Abstract
Objective/Aims
To determine whether early viral dynamics and evolution predict outcome of primary acute hepatitis C virus (HCV) infection.
Methods
HCV- and HIV-negative injection drug users were enrolled prospectively and followed monthly to identify acute HCV infection using RNA detection. Subjects with more than one month between HCV RNA negative and positive visits were excluded to ensure stringent acute infection. Differences in medians of log-transformed viral RNA levels and evolutionary rates in each gene of a 5’-hemigenomic amplicon were assessed using the Mann-Whitney Rank Sum test. Correlation coefficient was calculated using Spearman Rank Order.
Results
Initial viremia level was 50-fold higher in subjects with spontaneous clearance (compared with persistence) of primary acute HCV infection (median 7.1 versus 5.4 log10 IU/mL, P=0.002). Initial viremia level in subjects with IL-28B-C allele and clearance was higher than that in subjects with IL-28B-T allele and persistence (P=0.001). Evolutionary rates in the hypervariable region 1 (HVR1) region of E2 gene were significantly higher in self-resolvers than those in persistence subjects during early infection, whereas other genes/regions had comparable rates. All major substitutions in HVR1 in persistence subjects were convergent changes whereas over the same time interval clearance subjects displayed divergent evolution, indicating different immune responses between the two groups.
Conclusions
Spontaneous clearance of acute HCV infection is predicted by high initial viremia as well as favorable IL-28B genotype, and associated with rapid envelope sequence evolution. This linkage of host genetics, viral dynamics, and evolution provides new directions for mechanistic studies.
INTRODUCTION
Approximately 170 million people are currently infected with Hepatitis C virus (HCV) worldwide, with continued transmission mostly through blood supply in under-developed areas and needle sharing in developed countries.(1) The estimated infection incidence is 12.9 per 100 person-years among injection drug users (IDUs).(2) Following acute infection, which is usually asymptomatic, 60–80% of infected individuals progress to chronic infection, which is the leading cause of death from liver diseases and indication for liver transplantation in the United States.(3,4)
In spontaneously-resolving infections, patients usually experience clearance during the first year of infection, with the majority eliminating the virus within the first 6 months, during which complicated virus-host interactions, i.e. initiation of innate and adaptive immunity of the host and accordant adaptations of the virus, occur that may determine outcome in most infected patients.(5–9) However, investigation of virus-host interactions during this critical period of time has been greatly hampered by limited availability of study subjects and samples, as a majority of HCV infected individuals remains asymptomatic.(10,11) For symptomatic individuals, symptoms usually develop weeks or even months after infection. As a result, viral kinetics and viral evolution during early phase of acute HCV infection have rarely been investigated and their impacts on infection outcome remain poorly understood.
Two years ago, a strong association between variation in or near the IL-28B gene and the outcome of spontaneous or treatment-induced HCV clearance has been reported from separate study cohorts, though the mechanistic basis for these associations remains unknown.(12–15) IL-28B encodes IFN-λ3, a member of the IFN-λ family, with anti-HCV activity in vitro(16,17) and in vivo.(18) Separately, it has been reported that higher HCV RNA level is associated with persistence of acute infection.(19,20) We hypothesized that IL28B polymorphisms, early viral kinetics, adaptive immunity, and outcome are linked during acute HCV infection.
Adaptive immunity is crucial in determining the outcome of acute infection. Studies in human beings and chimpanzees suggest that clearance of viremia is associated with vigorous CD4 and CD8 T cell responses.(5,21,22) However, HCV often persists despite the detection of HCV-specific T cell responses during acute infection, indicating that initiation of cellular responses alone may not be sufficient for HCV clearance.(6,23) On the other hand, there has been controversy about whether humoral immune responses contribute to viral clearance. Studies in chimpanzees revealed weak and delayed humoral responses resulting in incomplete protection.(24,25) In vitro, nAbs do not block cell-to-cell spread of HCV.(26) In contrast, human studies using autologous envelope proteins detected nAb in spontaneous resolvers whereas chronically evolving subjects have delayed initiation of nAb responses.(27,28) Using the more sensitive autologous HCVpp method and evolutionary inference, several recent studies have demonstrated that nAbs drive sequence evolution in envelope proteins and thus contribute to clearance of HCV variants.(27,29,30) Therefore, we hypothesize that sequence evolution patterns are different between individuals who spontaneously clear compared with those who develop persistence during early acute HCV infection.
We investigated viral kinetics and evolution during early phase of primary acute infection in spontaneously-resolving and persisting acute HCV infections in human subjects. Here, we report for the first time that high initial viremia level is strongly correlated with spontaneous clearance of primary acute HCV infection. Rapid sequence evolution was found in resolving infection compared with slower and convergent evolution in patients who progressed to chronicity.
Materials and Methods
Study subjects
The Baltimore Before-and-After Acute Study of Hepatitis (BBAASH) cohort prospectively enrolls HCV-negative injection drug users in Baltimore and follows them monthly to detect HCV RNA and anti-HCV seroconversion in order to identify primary acute infection.(31) To investigate early viral dynamics and evolution with respect to outcome, the following criteria were employed to include only subjects with stringently acute primary infection, and in whom spontaneous clearance or persistence could be determined: (i) primary anti-HCV seroconversion; (ii) an interval no more than 1 month between HCV RNA negativity and positivity; (iii) sufficient follow-up to determine outcome as spontaneous clearance (HCV RNA negativity for at least 2 months within the first 18 months, with re-infection only when a genetically-distinct viral strain is identified) or persistence (viremic for at least 24 months with a phylogenetically-consistent strain); and (iv) anti-HIV and HBsAg negative. Written informed consent was obtained from each subject, and at each visit counseling is provided to reduce the risks of injection drug use. All participants with acute HCV infection were referred for evaluation for possible treatment. The BBAASH study protocol was approved by the Institutional Review Board of the Johns Hopkins University School of Medicine.
HCV testing protocol
A detailed HCV testing protocol has been described previously.(32) Briefly, HCV RNA was extracted from serum using a Qiagen MinElute column (Qiagen, Valencia, CA) and measured using a real-time RT-PCR assay (TaqMan HCV analyte-specific reagent, Roche Molecular Diagnostics, Indianapolis, IN). Amplification products were monitored on a COBAS TaqMan Analyzer (Roche Molecular Diagnostics), with detection limit of 50 IU/mL. Genetic similarity of viruses from sequential visits was determined by phylogenetic analysis on Core-E1 region as previously described,(32) with uncorrected genetic distance over 0.05 considered a different virus strain.
Genotyping of IL-28B gene polymorphism
Genotyping of IL-28B SNP rs12979860 was conducted using the ABI TaqMan Allelic Discrimination kit and the ABI7900HT Sequence Detection System (Applied Biosystems) as previously described.(13)
Amplification, cloning and sequencing of the 5’ hemigenome
HCV RNA was extracted, and the region from 5’-untranslated region to NS3/NS4A junction was reverse transcribed, nested-PCR amplified and cloned as previously described.(30,33,34) Superscript II reverse transcriptase (Invitrogen) was used with RT primer 6080G1R-16 (5‘-CCGGTTCATCCAYTGC-3’). Nested-PCR was performed using Platinum Taq Polymerase High-Fidelity (Invitrogen) and the same set of primers as described previously,(34) followed by gel purification, ligation, and transformation utilizing the TOPO XL PCR cloning kit (Invitrogen). Template resampling is avoided by this method.(33) Twenty-four clones were randomly selected, amplified using a high-fidelity polymerase (TempliPhi; GE Healthcare Products, Inc.) and sequenced as previously described,(30) producing a partial E1/E2 sequence of 603 nt, with 282 nt of E1 and 321 nt of E2, containing HVR1. Phylogenetic trees were built based on these sequences to determine representative clone(s) nearest the center of the tree.(35) Representative clones were sequenced across the entire 5.2 kb hemigenomic region.
Sequence alignment and phylogenetic analysis
Sequence contigs were assembled and aligned as previously described using CodonCode Aligner (version 2.0.6; http://www.codoncode.com/aligner/), ClustalX (version 2.0; http://www.clustal.org/clustal2/) and BioEdit (version 7.0.9.0; http://www.mbio.ncsu.edu/BioEdit/bioedit.html).(30) Reference sequences comprised 390 1a and 296 1b well defined human HCV complete genome sequences from GenBank. Maximum likelihood trees were built using PhyML (version 3.0). Divergence and rate of non-synonymous (dN) and synonymous (dS) evolution were calculated using MEGA (version 4.1; http://www.megasoftware.net). MargFreq (version 1.0.1,http://sray.med.som.jhmi.edu/SCRoftware/MargFreq) was used to generate consensus amino acid sequences. VarPlot (version 1.7, http://sray.med.som.jhmi.edu/SCRoftware/VarPlot) was used to detect directional evolution.
Statistical analysis
Correlation coefficient was calculated using the Spearman Rank Order as implemented in SigmaPlot (version 11.2). Viral RNA levels were analyzed and compared using Mann-Whitney U test. P values less than 0.05 were considered statistical significant.
Nucleotide sequence accession numbers
Nucleotide sequences described in this report have been submitted to GenBank and have been assigned accession numbers ___ through ___.
Results
Inclusion Criteria and Characteristics of subjects
Thirty subjects satisfied all inclusion criteria, 14 with clearance and 15 with persistent viremia, with similar baseline characteristics (Table. 1). As is typical for our cohort,(31) most subjects were self-identified as Caucasians and infected with genotype 1a HCV. Median (IQR) duration of viremia was 2.5 (1.25, 4) months for clearance subjects.
Table 1.
Characteristics of study subjects
| Clearance N=14 |
Persistence N=15 |
P value | |
|---|---|---|---|
| Female Gender (%) | 57 | 40 | ns |
| Caucasian Race (%) | 100 | 87 | ns |
| Genotype 1a (%) | 71 | 93 | ns |
| Age at seroconversion (mean±std) | 24.8 ± 2.6 | 25.5 ± 4.0 | ns |
| IL-28B genotype C/C (%) | 64 | 27 | ns |
Higher initial HCV RNA level correlated with spontaneous clearance of primary HCV infection
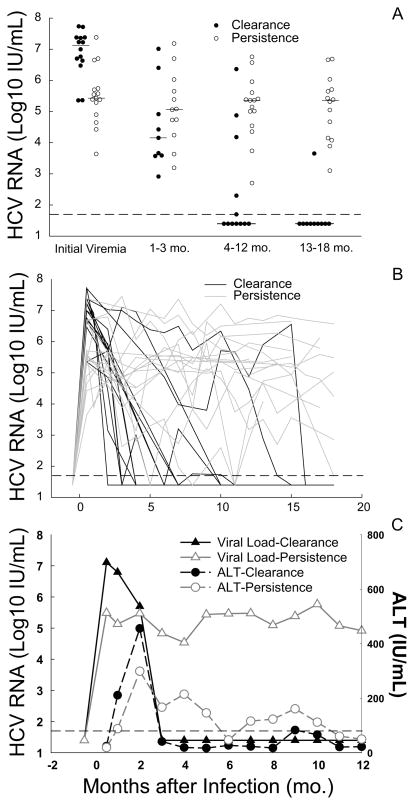
Initial viral RNA level in the clearance group was significantly higher than that in the persistence group (median 7.1 and 5.4 log10 IU/mL in clearance and persistence groups, respectively, P=0.002) (Fig.1A). Among the 14 clearance subjects, 12 (86%) had initial HCV RNA level higher than 6 log10 IU/mL, whereas among the 15 persistence subjects only 3 (20%) had initial viremia higher than 6 log10 IU/mL. Half of clearance subjects had initial HCV RNA over 7 log10 IU/mL, whereas only one out of 15 persistence subjects (6.7%) had values over 7 log10 IU/mL (Fig.1A). Individual and median viral RNA curves demonstrated early peak and fall of viral RNA levels in the clearance group compared with blunted peak and relatively stable viral RNA levels in the persistence group during the first year of infection (Fig. 1B, 1C). ALT levels peaked about 2 months after infection onset in both groups, later than the initial viral RNA peak (Fig.1C). ALT levels didn’t differ by outcome, and initial viremia level did not correlate with HCV genotype (P>0.05).
Figure 1. Comparison of initial viral RNA levels and viral kinetics in subjects with self-resolving and persistent infections.
(A) Median viral RNA of each individual during segmented period of time during acute infection. (B) Dynamic change of viral RNA levels in each individual during acute infection. (C) Median viral RNA and ALT curves for clearance and persistence groups. Dashed horizontal lines indicate lower limit of detection for HCV RNA (50 IU/mL).
Analysis of IL-28B genotype with respect to outcome and viremia
We examined IL-28B genotype in this cohort because recent reports indicated that the favorable treatment-response IL-28B genotype (C/C homozygosity at rs12979860) identified in persons with chronic infection (12) was also associated with spontaneous clearance during acute HCV infection (Table. 1).(13) There were more C/C homozygotes in the clearance group (9 of 14, 64%) compared with the persistence group (4 of 15, 27%), consistent with prior reports for spontaneous clearance, though this difference was not statistically significant in this relatively small cohort. Nevertheless, a strong correlation was observed between IL-28B genotype and initial viral RNA level, with C/C (C) genotype strongly associated with higher initial viremia and the C/T or T/T (T) with lower viremia (P=0.00074). To detect bias after the first visit (retention, management decisions, etc), we examined initial viral RNA level and IL-28B genotype data in all subjects (including those in whom spontaneous outcome is not known) in the BBAASH cohort who were strictly acutely infected (lapse between HCV RNA negativity and positivity less than 1 month) and whose IL-28B genotype data were available. In this larger group (n=44), strong association between IL-28B genotype and initial HCV RNA level was also observed (P=0.00005).
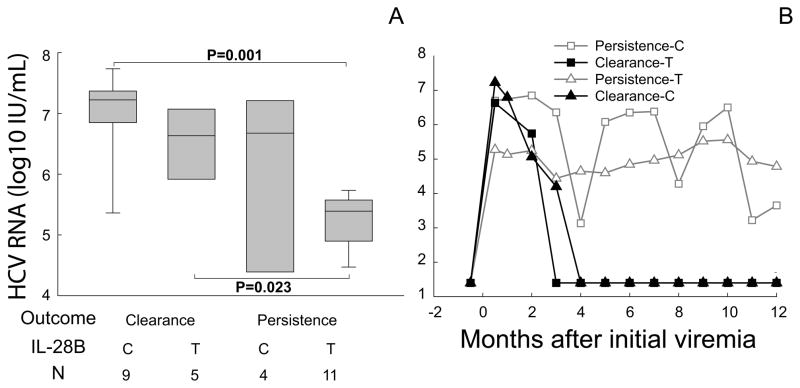
To examine heterogeneity within these groups, we classified subjects into four groups: Cleared subjects with IL-28B genotype C/C (clear-C); Cleared with genotype C/T or T/T (clear-T); Persistent with genotype C/C (persist-C); Persistent with genotype C/T or T/T (persist-T). Initial viral RNA level was significantly higher in clear-C subjects than in persist-T subjects (median 7.2 and 5.4 log10 IU/mL, respectively, P=0.001); however, the smaller clear-T and persist-C groups had highly variable but similarly intermediate viremia (median 6.6 and 6.7 log10 IU/mL, P> 0.05, Fig.2A). The clear-T group did have a significantly higher HCV RNA level at initial viremia than did the persist-T group (Fig.2A), suggesting an independent effect of HCV RNA level. Median viral RNA curves of the four groups demonstrated similar patterns of viral kinetics for clear-C and clear-T groups but slightly different viral dynamic pattern for persist-C and persist-T groups, where persist-C group had a high initial viremia peak followed by more fluctuation in median viral RNA than the persist-T group (Fig.2B). To extend our analysis of factors associated with outcome, we examined viral evolution.
Figure 2. Initial viral RNA and early viral kinetics in correspondence with IL-28B genotype and infection outcome.
(A) Initial viral RNA for four outcome/IL-28B groups. Boxes indicate median and inter-quartile ranges of log-transformed HCV RNA, with 5th and 95th percentiles indicated. IL-28B genotypes are indicated on X axis (C for C/C and T for C/T or T/T). Significant P values (Rank Sum) are shown. (B) Median viral RNA curves for each outcome/IL-28B group during the first year of infection.
Rapid evolution in HVR1 correlated with spontaneous clearance of primary HCV infection
To study viral evolution during acute HCV infection at matched intervals, we identified participants who met two additional criteria: (i) at least 2 amplifiable samples available during the 1st year of primary infection to allow calculation of evolutionary rates, and (ii) visit intervals between 2 and 6 months to minimize bias in evolutionary rate calculation. Thirteen (3 clearance and 10 persistence) subjects, all subtype 1a, satisfied both of these criteria, with median (range) sampling intervals 3 (2–3) and 4.5 (2–6) months, respectively.
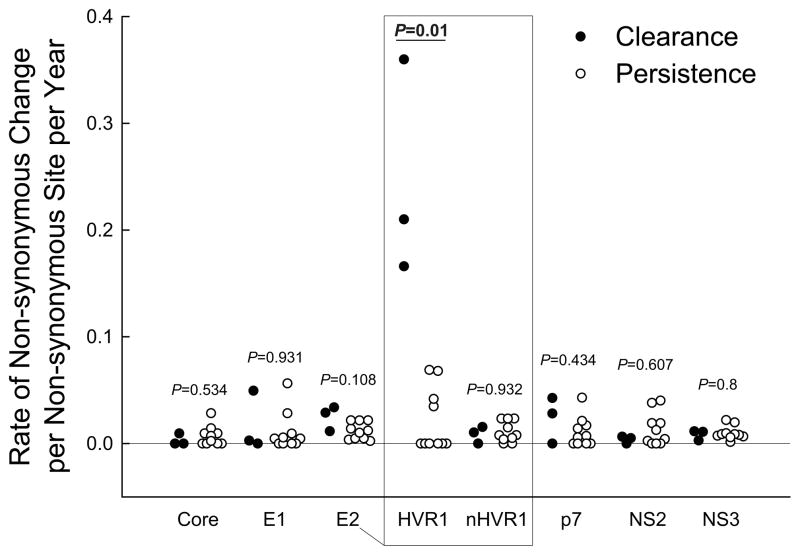
Because HVR1 evolution during acute infection is largely driven by nAb selective pressure,(30) and nAb responses have been detected earlier in cleared subjects than in subjects who develop persistent infection,(28) we hypothesized that the evolutionary rates in HVR1 would differ between outcome groups during early acute infection. The rate of genetic change overall (data not shown) and rate of nonsynonymous change (dN) were comparable between outcome groups in the whole hemigenomic regions. However, higher-resolution comparison of clearance versus persistence subjects’ rates of dN revealed that the rates in particular regions were very different. This is evident when E2 is divided into E2-HVR1 and E2-nonHVR1 segments (Fig.3). Significantly higher rates of change were observed in HVR1 in cleared subjects than in persistent subjects (P=0.01 for comparisons of rate of evolution as well as rate of dN), and comparable rates in all other regions.
Figure 3. Comparison of evolutionary rates for each gene/region of the 5’-hemigenome between cleared and persistent subjects.
Rate of nonsynonymous evolution is depicted as filled (clearance) or empty (persistence) circles for each gene/region and each individual. E2 is divided into E2-HVR1 (HVR1) and E2-nonHVR1 (nHVR1) regions. P values (rank sum) are shown for each comparison of rates.
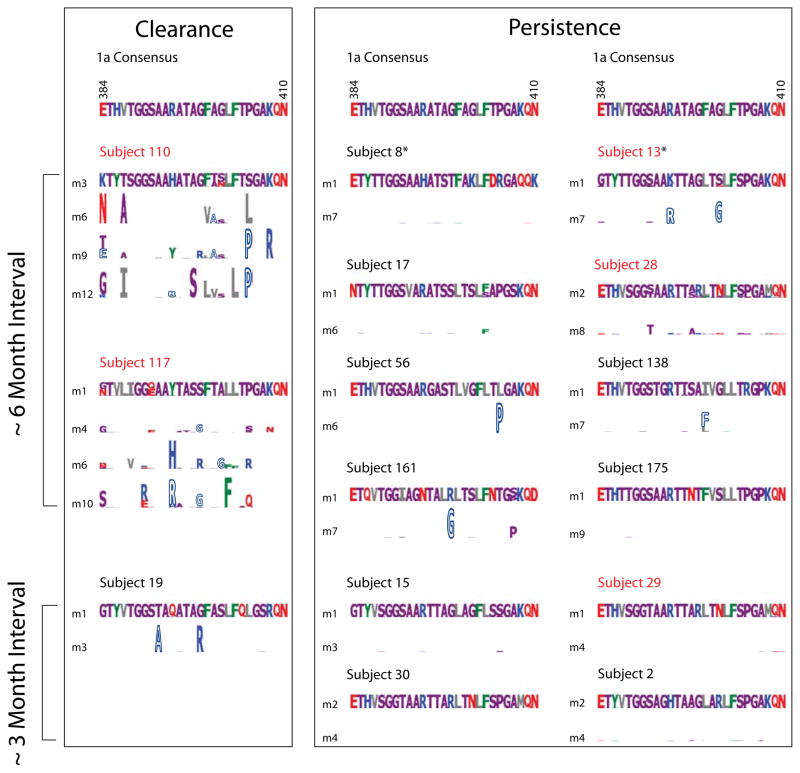
To investigate potential mechanisms linking sequence change in HVR1 with outcome, we characterized amino acid sequence changes in the HVR1 in both self-resolved and persistently infected subjects, some of whose nAb response profiles have been previously reported (Fig.4).(27) In self-resolved subjects amino acid sequences in HVR1 diverged rapidly from initial sequences in association with strong and early initiated nAb responses (subject 110, 117), whereas HVR1 aa sequences remained stable or changed slowly with the lack or late development of nAb responses in subjects who progressed to chronicity (subject 13, 28 and 29).(27,30)
Figure 4. Amino acid sequence evolution of HVR1 in clearance and persistence subjects.
Consensus sequences of HVR1 for genotype 1a HCV are shown above. Type 1 logos (with letter height proportional to frequency) illustrate initial viral quasispecies. Type 2 logos (51) compare amino acid sequences to initial sequences, with the height of each amino acid indicating the log2 unlikelihood of observing the amino acid at a given position. Amino acids that change toward the 1a consensus are depicted with hollow letters. Subjects with comparable visit intervals are depicted in parallel for comparison. Red colored subjects are those with nAb data reported previously (27). Subjects 8 and 13 (indicated by asterisks) who had intervals between HCV RNA negativity and positivity over 1 month but less than 3 months were also included. Amino acid sequence evolution with nAb titers of subject 29 have been shown elsewhere.(30)
As previously described, viral amino acid substitutions can be classified as either centripetal or centrifugal with respect to a worldwide consensus sequence, representing either purifying (negative) or positive selection pressures.(36–38) To determine whether the amino acid substitutions in HVR1 are centripetal or centrifugal, we compared each amino acid substitution with the worldwide genotype 1a consensus sequence. Notably, all major amino acid replacements in the HVR1 in persistence subjects were centripetal (substitutions that change toward the 1a worldwide consensus sequence), in contrast, every clearance subject examined had centrifugal replacements (Fig.4).
Discussion
Variations in IL-28B are associated with outcome of HCV infection,(12–14) but the mechanistic links between the protective genotype and spontaneous outcome remain unknown. Prospective monthly follow-up of HCV-uninfected subjects who became acutely infected revealed (i) a strong correlation between IL-28B genotype and initial HCV RNA level during primary acute HCV infection (P=0.00005), with the favorable IL-28B genotype (rs12979860-C homozygosity) correlated with higher initial viremia level, and (ii) a strong positive correlation between initial HCV RNA level and spontaneous clearance (P=0.00099). These findings are both counterintuitive and coherent with findings in other studies.
In this study, spontaneous resolution was more strongly predicted by initial HCV RNA level than by IL-28B genotype, with the former association reaching statistical significance even in this relatively small cohort. The association of protective IL28B genotype with high initial viremia resonates with recent findings from chronic infection that the protective IL-28B genotype is associated with higher (untreated) HCV RNA levels (12) and lower intrahepatic ISG levels.(39) Prior work demonstrated that lower baseline ISG expression predicts response to treatment.(40,41) It is apparent that IL28B genotype predisposes toward a phenotype that is associated with clinical outcome, and that the association between phenotype and outcome is likely to be stronger than for genotype because other factors are likely to contribute.(15) Taken together, the current and prior work suggest that the protective IL28B genotype is one factor that predisposes to high initial HCV RNA during acute infection and low baseline ISG during chronic infection, and that these represent measurable phenotypes in vivo that strongly predict the outcomes of interest, i.e. spontaneous resolution and treatment response, respectively. Our group recently assessed cytokine and chemokine levels in this cohort as potential markers of such a phenotype, but found no correlation between early levels of those factors and outcome or IL28B genotype.(42)
These data may appear to differ somewhat from previous findings showing higher HCV RNA level is correlated with persistence of acute infection.(19,20) Most studies investigating acute HCV infection have used either clinical symptoms, i.e. jaundice as well as other non-specific symptoms, or first clinical presentation/visit to identify acute infection.(19,20,43,44) The current study demonstrates that the initial HCV RNA peak preceded the ALT peak by about 2 months, and the HCV RNA level dropped rapidly after the initial peak in clearance subjects. Hence, it is likely that using symptoms to identify study subjects would result in missing this early viremia peak in clearance subjects. In addition, patients presenting symptomatically might include persons with prior cleared HCV infection (which we excluded), and reinfection is associated with brief, low-level viremia.(32)
The mechanisms linking IL-28B genotype, initial viremia level, viral evolution rate and outcome remain unknown. High-level HCV replication could trigger strong innate immune responses through pathways such as Toll like receptor 3 (TLR-3) (45) and retinoic-acid-inducible gene I (RIG-I) (46) and hence initiate strong adaptive immune responses that could eventually lead to eradication of the virus.(47) Lower initial viremia may limit inflammation in a manner analogous to preliminary evidence suggesting that small HBV inocula can result in higher rates of persistence in chimpanzees (48); in the current study, we could not assess inoculum size.
Accumulating data support a role for nAb responses in HCV control, though their role in spontaneous clearance remains unclear.(24-30) HCV sequence evolution is shaped by selective pressures, i.e. immune pressures (positive selection) and intrinsic viral fitness constraints (negative selection), reflected in evolutionary patterns.(9,27,30,33,37,38,49) We found that HVR1 was the only region with significantly different evolutionary rates between the two outcome groups and that these rates were significantly higher in clearance subjects than those in persistence subjects. The few sequence changes observed in HVR1 during the first year of persistent infection were convergent changes, consistent with reversion in the absence of immune pressure.(27) In clearance subjects rapid sequence evolution in HVR1 was accompanied by evidence of strong nAb responses.(30,50) Nonrandom evolution with respect to outcome suggests that pressure from nAb responses driving HVR1 evolution contribute to clearance of some viral variants.
In this study, we explored for the first time the potential linkage among IL-28B genotype, viral dynamics during early phase of HCV infection, early viral evolution patterns, and infection outcome. Detailed immunological results are not available because the inclusion criteria for this study were focused on studying viral evolution rather than the availability of large volume of blood draws.(6) Nonetheless, our prospective sampling, stringent inclusion criteria, high resolution of early viral dynamics, and detailed analysis of hemigenomic clone sequences make this the largest and highest-resolution study of viral dynamics and evolution and their correlation with infection outcome and host genetics in humans during early phase of acute HCV infection to date.
In summary, we demonstrate for the first time that initially high HCV RNA level is predictive of spontaneous clearance, that IL-28B genotype is associated with initial HCV RNA level, and that initial early evolutionary patterns in HVR1 are correlated with infection outcome. These new links are not explained by conventional models of acute HCV infection.
Supplementary Material
Acknowledgments
The authors thank all study participants for their contribution, Tom Parks for lab coordination, and David Hudson for recruitment and sample collection. This study was supported by NIH grants R01 DA024565 and U19 AI088791.
List of Abbreviations
- HVR1
hypervariable region-1
- IDU
Injection drug user
- nAb
neutralizing antibody
References
- 1.Ghany MG, Strader DB, Thomas DL, Seeff LB. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49(4):1335–1374. doi: 10.1002/hep.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brant LJ, Ramsay ME, Balogun MA, Boxall E, Hale A, Hurrelle M, et al. Diagnosis of acute hepatitis C virus infection and estimated incidence in low- and high-risk English populations. J Viral Hepat. 2008;15(12):871–877. doi: 10.1111/j.1365-2893.2008.01009.x. [DOI] [PubMed] [Google Scholar]
- 3.Maheshwari A, Ray S, Thuluvath PJ. Acute hepatitis C. Lancet. 2008;372(9635):321–332. doi: 10.1016/S0140-6736(08)61116-2. [DOI] [PubMed] [Google Scholar]
- 4.Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45(4):529–538. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Bowen DG, Walker CM. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature. 2005;436(7053):946–952. doi: 10.1038/nature04079. [DOI] [PubMed] [Google Scholar]
- 6.Cox AL, Mosbruger T, Lauer GM, Pardoll D, Thomas DL, Ray SC. Comprehensive analyses of CD8+ T cell responses during longitudinal study of acute human hepatitis C. Hepatology. 2005;42(1):104–112. doi: 10.1002/hep.20749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rutebemberwa A, Ray SC, Astemborski J, Levine J, Liu L, Dowd KA, et al. High-programmed death-1 levels on hepatitis C virus-specific T cells during acute infection are associated with viral persistence and require preservation of cognate antigen during chronic infection. J Immunol. 2008;181(12):8215–8225. doi: 10.4049/jimmunol.181.12.8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim AY, Schulze zur WJ, Kuntzen T, Timm J, Kaufmann DE, Duncan JE, et al. Impaired hepatitis C virus-specific T cell responses and recurrent hepatitis C virus in HIV coinfection. PLoS Med. 2006;3(12):e492. doi: 10.1371/journal.pmed.0030492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox AL, Mosbruger T, Mao Q, Liu Z, Wang XH, Yang HC, et al. Cellular immune selection with hepatitis C virus persistence in humans. J Exp Med. 2005;201(11):1741–1752. doi: 10.1084/jem.20050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koretz RL, Brezina M, Polito AJ, Quan S, Wilber J, Dinello R, et al. Non-A, non-B posttransfusion hepatitis: comparing C and non-C hepatitis. Hepatology. 1993;17:361–365. [PubMed] [Google Scholar]
- 11.Aach RD, Stevens CE, Hollinger FB, Mosley JW, Peterson DA, Taylor PE, et al. Hepatitis C virus infection in post-transfusion hepatitis. N Engl J Med. 1991;325:1325–1329. doi: 10.1056/NEJM199111073251901. [DOI] [PubMed] [Google Scholar]
- 12.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009 doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 13.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461(7265):798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rauch A, Kutalik Z, Descombes P, Cai T, di IJ, Mueller T, et al. Genetic variation in IL28B Is Associated with Chronic Hepatitis C and Treatment Failure - A Genome-Wide Association Study. Gastroenterology. 2010 doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 15.Balagopal A, Thomas DL, Thio CL. IL28B and the control of hepatitis C virus infection. Gastroenterology. 2010;139(6):1865–1876. doi: 10.1053/j.gastro.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, et al. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131(6):1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 17.Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79(6):3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muir AJ, Shiffman ML, Zaman A, Yoffe B, de la Torre A, Flamm S, et al. Phase 1b study of pegylated interferon lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology. 2010 doi: 10.1002/hep.23743. [DOI] [PubMed] [Google Scholar]
- 19.McGovern BH, Nagami EH, Birch CE, Bowen MJ, Reyor LL, Chung RT, et al. Rate of sustained virologic response in relation to baseline hepatitis C virus (HCV) RNA level and rapid virologic clearance in persons with acute HCV infection. J Infect Dis. 2009;200(6):877–881. doi: 10.1086/605444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis-Ximenez LL, Lauer GM, Schulze zur WJ, de Sousa PS, Ginuino CF, Paranhos-Baccala G, et al. Prospective follow-up of patients with acute hepatitis C virus infection in Brazil. Clin Infect Dis. 2010;50(9):1222–1230. doi: 10.1086/651599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lechner F, Wong DK, Dunbar PR, Chapman R, Chung RT, Dohrenwend P, et al. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med. 2000;191(9):1499–1512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thimme R, Oldach D, Chang KM, Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194(10):1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wedemeyer H, He XS, Nascimbeni M, Davis AR, Greenberg HB, Hoofnagle JH, et al. Impaired effector function of hepatitis C virus-specific CD8+ T cells in chronic hepatitis C virus infection. J Immunol. 2002;169(6):3447–3458. doi: 10.4049/jimmunol.169.6.3447. [DOI] [PubMed] [Google Scholar]
- 24.Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, et al. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc Natl Acad Sci USA. 1996;93(26):15394–15399. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bukh J, Thimme R, Meunier JC, Faulk K, Spangenberg HC, Chang KM, et al. Previously infected chimpanzees are not consistently protected against reinfection or persistent infection after reexposure to the identical hepatitis C virus strain. J Virol. 2008;82(16):8183–8195. doi: 10.1128/JVI.00142-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timpe JM, Stamataki Z, Jennings A, Hu K, Farquhar MJ, Harris HJ, et al. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology. 2008;47(1):17–24. doi: 10.1002/hep.21959. [DOI] [PubMed] [Google Scholar]
- 27.Dowd KA, Netski DM, Wang XH, Cox AL, Ray SC. Selection Pressure from Neutralizing Antibodies Drives Sequence Evolution during Acute Infection with Hepatitis C Virus. Gastroenterology. 2009;136(7):2377–2386. doi: 10.1053/j.gastro.2009.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pestka JM, Zeisel MB, Blaser E, Schurmann P, Bartosch B, Cosset FL, et al. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc Natl Acad Sci U S A. 2007;104(14):6025–6030. doi: 10.1073/pnas.0607026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Hahn T, Yoon JC, Alter H, Rice CM, Rehermann B, Balfe P, et al. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology. 2007;132(2):667–678. doi: 10.1053/j.gastro.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Liu L, Fisher BE, Dowd KA, Astemborski J, Cox AL, Ray SC. Acceleration of hepatitis C virus envelope evolution in humans is consistent with progressive humoral immune selection during the transition from acute to chronic infection. J Virol. 2010;84(10):5067–5077. doi: 10.1128/JVI.02265-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cox AL, Page K, Bruneau J, Shoukry NH, Lauer GM, Kim AY, et al. Rare birds in North America: acute hepatitis C cohorts. Gastroenterology. 2009;136(1):26–31. doi: 10.1053/j.gastro.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osburn WO, Fisher BE, Dowd KA, Urban G, Liu L, Ray SC, et al. Spontaneous Control of Primary Hepatitis C Virus Infection and Immunity Against Persistent Reinfection. Gastroenterology. 2010;138(1):315–324. doi: 10.1053/j.gastro.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang XH, Netski DM, Astemborski J, Mehta SH, Torbenson MS, Thomas DL, et al. Progression of fibrosis during chronic hepatitis C is associated with rapid virus evolution. J Virol. 2007;81(12):6513–6522. doi: 10.1128/JVI.02276-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z, Netski DM, Mao Q, Laeyendecker O, Ticehurst JR, Wang XH, et al. Accurate representation of the hepatitis C virus quasispecies in 5.2-kilobase amplicons. J Clin Microbiol. 2004;42(9):4223–4229. doi: 10.1128/JCM.42.9.4223-4229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nickle DC, Jensen MA, Gottlieb GS, Shriner D, Learn GH, Rodrigo AG, et al. Consensus and ancestral state HIV vaccines. Science. 2003;299(5612):1515–1518. doi: 10.1126/science.299.5612.1515c. [DOI] [PubMed] [Google Scholar]
- 36.Friedrich TC, Dodds EJ, Yant LJ, Vojnov L, Rudersdorf R, Cullen C, et al. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat Med. 2004;10(3):275–281. doi: 10.1038/nm998. [DOI] [PubMed] [Google Scholar]
- 37.Kuntzen T, Timm J, Berical A, Lewis-Ximenez LL, Jones A, Nolan B, et al. Viral sequence evolution in acute hepatitis C virus infection. J Virol. 2007;81(21):11658–11668. doi: 10.1128/JVI.00995-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ray SC, Fanning L, Wang XH, Netski DM, Kenny-Walsh E, Thomas DL. Divergent and convergent evolution after a common-source outbreak of hepatitis C virus. J Exp Med. 2005;201(11):1753–1759. doi: 10.1084/jem.20050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Honda M, Sakai A, Yamashita T, Nakamoto Y, Mizukoshi E, Sakai Y, et al. Hepatic Interferon-Stimulated Genes Expression Is Associated With Genetic Variation in Interleukin 28B and the Outcome of Interferon Therapy for Chronic Hepatitis C. Gastroenterology. 2010 doi: 10.1053/j.gastro.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 40.Chen L, Borozan I, Feld J, Sun J, Tannis LL, Coltescu C, et al. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128(5):1437–1444. doi: 10.1053/j.gastro.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 41.Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, Filipowicz W, et al. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci U S A. 2008;105(19):7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chattergoon MA, Levine JS, Latanich R, Osburn WO, Thomas DL, Cox AL. High plasma interleukin-18 levels mark the acute phase of hepatitis C virus infection. J Infect Dis. 2011;204(11):1730–1740. doi: 10.1093/infdis/jir642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beld M, Penning M, McMorrow M, Gorgels J, Van den Hoek A, Goudsmit J. Different hepatitis C virus (HCV) RNA load profiles following seroconversion among injecting drug users without correlation with HCV genotype and serum alanine aminotransferase levels. J Clin Microbiol. 1998;36(4):872–877. doi: 10.1128/jcm.36.4.872-877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villano SA, Vlahov D, Nelson KE, Cohn S, Thomas DL. Persistence of viremia and the importance of long-term follow-up after acute hepatitis C infection. Hepatology. 1999;29(3):908–914. doi: 10.1002/hep.510290311. [DOI] [PubMed] [Google Scholar]
- 45.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413(6857):732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 46.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454(7203):523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoebe K, Janssen E, Beutler B. The interface between innate and adaptive immunity. Nat Immunol. 2004;5(10):971–974. doi: 10.1038/ni1004-971. [DOI] [PubMed] [Google Scholar]
- 48.Asabe S, Wieland SF, Chattopadhyay PK, Roederer M, Engle RE, Purcell RH, et al. The size of the viral inoculum contributes to the outcome of hepatitis B virus infection. J Virol. 2009;83(19):9652–9662. doi: 10.1128/JVI.00867-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timm J, Lauer GM, Kavanagh DG, Sheridan I, Kim AY, Lucas M, et al. CD8 epitope escape and reversion in acute HCV infection. J Exp Med. 2004;200(12):1593–1604. doi: 10.1084/jem.20041006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartosch B, Verney G, Dreux M, Donot P, Morice Y, Penin F, et al. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J Virol. 2005;79(13):8217–8229. doi: 10.1128/JVI.79.13.8217-8229.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gorodkin J, Heyer LJ, Brunak S, Stormo GD. Displaying the information contents of structural RNA alignments: the structure logos. Comput Appl Biosci. 1997;13(6):583–586. doi: 10.1093/bioinformatics/13.6.583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.