Abstract
Prospective motion correction methods utilizing an optical system, DW-PACE or a free induction decay (FID) navigator, have recently been applied to correct for motion in DTI. These methods have some limitations and drawbacks. This paper describes a novel technique using a 3D-EPI navigator, of which the contrast is independent of the b-value, to perform prospective motion correction in diffusion weighted images, without having to reacquire volumes during which motion occurred, unless motion exceeded some pre-set thresholds. Water phantom and human brain data were acquired using the standard and navigated diffusion sequences, and the mean and whole brain histogram (WBH) of the fractional anisotropy (FA) and mean diffusivity (MD) were analysed. Our results show that adding the navigator does not influence the diffusion sequence. With head motion, the WBH-FA shows a shift toward lower anisotropy with a significant decrease in both the mean FA and the FA histogram peak location (p<0.01), while the WBH-MD shows a shift toward higher diffusivity with a significant increase in the mean MD (p<0.01), even after retrospective motion correction. These changes in the mean and the shape of the histograms are recovered substantially in the prospective motion corrected data acquired using the navigated sequence.
Keywords: diffusion tensor imaging (DTI), prospective motion correction, volumetric navigator, reacquisition, fractional anisotropy (FA), mean diffusivity (MD), whole brain histogram (WBH)
Introduction
Diffusion Tensor Imaging (DTI) provides information that has been widely used in mapping the architecture of the central nervous system (1), in studying development and aging (2–3), and in detecting diverse pathological conditions of the human brain due to its ability to identify microstructural abnormalities (4). DTI data differ from other imaging modalities in that each voxel contains not only a single value, but a 3×3 positive definite matrix also known as a diffusion tensor (5). The diffusion tensor is calculated by scanning a volume of the brain repeatedly: one scan without diffusion sensitization, and the others with diffusion sensitization in non-collinear directions. Head motion causes misalignment of the diffusion volumes and furthermore, individual voxels are exposed to a slightly different diffusion encoding direction/gradient than the desired one (6). Motion during diffusion sensitization gradients also causes signal dropouts in the images. Both uncorrected diffusion volumes and errors in diffusion encoding can cause serious artifacts in DTI, which can result in erroneous estimations of the diffusion tensor information. Generally, retrospective motion correction is used to correct for both the misalignment in the diffusion volumes as well as the b matrices. Retrospective techniques have many limitations in the co-registration of conventional MR images, including blurring artifacts (7) and the influence of through-plane motion on the local history of magnetization (8). Retrospective techniques face greater difficulties with diffusion weighted imaging (DWI), especially diffusion weighting with high b-values. A robust retrospective method has been implemented by Rohde et al. (9), in which mutual information is used to register the diffusion volumes and correct the b matrix. This method may not be able to correctly register diffusion volumes acquired with diffusion weightings greater than 1100 s mm−2 due to the fact that most of the tissue/air boundaries, which are some of the main features that guide the image registration process, are nearly eliminated at high b-values.
To avoid the limitations associated with retrospective motion correction, prospective motion correction in DTI has recently been proposed. Three studies have previously reported real-time motion correction in DTI. The optical system reported by Aksoy et al. (10) uses a camera and markers. The requirement for specialized hardware and software, which are not standard components of the scanner, is limiting. The second method or diffusion-weighted prospective acquisition correction (DW-PACE) was implemented by Benner et al. (11) to correct misalignment in diffusion data. DW-PACE is based on registering the diffusion volumes in real time to the first reference volume that has the same diffusion weighting using the prospective acquisition correction (PACE) algorithm for estimating the motion parameters (12). DW-PACE requires sufficient signal-to-noise ratio (SNR) in the diffusion images for accurate image co-registration, which limits the application of DW-PACE to low diffusion weightings, thus precluding applications such as q-space imaging (13), Q-ball imaging (14), and diffusion spectrum imaging (15). The third method proposed by Kober and colleagues (16) detects motion by performing a volume-wise comparison of the signal measured using a free induction decay (FID) navigator that is inserted after the slice rewinder gradient. While the authors report high sensitivity and specificity in detecting motion, the major disadvantage of this method is that the navigator contains no anatomical information, so that an extra b0 acquisition, two to allow time for feedback in the case of prospective motion correction, needs to be acquired each time motion is detected in order to compute the motion parameters, after which the diffusion volume is reacquired in the updated gradient coordinate system. As such, two (retrospective correction) to three (prospective correction) extra TR’s are added to the sequence each time motion is detected, which could result in impractical long scan times in young uncooperative subjects that move often. The authors also note increased noise using this method at higher b-values, which they attribute to eddy current history effects, and difficulties detecting slow movements.
Since most of the current self-navigating methods for motion correction in DTI rely on image contrast, the correction for the misalignment of the diffusion volumes is rendered inaccurate by the changes in the diffusion weighting, especially at high b-values. Navigator-based motion tracking methods typically take advantage of the k-space properties of rigid body transforms to subsample k-space in a time efficient manner. These include orbital (17–18), spherical (19) and cloverleaf (20) navigators. Despite these methods being very fast, imaging navigators, such as PROMO (21) and the echo planar imaging (EPI) volumetric navigator (22), are well suited to methods with long repetition times and have been implemented in spectroscopy. To date, imaging navigators have not been applied to DTI.
The current paper presents the first prospective motion correction technique in DTI that (1) uses a volumetric navigator that contains 3D anatomical information for direct computation of motion parameters, and (2) for which the accuracy of co-registration and motion estimates are not affected by the diffusion gradients even at high b-values. Using this method, prospective motion correction is performed without requiring reacquisition of volumes during which motion occurred. The additional scan time for the navigator and feedback is only 526 ms per TR (TR = 9500 ms). If selected in the protocol, reacquisition of volumes will only occur for large motions that exceed some pre-set threshold. In such cases, reacquisition of the motion-corrupted volume is performed in a single TR, thus minimising the extra scan time required. Also, the number of reacquisitions allowed is set at the start of the scan, thus limiting the increase in scan time.
Methods
Preparation of the 3D-EPI Navigator Sequence
The navigator is a three-dimensional multishot echo planar imaging (EPI) sequence element, with a very small flip angle of 2 degrees to minimize the impact of signal saturation on the diffusion sequence. The off-resonance distortions and T2 blurring effect are relatively insignificant due to the use of a multishot technique, high bandwidth in readout direction 3906 Hz/px, a very short echo spacing time of 310 µsec, and a very short TR of 14ms per shot. The acquisition matrix size is 32 × 32 × 28 (8 mm isotropic) to achieve both a very short scan time and good estimation of motion. The field-of-view (FOV) in all three directions was selected to cover the subject’s head. The navigator protocol is prepared and run on the scanner before starting the diffusion sequence. The aim of this process is to check and store the navigator protocol for future use in the modified diffusion sequence. Running the scan triggers the image reconstruction to store the navigator protocol and can be used to confirm that the brain is properly located in the navigator. This preliminary scan takes less than a second to run and once the protocol is saved does not need to be rerun unless the protocol changes.
Diffusion Pulse Sequence with 3D-EPI Navigator
A twice-refocused 2D diffusion pulse sequence (23) has been modified to acquire a 3D-EPI navigator following the acquisition of each diffusion volume. The navigator protocol, prepared in a preceding setup scan, is separate from the diffusion protocol. The diffusion sequence is enabled to read two separate protocols at the same time; one for the standard diffusion sequence and the other for the navigator. In order for the diffusion volumes to be spatially aligned in real time, each navigator volume, that follows the diffusion volume, has to be reconstructed and the transformation to a reference volume computed. The first navigator volume is chosen as the reference volume.
Both ICE (image calculation environment) and PACE (prospective acquisition correction) are Siemens software running on the scanner. Navigator image reconstruction and estimation of motion are performed in real time in ICE. PACE (12), which uses a least squares cost function for image alignment, is used to calculate the motion parameters for each navigator volume and provides the 6 parameter motion estimate. The navigator preparation process described in the previous step ensures that the navigator FOV covers the subject’s entire head, which is necessary for accurate estimation of motion by PACE. When subject motion in any direction is greater than ±20 mm translation or ±8 degrees rotation, PACE terminates completely because it is likely that the subject will have moved outside the image volume resulting in inaccurate motion estimates. When the motion exceeds these limits, the navigated diffusion sequence continues to run as a standard sequence without prospective motion correction. The navigator volumes will, however, still be acquired and stored.
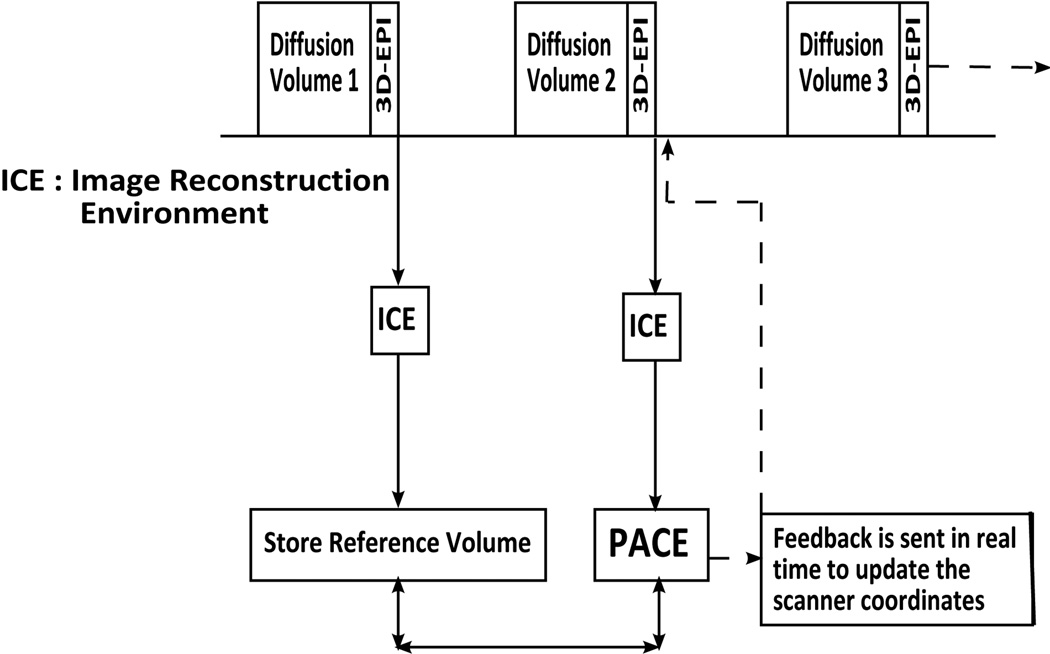
Insertion of the navigator increases the repetition time (TR) of the diffusion sequence by the total scan time of the navigator TRvNav. An additional waiting period for feedback from ICE, Tfeedback, is added to the modified TR to enable the sequence, in real time, to receive feedback and to update the slice positions and gradient coordinate system according to the new motion parameters in the TR immediately following the navigator. This feedback period is determined experimentally and depends on the matrix size of the navigator and diffusion calculation in ICE. Figure 1 summarizes the implementation of prospective motion correction in the volumetric navigated diffusion sequence.
Figure 1.
Flowchart of the modified diffusion pulse sequence with interleaved 3D-EPI navigator. Data is transferred to ICE, registration is performed by PACE, and the position and gradient system is updated before acquisition of the next diffusion volume.
Reacquisition
A limitation of prospective motion correction techniques, in general, is undetected motion. When ICE sends the motion parameters to the diffusion sequence, the sequence updates the slice orientation of the next diffusion volume, but the previously acquired volume is not corrected for motion that may have occurred during its acquisition. This uncorrected motion can cause an error in the estimation of the diffusion tensor if the motion is large and leads to low signal in certain slices of the volume during which the motion occurred. Furthermore, motion during the diffusion sensitization gradients may cause signal dropout in the corresponding images. For this reason, the diffusion pulse sequence was further modified to reacquire volumes during which the motion exceeded some pre-defined threshold. The diffusion protocol was modified to include this feature, which may be selected on the user interface and will prompt the user to specify the number of reacquisitions to be performed (Nreacq). In ICE the image reconstruction pipeline was modified to include a new function that compares the PACE motion parameters to certain thresholds. If the motion parameters exceed these thresholds, ICE sends a signal, in addition to the motion parameters, to the sequence to update the gradient system and reacquire this volume. The volume is reacquired in the next TR. The thresholds set in ICE are translation in any direction ≥ 2.5 mm or rotation in any direction ≥ 1 degree. These thresholds were defined according to the amount of motion that was found to typically lead to volumes with slices of low signal. Every time the sequence reacquires a volume the scan time increases by another TR. All the additional times that are added to the original diffusion scan time are given by Equation (1).
| [1] |
Here Tt is the total scan time of the whole sequence, TRbasic is the repetition time of the standard diffusion sequence (9500 ms in the current protocol), TRvNav is the total scan time of the navigator (406 ms in the current protocol), Tfeedback is the waiting period for the feedback (120 ms in the current protocol), Nreacq is the number of reacquisitions that were specified in the diffusion protocol (5 in the current protocol), Ndiff_directions is the total number of diffusion directions plus the low b value acquisitions (34 in the current protocol), and Nprep is the number of preparation or dummy scans (1 in the current protocol).
The number of reacquisitions Nreacq might change from one protocol to another depending on how much extra time can be tolerated in the total diffusion acquisition time. Specifying the number of reacquisitions at the start of the scan ensures that scans do not become impractically long in cases where subjects frequently move a lot. Should there be no (or too few) occurances of motion during the scan that trigger a reacquisition, the acquisition of the last diffusion volume will be repeated for each non-acquired reacquisition in order to keep the scan time correct. ICE is enabled to display the diffusion images for each TR. In the case where reacquisition has been enabled, ICE will generate both the uncorrected volume and the reacquired one, so that it is possible to study the effects of the modified sequence without or with reacquisition.
Feedback and Combination of Motion Parameters
The gradient pulses are programmed in the logical coordinate system defined by three directions: phase encoding, readout encoding and slice select encoding. The gradients in the logical coordinate system are transformed into the physical coordinate system using a rotation matrix which depends on the slice orientation. When the motion parameters are received from ICE in real time, the sequence updates the position and orientation of the next navigator and applies the updated navigator transformation matrix to adjust the position and the orientation for each diffusion slice. The gradients for both the navigator and the diffusion are adjusted to the new orientation by multiplying the gradients in the current logical coordinate system by the new rotation matrix.
Experimental Protocol
All scans were performed on a Siemens Allegra 3T (Siemens Healthcare, Erlangen, Germany) scanner at the Cape Universities Brain Imaging Centre. The navigator parameters were the same for all scans. The acquisition parameters for the navigator were: TR = 14 ms, TE = 6.6 ms, voxel size = 8 × 8 × 8 mm3, acquisition matrix size = 32 × 32 × 28, FOV 256 × 256 × 224 mm3, the bandwidth in the readout direction = 3906 Hz/px, flip angle = 2 degrees, and total scan time = 406 ms.
The acquisition parameters for the diffusion sequence were: TR = 9500 ms without the navigator (basic sequence), TR = 10026 ms with the navigator (vNav sequence), TE = 86 ms, 72 slices, matrix size = 112 × 112, single channel birdcage head coil, slice thickness = 2 mm, 30 non-collinear diffusion gradient directions, b-values 0 and 1000 s mm−2, and four low b-value scans. The waiting period for feedback (including all navigator-related computations in the sequence and in ICE) is 120 ms for each repetition time. For in vivo scans, reacquisition was enabled with five reacquisitions.
In order to ensure that insertion of the navigator in the diffusion sequence does not corrupt the diffusion data, we first performed DTI acquisitions of a stationary water phantom:
W_basic: water phantom stationary scans acquired using the basic diffusion sequence, repeated three times (W_basic1, W_basic2 and and W_basic3), and
W_vNav: scans of a stationary water phantom using the navigated (vNav) diffusion sequence with prospective motion correction, repeated three times (W_vNav1, W_vNav2, and W_vNav3).
Six healthy male subjects (24 to 30 years) were scanned. All subjects provided informed written consent prior to scanning according to protocols that had been approved by the Faculty of Health Sciences Human Research Ethics Committee of the University of Cape Town, South Africa. The experimental protocol comprised 4 different DTI acquisitions:
-
1)
NoMo_basic: an at rest (no motion - NoMo) scan acquired with the basic (standard) diffusion sequence,
-
2)
NoMo_vNav_NoCo: an at rest scan (NoMo) acquired using the navigated (vNav) diffusion sequence but without prospective motion correction enabled (no correction - NoCo),
-
3)
Mo_basic: a scan with motion (Mo) acquired using the basic diffusion sequence, and
-
4)
Mo_vNav_all: a scan with motion (Mo) acquired using the navigated (vNav) diffusion sequence with motion tracking and motion correction applied, and reacquisition enabled. The Mo_vNav_all data were analysed both without reacquisition (Mo_vNav_noReAq) and with reacquisition (Mo_vNav_ReAq) applied.
Two additional DTI acquisitions were performed for two subjects:
-
5)
Mo_vNav_NoCo: a scan with motion (Mo) acquired with the navigated (vNav) sequence but with no prospective motion correction applied (NoCo). Using this scan, motion parameters could be logged for an acquisition without prospective motion correction, allowing us to compare the navigator motion estimates by PACE with motion estimates determined using retrospective motion correction techniques (Mo_vNav_NoCo_retro), and
-
6)
NoMo_vNav_Co: an at rest scan (NoMo) acquired using the navigated (vNav) diffusion sequence with prospective correction (Co) applied.
During the ‘at rest’ (NoMo) scans, the subjects were asked to lie as still as possible. Since subjects were not anaesthetized, unexpected motion could occur. For the scans with motion, the subjects were instructed to change their head position upon verbal instruction. Five to six instructions (roughly one every 40 s) were given during the scan. The subjects were asked to repeat the same movements for each of the scans acquired with motion. The total scan time without the navigator is 5 minutes and 33 seconds, with the navigator it is 5 minutes and 50 seconds, and with five reacquisitions it is 6 minutes and 40 seconds.
Diffusion Data Processing
Diffusion volumes were processed after conversion from DICOM format to NifTi using Free surfer tools (Athinoula A. Martinos Center for Biomedical Imaging; http://surfer.nmr.mgh.harvard.edu/). The diffusion data of the six subjects were quantified using Diffusion Toolkit (24) (including TrackVis), which generates all the diffusion maps and the track bundles from the NifTi data. From the whole track bundles, the histograms of the DTI-derived indices (MD and FA maps) were computed. In the current studies the whole track bundles of the whole brain were selected. TrackVis then measured the mean FA and the mean MD for each track in the bundle. Each track therefore represents a single data point; the data points were binned to create the histogram. The FA and the MD histograms were transferred to Matlab to calculate the histogram-derived measurements (peak location, skewness and kurtosis). Each histogram contains 50 bins, for each of which the values were normalized by the total number of tracks that is estimated by TrackVis. The mean FA, the mean MD and the histogram-derived measurements were compared for the different acquisitions for the six subjects using a paired t-test implemented in Matlab. P-values less than 0.01 were considered statistically significant. The diffusion maps of the water phantom were processed in a similar way to the in vivo data also using Diffusion ToolKit, but the histogram parameters for the water phantom were calculated from the MD map, and normalized by the total number of pixels. Retrospective motion correction of the diffusion DICOM volumes was performed only for scans with motion that were acquired using the basic diffusion sequence (Mo_basic) and for the Mo_vNav_NoCo scan acquired using the navigated sequence without prospective motion correction. Two different registration methods were used to align all the diffusion volumes to the b0 volume, after which the corrected volumes were processed to extract all the diffusion maps in the same way as described above. The two methods include (1) the use of the linear image registration tool (FLIRT) in FSL (FMRIB Software Library; http://www.fmrib.ox.ac.uk/fsl) with a least squares cost function and 6 degrees of freedom, (2) statistical parametric mapping (SPM5 - http://www.fil.ion.ucl.ac.uk/spm/). We also investigated for three subjects whether eliminating motion corrupted volumes that have a low signal due to the motion improves retrospective motion correction.
Results
The Influence of the Navigator on the Diffusion Sequence
Water Phantom
Figure 2 shows the MD map of a single slice of the first baseline scan (W_basic1) of the water phantom, and the difference between the MD map of this slice for this acquisition and the same slice for each of the other acquisitions. The MD maps of all slices were averaged for the three basic scans and for the three prospectively corrected navigated scans. Shown in Figure 3 are the histograms of the averaged MD maps over the whole volume of the phantom for the two different sequences (W_basic and W_vNav). These figures demonstrate that the insertion of the navigator in the diffusion sequence does not corrupt the diffusion data of the water phantom and that there were no potential or residual errors from PACE, which would have introduced diffusion errors.
Figure 2.
(A) The MD map for slice 38 of a stationary water phantom scanned using the basic diffusion sequence, and the difference between the MD map of this slice and the MD map of the same slice for scan (B) W_basic2, (C) W_basic3, (D) W_vNav1, (E) W_vNav2, and (F) W_vNav3, where W denotes water phantom scans, basic denotes scans acquired using the basic diffusion sequence, and vNav denotes scans acquired using the navigated prospective motion corrected diffusion sequence. All color bars have units 10−3mm2s−1.
Figure 3.
Histograms of the averaged MD for the three scans over the whole volume of the water phantom for the navigated prospective motion corrected diffusion sequence (vNav) and the basic diffusion sequence (basic).
In Vivo Data
Figure 4 shows for two subjects (2 and 5) that there is no difference due to insertion of the navigator in the whole brain histograms (WBHs) of FA for the at rest (NoMo) scans acquired with the basic diffusion sequence and with the navigated sequence without prospective motion correction (vNav_NoCo). The bottom row (Figure 4 C,D) shows the motion parameters that were estimated in ICE by PACE during the NoMo_vNav_NoCo scans. These data show that even when a subject has been asked to lie as still as possible, there may still be some small residual motion. These motions were, however, very small and well below the limits that have been found to cause low signal and dropout slices.
Figure 4.
Normalized whole brain histograms (WBHs) of FA for two subjects (2 and 5) for the at rest (NoMo) scans acquired with the basic diffusion sequence and with the navigated sequence without prospective motion correction (vNav_NoCo). The plots in the bottom row are the corresponding motion parameters that were estimated in ICE by PACE during the NoMo_vNav_NoCo scans.
Subject Motion, Motion Correction and Reacquisition
In order to confirm that we are comparing diffusion data acquired under similar conditions of motion, we compare in figure 5 the motion parameter estimates for the first subject generated by PACE and retrospective motion correction. Figures 5A and 5D show the motion parameters that were estimated in ICE by PACE for the Mo_vNav_NoCo and Mo_vNav_all scans, respectively. Figures 5B and 5E show the retrospective motion estimates for the navigated sequence without prospective motion correction (Mo_vNav_NoCo) using SPM and FLIRT, respectively, and same for the basic sequence in figures 5C and 5F. In figure 5D, since reacquisition was enabled, the number of measurements was 39, compared to 34 in all the other acquisitions. These plots serve to demonstrate similar conditions of motion between the Mo_vNav_all scan and the Mo_vNav_NoCo scan, due to similar PACE motion estimates. Similarly, SPM retrospective motion estimates demonstrate similar motion between the Mo_vNav_NoCo scan and the Mo_basic scan. FLIRT retrospective motion correction is very sensitive to the choice of cost function and number of degrees of freedom and consistently overestimates the motion parameters.
Figure 5.
Comparison of motion parameter estimates generated by PACE and retrospective (retro) motion correction for the first subject. Figures 5A and 5D show the motion parameters that were estimated in ICE by PACE for the Mo_vNav_NoCo and Mo_vNav_all scans, respectively. Figures 5B and 5E show the retrospective motion estimates for the Mo_vNav_NoCo scan using SPM and FLIRT, respectively, and same for the basic diffusion sequence in figures 5C and 5F. Mo denotes a scan with motion, vNav the navigated diffusion sequence, NoCo without prospective motion correction, and all a scan with prospective motion correction and reacquisition enabled. Subjects moved upon verbal instruction, 5 to 6 times during the scan.
Shown in figure 6 are the FA and MD maps of a single slice for the first subject for the 5 different diffusion acquisitions, as well as the results of applying retrospective motion correction in the scan acquired using the basic sequence using SPM (Mo_basic_retro_SPM) and FLIRT (Mo_basic_retro_FLIRT), respectively. Data from the Mo_vNav_all scan have been analysed both without (Mo_vNav_noReAq) and with (Mo_vNav_ReAq) reacquisition applied. Figure 7 presents a comparison of the WBH-FA and the WBH-MD for the different acquisitions for the same subject. This figure clearly demonstrates how motion without prospective motion correction in the Mo_basic and Mo_vNav_NoCo acquisitions, even after applying retrospective motion correction (Mo_basic_retro, Mo_vNav_NoCo_retro), changes the distribution of the FA and the MD compared to both the at rest scans, namely the baseline scan (NoMo_basic) and the prospectively corrected navigated (NoMo_vNav_Co) scan. The distribution of the FA and the MD values are almost fully recovered when using the navigated prospective motion corrected sequence (Mo_vNav_noReAq and Mo_vNav_ReAq).
Figure 6.
FA and MD maps of slice 80 for the first subject for 5 different acquisitions, as well as results of retrospective motion correction in the scan acquired using the basic sequence using SPM (Mo_basic_retro_SPM) and FLIRT (Mo_basic_retro_FLIRT), respectively. Data acquired in the Mo_vNav_all scan have been analysed both without (Mo_vNav_noReAq) and with (Mo_vNav_ReAq) reacquisition. The two yellow circles on the FA maps demonstrate reduced blurring in the scan with reacquisition compared to the scan without reacquisition. All the maps are co-registered to the T1 space.
Figure 7.
Comparison of the normalized WBH-FA (A–D) and WBH-MD (E–H) for the first subject for different scans. (A,E) comparison of at rest scans acquired using both the basic and navigated sequence to scans with motion and no prospective motion correction acquired using both the basic and navigated sequence; (B,F) effect of retrospective motion correction using SPM on the scans acquired without prospective motion correction; (C,G) effect of retrospective motion correction using FLIRT on the scans acquired without prospective motion correction; and (D,H) prospective motion corrected scans acquired using the navigated sequence, both without and with reacquisition.
Tables 1 and 2 give the mean of the histogram parameters for all six subjects for FA and MD, respectively, for the different acquisitions, as well as the values obtained after applying retrospective motion correction. WBH parameters for each scan were compared with those of the at rest baseline (NoMo_basic) scan using a paired student’s t-test. The mean FA and the mean MD over the whole brain volume does not differ significantly from the baseline values for scans acquired at rest using the navigated sequence without prospective correction (NoMo_vNav_NoCo), paired Student’s t-test, p = 0.73 and p = 0.64, respectively. This result confirms that insertion of the navigator into the sequence does not corrupt the diffusion data.
Table 1.
Comparison of the mean of the WBH-FA parameters for all subjects for the different scans.
| WBH-FA Parameters |
NoMo_ basic |
NoMo_vNav _NoCo |
Mo_ basic |
Mo_basic _retro |
Mo_vNav _noReAq |
Mo_vNav _ReAq |
|
|---|---|---|---|---|---|---|---|
| SPM | FLIRT | ||||||
| Mean FA (SD) |
0.52 (0.02) |
0.52 (0.01) |
0.48* (0.02) |
0.41* (0.02) |
0.41* (0.02) |
0.51 (0.02) |
0.52 (0.01) |
| Peak Location (SD) |
0.50 (0.01) |
0.51 (0.01) |
0.31* (0.03) |
0.23* (0.03) |
0.2* (0.1) |
0.5 (0.1) |
0.5 (0.1) |
| Skewness (SD) |
0.3 (0.2) |
0.2 (0.2) |
0.6 (0.1) |
0.6 (0.1) |
0.7 (0.1) |
0.3 (0.2) |
0.3 (0.2) |
| Kurtosis (SD) |
1.4 (0.1) |
1.4 (0.1) |
1.6 (0.2) |
1.6* (0.1) |
1.9* (0.1) |
1.3 (0.1) |
1.4 (0.1) |
p<0.01 paired student’s t-test.
Table 2.
Comparison of the mean of the WBH-MD parameters for all subjects for the different scans
| WBH-MD Parameters |
NoMo_ basic |
NoMo_vNav _NoCo |
Mo_ basic |
Mo_basic _retro |
Mo_vNav _noReAq |
Mo_vNav _ReAq |
|
|---|---|---|---|---|---|---|---|
| SPM | FLIRT | ||||||
| Mean MD /10−3 mm2s−1 (SD) |
0.84 (0.02) |
0.83 (0.01) |
0.87* (0.02) |
0.78* (0.03) |
0.88* (0.03) |
0.85 (0.02) |
0.84 (0.01) |
| Peak Location /10−3 mm2s−1 (SD) |
0.73 (0.02) |
0.75 (0.02) |
0.74 (0.03) |
0.73 (0.03) |
0.73 (0.01) |
0.75 (0.04) |
0.74 (0.02) |
| Skewness (SD) |
3.3 (0.1) |
3.4 (0.2) |
2.8 (0.4) |
2.7 (0.5) |
2.8 (0.4) |
3.4 (0.1) |
3.5 (0.1) |
| Kurtosis (SD) |
14 (1) |
14 (1) |
10 (2) |
10 (3) |
11 (3) |
14 (1) |
15 (1) |
p<0.01 paired student’s t-test.
There are no significant differences in any of the WBH-FA (Table 1) and WBH-MD (Table 2) parameters between any of the navigated prospectively motion corrected sequences (Mo_vNav_noReAq and Mo_vNav_ReAq) and the at rest baseline scan (NoMo_basic). By contrast, mean FA and peak location differ significantly (p<0.01) for acquisitions with the basic sequence in the presence of motion, even after retrospective motion correction using SPM (Mo_basic_retro_SPM) and FLIRT (Mo_basic_retro_FLIRT). Although the FA histogram skewness and kurtosis are not altered significantly by motion in the basic sequence, the mean kurtosis is significantly higher after performing retrospective motion correction using both SPM and FLIRT. Mean MD differs significantly (p<0.01) from the at rest baseline value for data acquired with the basic sequence in the presence of motion (Mo_basic), even after retrospective motion correction with FLIRT and SPM. Although not significant, motion reduces the MD histogram skewness and kurtosis in the basic sequence, even after retrospective motion correction.
In order to investigate whether elimination of motion corrupted volumes in the basic acquisition improves retrospective motion correction, we eliminated for three of the subjects (2, 4, and 6) volumes that have a low signal due to motion. Figure 8 shows the WBH-FA for the three subjects for retrospective motion corrected data before elimination (BE) and after elimination (AE) of motion corrupted volumes. Shown also are the at rest baseline scans (NoMo_basic). Elimination of the corrupted volumes does not improve the diffusion data; there is still a significant reduction in the mean FA and in the peak location. These results demonstrate that retrospective motion correction completely fails to recover the diffusion data without prospective motion correction, even when the corrupted diffusion volumes with low signal are eliminated.
Figure 8.
Comparison of the normalized WBH-FA in three subjects for an at rest baseline scan (NoMo_basic) compared to a scan with motion and retrospective motion correction where the suffices BE and AE, respectively, denote before and after elimination of corrupted volumes that have low signal due to motion.
Subject 6 was very restless with motion that caused PACE to terminate in the Mo_vNav_all acquisition. This acquisition was repeated but with only three reacquisitions in order to shorten the scan time. The repeated Mo_vNav_all acquisition had 11 corrupted noisy volumes, only 3 of which were reacquired. With this data we could explore the improvement offered by elimination of motion corrupted volumes in prospectively corrected data. While significantly better than scans acquired without prospective motion correction (Figure 9A), the prospectively corrected navigated scan in this case does not fully recover the diffusion data (Figure 9B), even with reacquisition, due to the presence of uncorrected corrupted volumes. The WBH-FA was recalculated for this subject after elimination of the noisy corrupted diffusion volumes for both the Mo_vNav_noReAq (11 corrupted volumes) and the Mo_vNav_ReAq (8 corrupted volumes) data, respectively. The results presented in Figure 9C show that both without and with reacquisition the diffusion data improved after elimination of the corrupted volumes, but that recovery of the FA peak location was slightly better for the data with reacquisition than without. The FA peak location for both Mo_vNav_noReAq and Mo_vNav_ReAq before elimination was 0.2; after elimination it was 0.3 and 0.5 for Mo_vNav_noReAq and Mo_vNav_ReAq, respectively.
Figure 9.
(A) The effect of motion on the normalized WBH-FA of the basic diffusion sequence (both before and after retrospective motion correction with FLIRT and SPM) for a particularly restless subject, (B) normalized WBH-FA of the navigated sequence with prospective motion correction, both without and with reacquisition, for this data with many uncorrected corrupted volumes, (C) normalized WBH-FA of the navigated sequence after elimination (AE) of uncorrected corrupted volumes.
Discussion
The quantitative application of DTI in clinical or research areas relies on computation of FA and mean diffusivity (MD). The whole brain histogram (WBH) of FA and MD has been widely used to study different clinical diseases (25, 26, 27–28). Subject motion influences the diffusion maps in a way that can confound the interpretation of DTI results. The accurate calculation of the diffusion tensor and the diffusion maps requires that the diffusion weighted volumes for different diffusion directions be co-aligned. We have presented a method using a 3D-EPI navigator to track and correct in real time changes in position that result from rigid body motion.
The only increase in scan time is due to the navigator (406 ms) and waiting period for feedback (120 ms), both of which occur once per TR, increasing the TR from 9500 ms to 10026 ms. While intra-volume motion in self-navigation techniques (11,16) renders the 3D volume used for co-registration unusable, the advantage of such a short additional navigator (406 ms) is that motion at any time during the TR is unlikely to affect the navigator, and correction is applied immediately after the navigator. Only when the motion exceeds a pre-defined threshold will the motion corrupted volume be reacquired adding an additional TR. The total number of reacquisitions are, however, limited and is specified when preparing the diffusion protocol. Therefore the total acquisition time of the diffusion sequence is also limited to the value given by Equation (1).
The Influence of the Navigator on the Diffusion Sequence
Using both a water phantom and at rest scans in six subjects, we have confirmed that the insertion of the 3D-EPI navigator has negligible effect on the diffusion data. Figure 2 demonstrates that there are no artifacts or distortions in the mean diffusivity map of the water phantom that is acquired using the navigated prospective motion corrected diffusion sequence compared to the standard diffusion sequence. The results are the same for the other slices. Furthermore, the histograms of the averaged MD for the three scans over the whole volume of the water phantom for the navigated sequence and the basic sequence are similar, with no differences in peak location, skewness or kurtosis (Fig. 3). These results also confirm that there are no residual errors in the motion estimates derived by PACE which could corrupt the diffusion data.
For the in vivo data, the WBH-FA is generated from images containing gray and white matter, giving rise to 2 distinct peaks, while the WBH-MD shows a single normal distribution due to the fact that normal gray and white matter have very similar MD values. The plots in figure 4 show that the insertion of the navigator into the diffusion sequence does not shift or change the properties of the FA or MD histograms for in vivo data acquired at rest. The results are similar for the other subjects. The average of the mean FA and the MD for all six subjects do not differ significantly between the baseline scan and the scan acquired at rest using the navigated sequence without prospective correction (Tables 1 and 2). The excellent agreement between the WBH-FA and WBH-MD shown in figure 7 for a single subject for the NoMo_basic and NoMo_vNav_Co acquisitions, provides further evidence for the fact that there are no residual errors in the motion estimates derived by PACE that could introduce errors in the diffusion data.
Subject Motion, Correction and Reacquisition
In the current studies there was no way to monitor the head pose inside the scanner when using the basic diffusion sequence. Even in stationary scans where subjects are instructed to remain still, they may move unexpectedly with resulting changes in the diffusion data. The navigated diffusion sequence generates the 6 parameter motion estimates that reflect the motion between every two successive diffusion volumes (Fig. 5). Retrospective motion correction with FLIRT yielded different motion estimates compared to PACE. In the current study, FLIRT was implemented using a least squares cost function with 6 degrees of freedom. FLIRT showed overestimation of motion parameters (Fig. 5E,F), while SPM motion estimates showed good agreement with PACE. Using a mutual information-based cost function and a higher number of degrees of freedom did not improve the accuracy of the FLIRT motion estimates. Using the SPM and PACE motion estimates, we could confirm that we are comparing diffusion data acquired under similar conditions of motion.
The degradation of the FA and MD maps in the presence of motion are apparent in the Mo_basic and Mo_vNav_NoCo data shown in figure 6. The motion adds blurring and edge artifacts, rendering the finer details of the maps inaccurate. Retrospective motion correction does reduce blurring marginally, but does not recover all the finer details. These artifacts in the FA and the MD maps are eliminated substantially with the navigated diffusion sequences Mo_vNav_noReAq and Mo_vNav_ReAq.
Motion causes a significant reduction in the mean FA of the whole brain volume for all six subjects (Table 1). Retrospective motion correction does not improve the mean FA, but rather causes a further significant reduction in the mean FA for both retrospective motion correction methods (Mo_basic_retro_FLIRT and Mo_basic_retro_SPM). The mean FA is recovered substantially for the navigated prospective motion corrected acquisitions Mo_vNav_noReAq and Mo_vNav_ReAq (Table 1). In contrast, motion causes a significant increase in the mean MD of the whole brain volume for the six subjects. Retrospective motion correction with FLIRT yields no improvement in the mean MD. The mean MD is recovered substantially for the navigated scans Mo_vNav_noReAq and Mo_vNav_ReAq (Table 2).
In the presence of motion the WBH-FA for all subjects shows changes in FA toward lower anisotropy and a shift in the location of the high anisotropy peak (Fig. 7A). This decrease of anisotropy in the whole brain changes the unique shape of the WBH-FA with an increase in both the kurtosis (the curve becomes more peaked) as well as the skewness (the curve becomes more asymmetric) (Table 1). In Figure 7A, the double bell-shaped curve of the WBH-FA becomes distorted due to motion and the distribution moves towards a single bell-shaped curve due to a reduction in the anisotropy of the white matter and a shift towards higher diffusivity. In contrast, with subject motion the WBH-MD changes toward more diffusivity with a decrease in kurtosis (the curve becomes less peaked) as well as in skewness (the curve becomes less asymmetric) (Table 2, Fig. 7E). Retrospective motion correction does not recover the distinct peaks of the WBH-FA and moreover it causes a big increase in the height of the lower FA peak in the case of FLIRT (Fig. 7C) or a reduction in the lower FA peak in the case of SPM (Fig. 7B). Even eliminating the corrupted uncorrected volumes in the Mo_basic scan, does not improve the retrospective motion correction (Fig. 8). The navigated prospective motion corrected acquisition (Mo_vNav_noReAq and Mo_vNav_ReAq) recovers the properties of both the WBH-FA and the WBH-MD.
The incremental improvement of reacquisition on the diffusion data is slight, especially if there are no uncorrected diffusion volumes with low signal due to motion. The Mo_vNav_all scan for subject 6 had many corrupted noisy volumes, only 3 of which were reacquired. While significantly better than scans acquired without prospective motion correction (Figure 9A), the prospectively corrected navigated scan does not fully recover the diffusion data in this case (Figure 9B), even with reacquisition, due to the presence of uncorrected corrupted volumes. Elimination of the corrupted diffusion volumes in both the Mo_vNav_noReAq and the Mo_vNav_ReAq acquisitions, almost fully recovers the diffusion data, but with better recovery of the FA peak location for the data with reacquisition than without.
Limitations of the 3D-EPI Navigator
Due to the long acquisition time (TR = 9500 ms) of the diffusion volume in the current protocol, real-time motion tracking by the 3D-EPI navigator once per diffusion volume might not be sufficient to detect brief fast intra-volume movements if the subject returns to their original position. Acquiring additional 3D-EPI navigators with a smaller acquisition matrix size (e.g. by partial Fourier) in between certain diffusion slices of the same volume, and not only at the end of the diffusion volume, could improve the sensitivity of the navigator. Another option would be to combine the current method with the FID navigator method proposed by Kober et al. (2011), since their volume-wise evaluation of the FID navigator is sensitive to motion throughout the volume and requires that the head position be different to the preceding repetition in at least five slices.
It is not ideal in the current implementation of the sequence that unused reacquisitions need to be acquired at the end of the scan in order to keep the scan time correct. It would be better if the sequence could terminate the scan after all the diffusion volumes have been acquired, even if there are remaining unused reacquisitions.
Finally, the allowable range of motion that can be corrected prospectively using this technique is limited by the Siemens implementation of PACE, which terminates for translations in any direction greater than 20 mm and rotations in any direction greater than 8 degrees. If the motion exceeds these limits, PACE will terminate and subsequent diffusion volumes will not be prospectively motion corrected. The navigator images will, however, still be acquired enabling offline estimation of motion parameters and retrospective motion correction.
Conclusion
In this paper we present a volumetric navigated DTI sequence that allows real-time tracking of head pose and adjustment in real time of the RF pulses and all the diffusion gradients to correct for changes in head pose. If enabled, the system also reacquires volumes during which excessive motion occurred. Apart from the reacquisition time, the only additional scan time is 526 ms per TR, which is minimal in view of the long diffusion TR of 9500 ms that is widely used in DTI protocols.
Acknowledgements
The South African Research Chairs Initiative of the Department of Science and Technology and National Research Foundation of South Africa, Medical Research Council of South Africa, NIH grants R21AA017410, R21EB008547, R21MH096559, R01HD071664, R01NS05574, P41RR014075, the Ellison Medical Foundation, and the University of Cape Town.
References
- 1.Basser PJ, Pierpaoli C. Microstructural and physiological features of tissues elucidated by quantitative-diffusion-tensor MRI. J Magn Reson B. 1996;111(3):209–219. doi: 10.1006/jmrb.1996.0086. [DOI] [PubMed] [Google Scholar]
- 2.Hasan KM, Iftikhar A, Kamali A, Kramer LA, Ashtari M, Cirino PT, Papanicolaou AC, Fletcher JM, Ewing-Cobbs L. Development and aging of the healthy human brain uncinate fasciculus across the lifespan using diffusion tensor tractography. Brain Research. 2009;1276:67–76. doi: 10.1016/j.brainres.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snook L, Paulson LA, Roy D, Phillips L, Beaulieu C. Diffusion tensor imaging of neurodevelopment in children and young adults. NeuroImage. 2005;26(4):1164–1173. doi: 10.1016/j.neuroimage.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Chung S, Pelletier D, Sdika M, Lu Y, Berman JI, Henry RG. Whole brain voxel-wise analysis of single-subject serial DTI by permutation testing. NeuroImage. 2008;39(4):1693–1705. doi: 10.1016/j.neuroimage.2007.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartzman A, Dougherty RF, Taylor JE. Cross-subject comparison of principal diffusion direction maps. Magn Reson Med. 2005;53(6):1423–1431. doi: 10.1002/mrm.20503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aksoy M, Liu C, Moseley ME, Bammer R. Single-step nonlinear diffusion tensor estimation in the presence of microscopic and macroscopic motion. Magn Reson Med. 2008;59(5):1138–1150. doi: 10.1002/mrm.21558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong R, Cox RW. Rotation of NMR images using the 2D chirp-z transform. Magn Reson Med. 1999;41(2):253–256. doi: 10.1002/(sici)1522-2594(199902)41:2<253::aid-mrm7>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 8.Muresan L, Renken R, Roerdink JB, Duifhuis H. Automated correction of spin-history related motion artifacts in fMRI : simulated and phantom data. IEEE Trans Biomed Eng. 2005;52(8):1450–1460. doi: 10.1109/TBME.2005.851484. [DOI] [PubMed] [Google Scholar]
- 9.Rohde GK, Barnett AS, Basser PJ, Marenco S, Pierpaoli C. Comprehensive approach for correction of motion and distortion in diffusion-weighted MRI. Magn Reson Med. 2004;51(1):103–114. doi: 10.1002/mrm.10677. [DOI] [PubMed] [Google Scholar]
- 10.Aksoy M, Forman C, Straka M, Skare S, Holdsworth S, Hornegger J, Bammer R. Real-time optical motion correction for diffusion tensor imaging. Magn Reson Med. 2011;66(2):366–378. doi: 10.1002/mrm.22787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benner T, van der Kouwe AJW, Sorensen AG. Diffusion imaging with prospective motion correction and reacquisition. Magn Reson Med. 2011;66(1):154–167. doi: 10.1002/mrm.22837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thesen S, Heid O, Mueller E, Schad LR. Prospective acquisition correction for head motion with image-based tracking for real-time fMRI. Magn Reson Med. 2000;44(3):457–465. doi: 10.1002/1522-2594(200009)44:3<457::aid-mrm17>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 13.Assaf Y, Cohen Y. Structural information in neuronal tissue as revealed by q-space diffusion NMR spectroscopy of metabolites in bovine optic nerve. NMR Biomed. 1999;12(6):335–344. doi: 10.1002/(sici)1099-1492(199910)12:6<335::aid-nbm581>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 14.Tuch DS, Reese TG, Wiegell MR, Wedeen VJ. Diffusion MRI of complex neural architecture. Neuron. 2003;40(5):885–895. doi: 10.1016/s0896-6273(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 15.Wedeen VJ, Hagmann P, Tseng W-YI, Reese TG, Weisskoff RM. Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn Reson Med. 2005;54(6):1377–1386. doi: 10.1002/mrm.20642. [DOI] [PubMed] [Google Scholar]
- 16.Kober T, Gruetter R, Krueger G. NeuroImage Prospective and retrospective motion correction in diffusion magnetic resonance imaging of the human brain. NeuroImage. 2012;59(1):389–398. doi: 10.1016/j.neuroimage.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Fu ZW, Wang Y, Grimm RC, Rossman PJ, Felmlee JP, Riederer SJ, Ehman RL. Orbital navigator echoes for motion measurements in magnetic resonance imaging. Magn Reson Med. 1995;34(5):746–753. doi: 10.1002/mrm.1910340514. [DOI] [PubMed] [Google Scholar]
- 18.Ward HA, Riederer SJ, Grimm RC, Ehman RL, Felmlee JP, Jack CR., Jr Prospective multiaxial motion correction for fMRI. Magn Reson Med. 2000;43(3):459–469. doi: 10.1002/(sici)1522-2594(200003)43:3<459::aid-mrm19>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 19.Welch EB, Manduca A, Grimm RC, Ward HA, Jack CR., Jr Spherical Navigator Echoes for Full 3D Rigid Body Motion Measurement in MRI. Magn Reson Med. 2002;47(1):32–41. doi: 10.1002/mrm.10012. [DOI] [PubMed] [Google Scholar]
- 20.Kouwe JWVD, Benner T, Dale AM. Real-Time Rigid Body Motion Correction and Shimming Using Cloverleaf Navigators. Magn Reson Med. 2006;56(5):1019–1032. doi: 10.1002/mrm.21038. [DOI] [PubMed] [Google Scholar]
- 21.White N, Roddey C, Shankaranarayanan A, Han E, Rettmann D, Santos J, Kuperman J, Dale A. Real-time prospective motion correction in MRI using image-based tracking. Magn Reson Med. 2010;63(1):91–105. doi: 10.1002/mrm.22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hess AT, Tisdall MD, Andronesi OC, Meintjes EM, van der Kouwe AJW. Real-time motion and B(0) corrected single voxel spectroscopy using volumetric navigators. Magn Reson Med. 2011;66(2):314–323. doi: 10.1002/mrm.22805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reese TG, Heid O, Weisskoff RM, Wedeen VJ. Reduction of eddy-current-induced distortion in diffusion MRI using a twice-refocused spin echo. Magn Reson Med. 2003;49(1):177–182. doi: 10.1002/mrm.10308. [DOI] [PubMed] [Google Scholar]
- 24.Wang R, Benner T, Sorensen AG, Wedeen VJ. Proc Int Soc Magn Reson Med. 2007;15:3720. Available at: http://trackvis.org/ [Google Scholar]
- 25.Bozzali M, Franceschi M, Falini A, Pontesilli S, Cercignani M, Magnani G, Scotti G, Comi G, Filippi M. Quantification of tissue damage in AD using diffusion tensor and magnetization transfer MRI. Neurology. 2001;57(6):1135–1137. doi: 10.1212/wnl.57.6.1135. [DOI] [PubMed] [Google Scholar]
- 26.Della Nave R, Foresti S, Pratesi A, Ginestroni A, Inzitari M, Salvadori E, Giannelli M, Diciotti S, Inzitari D, Mascalchi M. Whole-brain histogram and voxel-based analyses of diffusion tensor imaging in patients with leukoaraiosis: correlation with motor. AJNR Am J Neuroradiol. 2007;28(7):1313–1319. doi: 10.3174/ajnr.A0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mori N, Miki Y, Fushimi Y, Kikuta K, Urayama S, Okada T, Fukuyama H, Hashimoto N, Togashi K. Cerebral infarction associated with moyamoya disease: histogram-based quantitative analysis of diffusion tensor imaging -- a preliminary study. Magn Reson Imaging. 2008;26(6):835–840. doi: 10.1016/j.mri.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Y, Lin FC, Zhu J, Zhuang ZG, Li YS, Tao J, Qian LJ, Xu JR, Lei H. Whole brain diffusion tensor imaging histogram analysis in vascular cognitive impairment. J Neurol Sci. 2008;268(1–2):60–64. doi: 10.1016/j.jns.2007.11.005. [DOI] [PubMed] [Google Scholar]