fig. 3). The average distance to the TSS was significantly shorter for hypermethylated (mean=332bp, SD=312bp) compared with hypomethylated sites (mean=437bp, SD=377bp, p=3.95X10−10). Supporting Table 5 and Supporting Fig. 4 show that within CpG islands, more sites are significantly hypermethylated in tumors, while within non CpG island regions, more sites are significantly hypomethylated in tumors. The pattern does not vary whether the sites are in promoter regions or not.
Through PANTHER ontology analysis, we found 12 significant pathways for hypermethylated and 11 pathways for hypomethylated genes (Supporting Table 6). A number of potentially important cellular pathways involved in tumorigenesis were observed, such as the pathways of heterotrimeric G-protein signaling, endothelin signaling, PI3 kinase, interleukin signaling, inflammation mediated by chemokine/cytokine signaling and insulin/IGF, etc. For the first time, Wnt and 5HT4 type receptor mediated signaling pathways were identified.
Methylation Profiles Altered by HCC Risk Factors
Two-sample t-test was used to compare methylation levels among tumor and adjacent tissues separately for several HCC risk factors. No site was identified that was significantly differentially methylated by gender, HBV status, HCV status or AFB1-DNA adduct levels (high/medium vs low) (data not shown). However, the results may be partially due to small numbers of females, viral status and missing adduct data in some adjacent tissues. For alcohol consumption status, within adjacent tissues, methylation level at one CpG site in VPREB1 significantly differed between drinkers and non-drinkers while within tumor tissues 7 CpG sites in CRISPLD1, PCDHB2, PCSK1, LXH1, KCTD8, TSHD3 and CXCL12 were identified after Bonferroni adjustment. Further unsupervised hierarchic cluster analysis clearly suggested an even better separation of drinkers from non-drinkers using the top differentially methylated sites among tumor tissues (Supporting Fig. 5A) compared to non-tumor tissues (Supporting Fig. 5B).
Selection of Candidate Genes and Validation of Methylation by Pyrosequencing
To select the list of candidate CpG sites for confirmatory analysis, method A with the complete data set of 62 pairs resulted in a list of 24 sites in 18 genes (Supporting Table 7). Supporting Fig. 6, the heatmap of the selected 24 CpG sites, shows good separation of tumor and adjacent tissues in general. Method B based on 1,000 three-fold cross-validations of training set with 40 pairs results in a list of 24 top CpG sites that were most frequently selected (all ≥ 98% of times out of 1,000 three-fold cross-validations) (Table 3). The two panels of 24 CpG sites have 20 overlapping sites (Table 3 and supporting Table 7). Fig. 4 shows the heatmap of the selected 24 CpG sites using method B. The two heatmaps show similar separations. Using the testing set, the selected panel of 24 CpG sites (method B) has high prediction accuracy in the testing set: 0.886 (SD=0.044) based on diagonal linear discriminant analysis, 0.918 (SD=0.044) based on support vector machines, and 0.877 (SD=0.038) based on k-nearest neighbor. This suggests that the selected list of 24 CpG sites using the three-fold cross-validation for second stage confirmatory analysis is robust. Furthermore, compared to Fig. 2 which displays the top 1,000 differentially methylated sites with both hyper- and hypomethylated, almost the same set of tumor tissues are misclassified.
Table 3.
Selected 24 CpG Sites Based on 1,000 three-fold Cross-validations Distinguishing HCC Tumors from Adjacent Tissues with their Mean Methylation Levels in Tumor and Adjacent Tissues and the Differences*
| Symbol | Target ID | Consensus frequency | Mean β in Tumor | Mean β in Adjacent | Mean β difference | Adjusted P Value † | Function |
|---|---|---|---|---|---|---|---|
| BMP4 | cg14310034 | 1,000 | 0.55 | 0.14 | 0.41 | 5.36E-16 | Bone morphogenetic protein 4 |
| C6orf206 | cg04600618 | 985 | 0.44 | 0.16 | 0.28 | 4.08E-09 | Radial spoke head 9 homolog |
| CCDC37 | cg00891278 | 980 | 0.60 | 0.31 | 0.29 | 5.07E-12 | Coiled-coil domain containing 37 |
| CDKL2 | cg24432073 | 1,000 | 0.46 | 0.11 | 0.35 | 8.03E-13 | Cyclin dependent |
| CDKL2 | cg14988503 | 999 | 0.39 | 0.04 | 0.35 | 1.31E-10 | kinase |
| CDKN2A | cg07752420 | 1,000 | 0.49 | 0.20 | 0.30 | 1.92E-11 | Cyclin dependent |
| CDKN2A | cg09099744 | 1,000 | 0.50 | 0.08 | 0.42 | 8.69E-14 | kinase inhibitor |
| CDKN2A | cg10895543 | 1,000 | 0.48 | 0.14 | 0.34 | 2.36E-11 | |
| CDKN2A | cg12840719 | 1,000 | 0.39 | 0.14 | 0.25 | 2.26E-10 | |
| CDKN2A | cg11653709 | 994 | 0.46 | 0.21 | 0.25 | 3.17E-10 | |
| CFTR | cg25509184 | 1,000 | 0.59 | 0.31 | 0.28 | 4.23E-11 | Cystic fibrosis transmembrane conductance regulator |
| DAB2IP | cg05684891 | 1,000 | 0.61 | 0.21 | 0.40 | 3.58E-17 | GTPase-activating protein |
| DNM3 | cg23391785 | 993 | 0.55 | 0.18 | 0.36 | 2.75E-10 | Dynamin 3 |
| HIST1H3G | cg02909790 | 1,000 | 0.49 | 0.17 | 0.32 | 5.28E-10 | H3 histone family |
| HIST1H3J | cg17718302 | 1,000 | 0.37 | 0.05 | 0.31 | 2.36E-08 | H3 histone family |
| NKX6-2 | cg09260089 | 1,000 | 0.43 | 0.13 | 0.30 | 2.59E-10 | NK6 related transcription factor |
| PBX4 | cg19996355 | 999 | 0.47 | 0.15 | 0.32 | 2.46E-09 | Pre-B-cell leukemia homeobox 4 |
| RAB31 | cg17982102 | 988 | 0.38 | 0.14 | 0.24 | 7.14E-10 | Member Ras oncogene family |
| SPDY1 | cg04786857 | 1,000 | 0.55 | 0.19 | 0.36 | 1.79E-14 | Speedy homologue 1 |
| STEAP4 | cg00564163 | 999 | 0.49 | 0.19 | 0.31 | 1.68E-09 | Tumor necrosis factor |
| ZFP41 | cg12680609 | 1,000 | 0.51 | 0.12 | 0.39 | 3.25E-15 | Zinc finger protein 41homologue |
| ZNF154 | cg08668790 | 1,000 | 0.51 | 0.12 | 0.39 | 4.67E-12 | Zinc finger |
| ZNF154 | cg21790626 | 1,000 | 0.51 | 0.08 | 0.43 | 1.14E-12 | protein 154 |
| ZNF540 | cg03975694 | 1,000 | 0.53 | 0.24 | 0.30 | 2.03E-12 | Zinc finger protein 540 |
Sites were selected based on a mean difference in methylation levels between tumor and adjacent tissues of at least 20%; more than 60% of tumors have methylation levels greater than 2 standard deviation above the mean in all non tumor tissues and with mean levels of methylation in adjacent tissues <40%
After Bonferroni adjustment
Because prior studies have found a good correlation between Illumina array %methylation and that by pyrosequencing,28,36,37 we randomly selected just five genes (CDKL2, STEAP4, HIST1H3G, CDKN2A and ZNF154) from the top 18 candidates for validation in 42 paired tissues. Data were analyzed looking at the correlation between array and pyrosequencing data for both the specific CpG site on the array as well as the mean of all the CpG sites analyzed by pyrosequencing. Excellent correlations were found between array data and pyrosequencing results for both specific sites and the mean of all CpG sites ranging from 0.921 to 0.971 (Table 4, Supporting Fig. 7).
Table 4.
Correlations Between Array Data and Pyrosequencing Data within 86 Tumor and Non-tumor Tissues at the Specific CpG Sites on the Array as well as the Mean of All CpG Sites Per Gene Aassayed
| Gene | Array CpG site | Mean of all CpG sites (Number of CpG sites) |
|---|---|---|
| STEAP4 | 0.936 | 0.940 (4) |
| CDKL2 | 0.942 | 0.944 (3) |
| CDKN2A | 0.938 | 0.925 (5) |
| HIST1H3G | 0.954 | 0.921 (5) |
| ZNF154 | 0.971 | 0.967 (3) |
Analysis of Methylation of Candidate Genes in Plasma Samples
We next determined the feasibility of measuring methylation in the 5 randomly selected genes in plasma DNA available for a subset of 30 of the cases with tissue data plus 8 plasma samples from additional cases. The characteristics of these additional 8 cases are similar to those 62 with tissue data (Table1). The success rate of pyrosequencing ranged from 63–100% (Table 5). Detailed data on % methylation for each sample is given in Supporting Table 7. The frequency of hypermethylated DNA in plasma (defined as methylation level by pyrosequencing ≥5%) ranged between 37% and 63% (Table 5). With available data, 33 (87%) subjects had at least one gene positive while 2 subjects had all 5 genes. However, data were complete for only 20 (53%) subjects. Five subjects were negative for all genes but none had complete data.
Table 5.
Plasma DNA Methylation Using Pyrosequencing and Number of Samples with Available Pyrosequencing Data
| Gene | Samples positive* N (%†) | Samples with data N (%) |
|---|---|---|
| CDKL2 | 14 (37) | 38 (100) |
| CDKN2A | 13 (48) | 27 (71) |
| HIST1H3G | 9 (38) | 24 (63) |
| STEAP4 | 20 (63) | 32 (84) |
| ZNF154 | 16 (47) | 34 (89) |
Defined as ≥5% methylation
Based on successful pyrosequencing analysis
Discussion
We screened 62 paired tumor and adjacent tissues at 26,486 autosomal CpG sites. After Bonferroni adjustment, we found 2,324 CpG sites to significantly differ in methylation level; 684 were significantly hypermethylated and 1,640 were significantly hypomethylated. Since our goal is to identify methylation biomarkers in plasma DNA, mostly derived from necrotic or apoptotic cells,23 for early identification of HCC in high risk populations, we limited further study to hypermethylated sites. To select candidate CpG sites for confirmatory analysis, we used both the full data set and a training set of 40 pairs from the three-fold cross-validation. Two panels of 24 hypermethylated CpG sites in 18 genes with 20 CpG sites overlapping were selected. This suggests that the selected panel of CpG sites based on the training set from the three-fold cross-validation is robust. Further analysis of prediction accuracy using the testing data with 22 pairs suggested good prediction power in the testing set to separate tumor and adjacent non-tumor tissues. With the largest sample size thus far, we identified more significant CpG sites that differentiate tumor from adjacent tissue than previous methylation array studies in HCC.24–26
Only one prior study by Ammerpohl et al reported the use of Illumina Human Methylation 27K arrays to investigate DNA methylation in 12 HCC paired tissues; alcohol was likely the major etiologic agent for half of the cases.26 All 24 sites we selected (Table 3) were also identified by Ammerphol et al as being significantly hypermethylated in HCC compared to cirrhosis. Among all sites they identified as having a >20% difference in methylation, there is an overlap of 823 sites (63%) with our significant sites. These overlapping sites are 100% consistent in the direction of the methylation change. The magnitude of methylation levels are also significantly correlated (R2 from 0.76 to 0.99, p<0.0001). In addition to identifying two novel pathways (Wnt and 5HT4 type receptor mediated signaling), ten cellular pathways overlap with those identified by Ammerphol.
Two other studies have used the Illumina 1,500 Golden Gate Methylation Assay containing to evaluate 5 paired samples from Korea25 and 30 from France.24 In the Korean study, 24 new genes were identified as significantly hypermethylated in tumor.25 Nine (ADCYAP1, FLT3, HOXA9, IRAK3, MLF1, NPY, SH3BP2, TAL1, and TNFRSF10C) were also significantly hypermethylated in our tumor tissues. The remaining genes were nonsignificantly weakly hypermethylated in our tumors except for HIC2, NOTCH3 and PTCH2 which showed no hypermethylation. These three genes were also not hypermethylated in Ammerphol et al26 and thus are unlikely to be significantly hypermethylated in HCC. The second study 24 identified 27 genes as hypermethylated. Fourteen overlap with those we identified, including APC, BMP4, CDKN2A, F2R, FLT4, GSTP1, HOXA9, IGF1R, IRAK3, MYOD1, RASSF1, SH3BP2, TERT and ZMYND10 (Supporting Table 2). Ninety six of their 124 significant CpG sites overlap with ours with 92% consistency in the direction of methylation change.
Using pyrosequencing we confirmed methylation data for the 5 genes analyzed. Array data was highly correlated with both the specific CpG site and the mean of the 3 to 5 CpG sites assayed within a gene (Table 4 and Supporting Fig. 7).
We attempted to determine if methylation changes in specific CpG sites were associated with certain risk factors such as gender, viral infection, alcohol consumption and AFB1-DNA adduct levels. We identified sites that differed significantly after Bonferroni adjustment only for alcohol consumption. However, these results did not match prior data.24 Most of our cases were virus infected while the prior study was able to look at noninfected cases in which alcohol was the major risk factor. This may explain the discrepant results. Data on survival was not available for most of our cases so we were unable to investigate methylation profile and survival.
We also determined whether methylation of a randomly subset of 5 genes could be detected in plasma DNA by pyrosequencing. Not all samples were successfully amplified for all 5 genes with HIST1H3G having the lowest frequency of usable data (63%). This may be due to the larger PCR product for this gene (248bp) compared to the other 4 genes (<200bp). Future studies should consider PCR product size when designing pyrosequencing assays for plasma DNA. Using ≥5% methylation as the cutoff for positivity, the frequency of positive plasma DNA samples ranged from 37 to 63%. When any one gene positive was used to define a positive case, 87% were positive. These results, in conjunction with our prior study of plasma from controls22 suggest that analysis of plasma DNA is feasible and may be useful for diagnosis of HCC. However, the quality of the bisulfite treated plasma DNA will be a key component of a successful screening assay.
Among the strengths of our study is that it is the largest sample size methylation array study of HCC to date. Among the limitations is the lack of information on AFB1-DNA in adjacent non-tumor tissue for some cases. In addition, data on alcohol consumption and cigarette smoking was missing for about 20% of the cases. These missing data limited our ability to investigate relationships between methylation profiles and these factors. In addition, almost all our cases were infected with either HBV or HCV or both. Thus, we could not investigate the role of viral infection on methylation. Another limitation was the lack of healthy tissue from unaffected controls as a comparison group for our array studies. Our tumor adjacent tissues are primarily cirrhotic. Thus, we identified genes whose methylation was increased in progression from cirrhosis to HCC. Since our goal was to identify genes whose methylation is associated with HCC but not cirrhosis, this comparison is appropriate but tells us nothing about progression from normal tissue. A limitation of our plasma DNA analysis is that only samples from cases were available. Thus, while the frequency of methylation was high, we have no data on controls. In our prior prospective study of plasma DNA analyzing 3 genes using methylation specific PCR, we found 2/50 (4%) controls with CDKN2A methylation and comparable cases positive (44 vs 48%).22
In summary, we have used genome-wide methylation arrays to identify genes methylated in HCC from primarily HBV-infected Taiwanese cases. Pyrosequencing of candidate genes validated the array data and analysis of plasma DNA suggests that these genes may be appropriate to apply as biomarkers of early HCC diagnosis. We are in the process of testing custom arrays for analyzing larger numbers of CpG sites then pyrosequencing that can be applied to small amounts of plasma DNA. We will then use this methodology in our prospective study that includes HCC cases and controls, as required to further determine the utility of this approach.
Supplementary Material
Fig. 3.
Hierachical cluster analysis of the selected 24 hypermethylated CpG sites selected by 1,000 three-fold cross-validations. Blue represents adjacent non-tumor tissue, and row represents tumor tissue.
Acknowledgments
This work was supported by NIH grants R01 ES005116, P30 ES009089, P30 CA013696 and R03 CA150140. We thank Dr. Abby Siegel for careful reading of the manuscript.
References
- 1.Imbeaud S, Ladeiro Y, Zucman-Rossi J. Identification of novel oncogenes and tumor suppressors in hepatocellular carcinoma. Semin Liver Dis. 2010;30:75–86. doi: 10.1055/s-0030-1247134. [DOI] [PubMed] [Google Scholar]
- 2.Woo HG, Park ES, Thorgeirsson SS, Kim YJ. Exploring genomic profiles of hepatocellular carcinoma. Mol Carcinog. 2011;50:235–43. doi: 10.1002/mc.20691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tischoff I, Tannapfe A. DNA methylation in hepatocellular carcinoma. World J Gastroenterol. 2008;14:1741–8. doi: 10.3748/wjg.14.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nomoto S, Kinoshita T, Kato K, Otani S, Kasuya H, Takeda S, et al. Hypermethylation of multiple genes as clonal markers in multicentric hepatocellular carcinoma. Br J Cancer. 2007;97:1260–5. doi: 10.1038/sj.bjc.6604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kondo Y, Shen L, Suzuki S, Kurokawa T, Masuko K, Tanaka Y, et al. Alterations of DNA methylation and histone modifications contribute to gene silencing in hepatocellular carcinomas. Hepatol Res. 2007;37:974–83. doi: 10.1111/j.1872-034X.2007.00141.x. [DOI] [PubMed] [Google Scholar]
- 6.Harder J, Opitz OG, Brabender J, Olschewski M, Blum HE, Nomoto S, et al. Quantitative promoter methylation analysis of hepatocellular carcinoma, cirrhotic and normal liver. Int J Cancer. 2008;122:2800–4. doi: 10.1002/ijc.23433. [DOI] [PubMed] [Google Scholar]
- 7.Su H, Zhao J, Xiong Y, Xu T, Zhou F, Yuan Y, et al. Large-scale analysis of the genetic and epigenetic alterations in hepatocellular carcinoma from Southeast China. Mutat Res. 2008;641:27–35. doi: 10.1016/j.mrfmmm.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Su PF, Lee TC, Lin PJ, Lee PH, Jeng YM, Chen CH, et al. Differential DNA methylation associated with hepatitis B virus infection in hepatocellular carcinoma. Int J Cancer. 2007;121:1257–64. doi: 10.1002/ijc.22849. [DOI] [PubMed] [Google Scholar]
- 9.Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47:908–18. doi: 10.1002/hep.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen L, Ahuja N, Shen Y, Habib NA, Toyota M, Rashid A, et al. DNA methylation and environmental exposures in human hepatocellular carcinoma. Journal of the National Cancer Institute. 2002;94:755–61. doi: 10.1093/jnci/94.10.755. [DOI] [PubMed] [Google Scholar]
- 11.Zhang C, Guo X, Jiang G, Zhang L, Yang Y, Shen F, et al. CpG island methylator phenotype association with upregulated telomerase activity in hepatocellular carcinoma. Int J Cancer. 2008;123:998–1004. doi: 10.1002/ijc.23650. [DOI] [PubMed] [Google Scholar]
- 12.Calvisi DF, Ladu S, Gorden A, Farina M, Lee JS, Conner EA, et al. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117:2713–22. doi: 10.1172/JCI31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hui AM, Sakamoto M, Kanai Y, Ino Y, Gotoh M, Yokota J. Inctivation of p16INK4 in hepatocellular carcinoma. Hepatol. 1996;24:575–9. doi: 10.1002/hep.510240319. [DOI] [PubMed] [Google Scholar]
- 14.Matsuda Y, Ichida T, Matsuzawa J, Sugimura K, Asakura H. p16(INK4) is inactivated by extensive CpG methylation in human hepatocellular carcinoma. Gastroenterology. 1999;116:394–400. doi: 10.1016/s0016-5085(99)70137-x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang YJ, Ahsan H, Chen Y, Lunn RM, Wang LY, Chen SY, et al. High frequency of promoter hypermethylation of the RASSF1A and p16 genes and its relationship to aflatoxin B1-DNA adducts level in human hepatocellular carcinoma. Mol Carcinogenesis. 2002;35:85–92. doi: 10.1002/mc.10076. [DOI] [PubMed] [Google Scholar]
- 16.Zhang YJ, Rossner P, Chen Y, Agrawal M, Wang Q, Wang L, et al. Aflatoxin B1 and polycyclic aromatic hydrocarbon adducts, p53 mutations and p16 methylation in liver tissue and plasma of hepatocellular carcinoma patients. International Journal of Cancer. 2006;119:985–91. doi: 10.1002/ijc.21699. [DOI] [PubMed] [Google Scholar]
- 17.Yeo W, Wong N, Wong WL, Lai PB, Zhong S, Johnson PJ. High frequency of promoter hypermethylation of RASSF1A in tumor and plasma of patients with hepatocellular carcinoma. Liver Int. 2005;25:266–72. doi: 10.1111/j.1478-3231.2005.01084.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang YJ, Chen Y, Ahsan H, Lunn RM, Chen SY, Lee PH, et al. Silencing of glutathione S-transferase P1 by promoter hypermethylation and its relationship to environmental chemical carcinogens in hepatocellular carcinoma. Cancer Lett. 2005;221:135–43. doi: 10.1016/j.canlet.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 19.Zhong S, Tang MW, Yeo W, Liu C, Lo YM, Johnson PJ. Silencing of GSTP1 gene by CpG island DNA hypermethylation in HBV-associated hepatocellular carcinomas. Clin Cancer Res. 2002;8:1087–92. [PubMed] [Google Scholar]
- 20.Yang B, Guo M, Herman JG, Clark DP. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. American Journal of Pathology. 2003;163:1101–7. doi: 10.1016/S0002-9440(10)63469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang YJ, Chen Y, Ahsan H, Lunn RM, Lee PH, Chen CJ, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation and its relationship to aflatoxin B1-DNA adducts and p53 mutations in hepatocellular carcinoma. Int J Cancer. 2003;103:440–4. doi: 10.1002/ijc.10852. [DOI] [PubMed] [Google Scholar]
- 22.Zhang YJ, Wu HC, Shen J, Ahsan H, Tsai WY, Yang HI, et al. Predicting hepatocellular carcinoma by detection of aberrant promoter methylation in serum DNA. Clin Cancer Res. 2007;13:2378–84. doi: 10.1158/1078-0432.CCR-06-1900. [DOI] [PubMed] [Google Scholar]
- 23.Lo YM, Tein MS, Lau TK, Haines CJ, Leung TN, Poon PM, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62:768–75. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez-Vargas H, Lambert MP, Calvez-Kelm F, Gouysse G, McKay-Chopin S, Tavtigian SV, et al. Hepatocellular carcinoma displays distinct DNA methylation signatures with potential as clinical predictors. PLoS ONE. 2010;5:e9749. doi: 10.1371/journal.pone.0009749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin SH, Kim BH, Jang JJ, Suh KS, Kang GH. Identification of novel methylation markers in hepatocellular carcinoma using a methylation array. J Korean Med Sci. 2010;25:1152–9. doi: 10.3346/jkms.2010.25.8.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ammerpohl O, Pratschke J, Schafmayer C, Haake A, Faber W, von Kampen O, et al. Distinct DNA methylation patterns in cirrhotic liver and hepatocellular carcinoma. Int J Cancer. 2011 doi: 10.1002/ijc.26136. [DOI] [PubMed] [Google Scholar]
- 27.Gao W, Kondo Y, Shen L, Shimizu Y, Sano T, Yamao K, et al. Variable DNA methylation patterns associated with progression of disease in hepatocellular carcinomas. Carcinogenesis. 2008 doi: 10.1093/carcin/bgn170. [DOI] [PubMed] [Google Scholar]
- 28.Kim YH, Lee HC, Kim SY, Yeom YI, Ryu KJ, Min BH, et al. Epigenomic analysis of aberrantly methylated genes in colorectal cancer identifies genes commonly affected by epigenetic alterations. Ann Surg Oncol. 2011;18:2338–47. doi: 10.1245/s10434-011-1573-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikeska T, Felsberg J, Hewitt CA, Dobrovic A. Analysing DNA methylation using bisulphite pyrosequencing. Methods Mol Biol. 2011;791:33–53. doi: 10.1007/978-1-61779-316-5_4. [DOI] [PubMed] [Google Scholar]
- 30.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–93. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kibriya MG, Raza M, Jasmine F, Roy S, Paul-Brutus R, Rahaman R, et al. A Genome-wide DNA Methylation Study in Colorectal Carcinoma. BMC Med Genomics. 2011;4:50. doi: 10.1186/1755-8794-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–6. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 33.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lunn RM, Zhang YJ, Wang LY, Chen CJ, Lee PH, Lee CS, et al. p53 Mutations, chronic hepatitis B virus infection, and aflatoxin exposure in hepatocellular carcinoma in Taiwan. Cancer Res. 1997;57:3471–7. [PubMed] [Google Scholar]
- 35.Zhang YJ, Chen CJ, Lee CS, Haghighi B, Yang GY, Wang LW, et al. Aflatoxin B1-DNA adducts and hepatitis B virus antigens in hepatocellular carcinoma and non-tumorous liver tissue. Carcinogenesis. 1991;12:2247–52. doi: 10.1093/carcin/12.12.2247. [DOI] [PubMed] [Google Scholar]
- 36.Rajendram R, Ferreira JC, Grafodatskaya D, Choufani S, Chiang T, Pu S, et al. Assessment of methylation level prediction accuracy in methyl-DNA immunoprecipitation and sodium bisulfite based microarray platforms. Epigenetics. 2011;6:410–5. doi: 10.4161/epi.6.4.14763. [DOI] [PubMed] [Google Scholar]
- 37.Yuen RK, Penaherrera MS, von Dadelszen P, McFadden DE, Robinson WP. DNA methylation profiling of human placentas reveals promoter hypomethylation of multiple genes in early-onset preeclampsia. Eur J Hum Genet. 2010;18:1006–12. doi: 10.1038/ejhg.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.