Abstract
UK NHS diagnostic service sequence analysis of genes generally examines and reports on variations within a designated region 5′ and 3′ of each exon, typically 30 bp up and downstream. However, because of the degenerate nature of the splice sites, intronic variants outside the AG and GT dinucleotides of the acceptor and donor splice sites (ASS and DSS) are most often classified as being of unknown clinical significance, unless there is some functional evidence of their pathogenicity. It is now becoming clear that mutations deep within introns can also interfere with normal processing of pre-mRNA and result in pathogenic effects on the mature transcript. In diagnostic laboratories, these deep intronic variants most often fall outside of the regions analysed and so are rarely reported. With the likelihood that next generation sequencing will identify more of these unclassified variants, it will become important to perform additional studies to determine the pathogenicity of such sequence anomalies. Here, we analyse variants detected in either COL2A1 or COL11A1 in patients with Stickler syndrome. These have been analysed both in silico and functionally using either RNA isolated from the patient's cells or, more commonly, minigenes as splicing reporters. We show that deep intronic mutations are not a rare occurrence, including one variant that results in multiple transcripts, where both de novo donor and ASS are created by the mutation. Another variant produces transcripts that result in either haploinsufficiency or a dominant negative effect, potentially modifying the disease phenotype.
Keywords: Stickler syndrome, splicing, mutations
Introduction
The arrangement of genes into coding and non-coding regions or exons and introns has evolutionary advantageous effects. First, genomic rearrangements can result in novel genes, creating new products that can potentially benefit an organism via natural selection.1, 2 In addition, alternative splicing of mRNA, whereby selected exons can be expressed or repressed in different tissues, increases the diversity of the proteome without the need to increase the number of genes.3, 4 However, pre-mRNA processing needs to be accurately regulated, and one consequence of this is that missplicing is a major cause in inherited genetic disorders.5, 6
In the clinical setting, the degenerate nature of the splicing regulatory signals results in uncertainty as to whether some sequence variants that may affect mRNA processing are pathogenic. Consequently, within the UK NHS diagnostic laboratories only those mutations that alter the highly conserved AG and GT dinucleotides of the acceptor and donor splice sites (ASS and DSS) will be officially reported as pathogenic, unless there is additional functional evidence that the variant alters normal splicing.
Types 1 and 2 Stickler syndrome (MIM nos. 108300 604841) are dominantly inherited disorders due to mutations in the genes for collagen expressed in cartilage and vitreous, namely COL2A1 and COL11A1.7, 8 Both are multi-exon genes with a large number of characterised mutations, many of which affect consensus splice sites.9, 10, 11, 12
Stickler syndrome can present with high myopia, midline clefting, midfacial hypoplasia, hearing loss, premature osteoarthritis and a high incidence of retinal detachment.13 With an incidence of between 1 in 7500–1 in 9000 newborns,14 it is phenotypically highly variable both between and within families, and patients often remain undiagnosed for many years, until the suggestion of the condition is raised clinically and later confirmed by genetic testing. Type 1 Stickler syndrome due to mutations in COL2A1 is usually but not exclusively due to haploinsufficiency.11, 12 Because most of the exons in this gene consist of complete codons, it is presumed that many of the splice site mutations in type 1 Stickler syndrome result in the use of cryptic splice sites that introduce premature termination codons into the COL2A1 mRNA, but this has been demonstrated in only a small number of cases.10, 15 We have also previously described one example of a deep intronic mutation c.1527+135G>A that creates a de novo ASS in intron 23, which inserts additional mRNA sequence and alters the reading frame.10 In contrast, exon skipping in COL2A1 typically leaves the message in frame leading to dominant negative transcripts that result in more severe phenotypes, usually Kniest dysplasia16 (MIM #156550), but also Spondyloepiphyseal dysplasia congenita, SEDC (MIM # 183900) and achondrogenesis type 2 (MIM # 200610). However, it is possible that some of the phenotypic variation seen in type 1 Stickler syndrome results from a mixture of misspliced transcripts, resulting from a single mutation that can lead either to nonsense-mediated decay or alternatively a dominant negative effect via exon skipping.17 In type 2 Stickler syndrome, which is due to mutations in COL11A1, dominant negative mutations are the norm, most commonly manifesting as exon skipping.9, 11 Here, we demonstrate that deep intronic mutations in COL2A1 are not a rare occurrence and use both in silico analysis and functional studies to characterise their effect on pre-mRNA processing, along with other splice site mutations outside of the highly conserved AG-GT dinucleotides.
Materials and methods
Gene sequencing
One patient was previously screened with the long range PCR multi-exon approach.10 However, for the majority of patients, all of the exons of either COL2A1 or COL11A1 were sequenced as previously described11 by the East Anglian Regional Molecular Genetics Laboratory, Addenbrooke's Hospital, Cambridge UK, using the high throughput approach of amplifying multiple small products, containing only 1–3 exons, tagged to allow sequencing of all products with an M13-derived primer. Patient DNA sequences were compared with a reference sequence (COL2A1, NG_008072; COL11A1, NG_008033) using Mutation Surveyor (Soft Genetics, State College, PA, USA). Each exon was analysed within a defined region of interest that included 30 bp either side of each exon, except for exon 23 that had a region of interest that extended into intron 23 to include the previously characterised c.1527+135G>A mutation.10 When no mutation was detected within the regions of interest, additional intron sequences were inspected for novel base changes. This was limited to the intronic sequences that occurred within the amplicons designed for the high throughput system, and therefore was not able to examine most introns in any great depth. The current dbSNP database (http://www.ncbi.nlm.nih.gov/snp/) was utilised to determine whether unusual variants detected in patient sequences were merely rare polymorphisms. Those variants that were present in dbSNP at a frequency >1% were not analysed further, given the incidence of Stickler syndrome in the population and the individual nature of mutations in most families. In contrast, the variants tested here were unique. The present version of dbSNP is build 135.
In silico analysis
Sequence variations were analysed using Alamut version 2.0. (Interactive Biosoftware, Rouen, France), a software package that in addition to protein sequence analysis, utilises different splice site prediction programmes to compare the normal with variant sequences for differences in potential regulatory signals, including donor, ASS and branch splice sites, as well as differences in potential exon splicing enhancers. The splicing prediction tools within Alamut that are freely available as separate stand alone utilities include MaxEntScan (http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html), NNsplice (http://www.fruitfly.org/seq_tools/splice.html), human splicing finder (http://www.umd.be/HSF/), ESE finder (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home) and Rescue-ESE (http://genes.mit.edu/burgelab/rescue-ese/).
RNA analysis
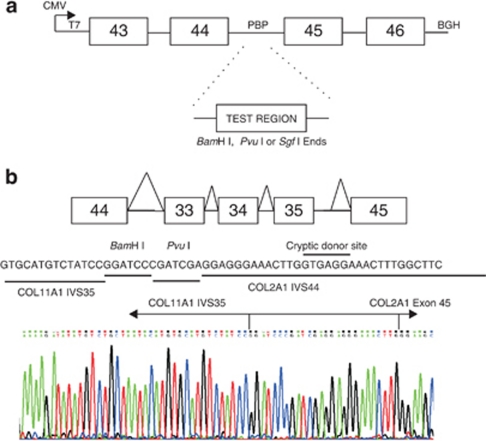
Effects on RNA splicing were analysed either by RT-PCR using RNA from a patient's cultured dermal fibroblasts, essentially as previously described,18 using superscript II (Invitrogen, Paisley, UK). More commonly, minigene constructs19 were used to transfect an immortalised Muller cell line, MIO-M1,20 followed by RT-PCR, as previously described.21 For minigene analysis sequence variants were cloned in their natural context, with flanking exons, into the expression vector pcDNA3.1 myc-His(−) A (Invitrogen). Alternatively, a universal splicing reporter (USR13) was used that consisted of pcDNA3.1 myc-His(−) A, including exons 43–46 of the COL2A1 gene with an engineered BamHI/PvuI cloning site inserted in intron 44 (Figure 1a). This hybrid minigene system was used when the gene structure was too large to allow the test exon/variant to be cloned with its natural flanking exons. A vector-specific primer (5′-GTCTCCAGAAGGACCAGGAGG-3′) complementary to the bovine growth hormone polyadenylation site was used for reverse transcription, and the T7 sequence (5′-TAATACGACTCACTATAGGGAGACC-3′) was used as a primer for PCR along with a minigene-specific primer. Sequences of primers used for minigene construction are available on request.
Figure 1.
The universal splicing reporter (USR13). The COL2A1 gene region consisting of exons 43–46 was cloned into the vector pcDNA3.1A (a) in two separate sections using primers with additional sequences for BamH I and Pvu I at their 5′ ends (PBP). This created a site in intron 44 into which test exons could be ligated using either BamH I, Pvu I or Sgf I restriction sites incorporated into the amplified product. The positions of the vector sequences for the CMV promoter, T7 promotor and bovine growth hormone (BGH) polyadenylation site are indicated. The COL11A1 IVS35 variant c. 2755+5G>A was analysed by inserting exons 33, 34 and 35 into this reporter (b). The variant was misspliced with a cryptic donor splice site in the hybrid construct used instead of the altered donor splice site in COL11A1 intron 35. Resulting in intron 35, the cloning site and part of intron 44 spliced to COL2A1 exon 45. The sequence of the DNA construct is shown as text and the cDNA as a chromatogram.
Products obtained from the c.1527+104T>G minigene were also sequenced with primers that specifically bound to cDNAs that included either the 5′- (5′-CCCCTGGAGAAAGAGTTCCGT-3′) or the 3′- end of intron 23 (5′-GACTATTTCATGTCAGTCTGGTGG-3′).
Results
In silico analysis indicated that the majority of sequence variants were predicted to either create or disrupt ASS and DSS (Table 1). Two of the variants were predicted to merely alter possible binding sites for splicing factors. Typical outputs from Alamut are illustrated in Supplementary Figure 1.
Table 1. In silico prediction.
| Variant | Alamut prediction | Functional result |
|---|---|---|
| COL2A1 | ||
| Intron 11 c.762+3G>C | Weakens normal DSS | Mixture of normal splicing and exon skipping (minigene) |
| Intron 12 c.817-9G>A | Disrupts normal ASS, possible de novo ASS | Mixture of insertion of intronic sequence and exon skipping (minigene) |
| Intron 19 c.1222-98A>G | Disrupts SC35 site | Normal splicing (minigene) |
| Intron 23 c.1527+104T>G | Possible de novo DSS and ASS | Multiple misspliced products (minigene) |
| Intron 35 c.2356-62C>A | Possible de novo ASS | Normal splicing (minigene) |
| Intron 48 c.3435+18C>A | Substitutes SRp40 site for SRp55 | Normal splicing (minigene) |
| Intron 48 c.3435+79A>T | Possible de novo DSS | Insertion of intronic sequence (minigene) |
| Intron 48 c.3435+83C>G | Possible de novo DSS | Insertion of intronic sequence (minigene) |
| COL11A1 | ||
| Intron 35 c.2755+5G>A | Disrupts normal DSS | Missplicing (hybrid minigene) |
| Intron 43 c.3385-6T>G | Disrupts normal ASS, possible de novo ASS | Exon skipping (illegitimate) |
Abbreviations: ASS, acceptor splice sites; DSS, donor splice sites.
Variants were analysed using the splicing function in Alamut. Predicted effects to ASS and DSS are shown, along with the functional results and the type of assay used.
COL2A1 intron 11 c.762+3G>C
This variant was identified in a patient with type 1 Stickler syndrome, with the membranous vitreous anomaly,11 as it occurred within the donor splice site it was considered highly likely to be pathogenic. No other potentially pathogenic variants were detected. Alamut predicted that the change weakened the donor splice site, but did not fully destroy it (Supplementary Figure 1A). Minigene analysis of the variant cloned within exons 9–12, demonstrated both normal splicing and skipping of exon 11 (sequence data not shown).
COL2A1 intron 12 c 817-9G>A
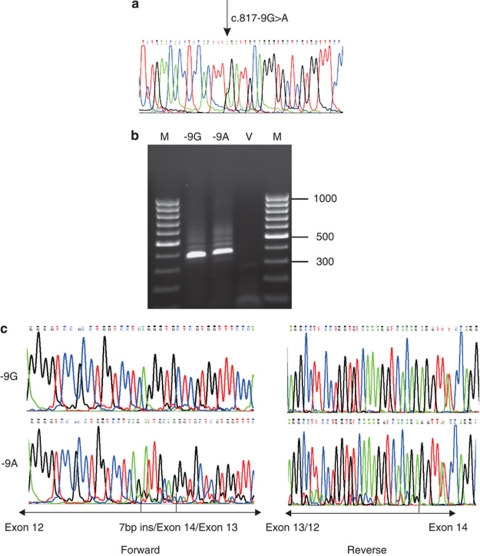
This variant was identified in a patient with type 1 Stickler syndrome. No other potentially pathogenic mutations were found in COL2A1. Alamut predicted both disruption to the normal ASS and creation of a de novo ASS in the −9 and −8 positions. Minigene analysis, consisting of exons 12–16, identified two missplicing events. The major product had 7 bp inserted into the cDNA sequence upstream of exon 13 and corresponded to utilisation of the de novo ASS (Figure 2). In addition, the sequencing chromatogram showed a minor product that corresponded to skipping of exon 13. Unlike the 7 bp insertion, the exon 13 skip removed 54 bp and left the message inframe.
Figure 2.
Minigene analysis for COL2A1 c.817-9G>A. The genomic variant c.817-9G>A (a), was expressed as a minigene in cultured cells and analysed by RT-PCR (b) and sequencing (c) in both the forward and reverse directions. This revealed a major product consisting of a 7-bp insertion 5′ of exon 13, and a minor product where exon 13 was skipped. A vector only negative control (V) and standard size (bp) markers (M) in are indicated.
COL2A1 intron 19 c 1222-98A>G and intron 35 c.2356-62C>A
These two variants were seen in the same Stickler syndrome patient with a hyploplastic vitreous phenotype.11 COL11A1 was also screened but no potential mutations were found in that gene. Alamut predictions are shown in Table 1. Both variants were analysed using minigenes (exons 19–22 and 34–37). Identical splicing profiles were seen between the normal and variant alleles, so these two changes remain as variants of unknown clinical significance (data not shown).
COL2A1 intron 23 c.1527+104T>G
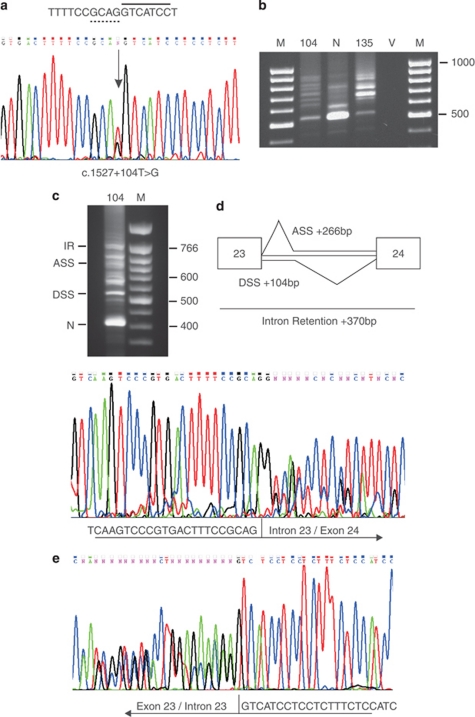
This variant was seen in a patient with Stickler syndrome. Vitreous phenotyping was impossible owing to previous surgery. The COL2A1 variant was upstream of a previously identified mutation c.1527+135G>A, deep in intron 23 that using illegitimate transcripts was demonstrated to create a de novo ASS.10 Alamut also predicted this effect for the +104T>G variant, as well as the creation of a possible de novo DSS (Table 1, Supplemetary Figure 1B). Minigene analysis using both variants, within exons 21–25, demonstrated that the +104G variant produced a more complex splicing pattern than that seen with the known positive control +135G>A mutation (Figure 3). More detailed analysis and sequencing using intron 23 specific primers (see Materials and Methods) demonstrated that in addition to the variant being utilised as both ASS and DSS there was also complete intron retention, as well as normal splicing. All three missplicing events would result in a premature termination codon being inserted into the reading frame. Intron retention was not detected in either of the normal or c.1527+135G>A minigenes, but the use of the +35G>A mutation as an ASS, previously identified in illegitimate transcripts,10 was confirmed using the minigene (sequence data not shown).
Figure 3.
Minigene analysis for COL2A1 c.1527+104T>G. The genomic sequence is shown (a) with potential de novo ASS (dashed line) and DSS (solid line), indicated as predicted by Alamut. RT-PCR products resulting from minigene analysis are shown (b) for constructs for the variant allele (104), the normal allele (N), a known pathogenic variant (135), and a vector only control (V) along with size standard markers (M). The cDNA for the 104G variant was reamplified to produce shorter products and analysed on a Nusieve 3:1 agarose (Lonza, Rockland, ME, USA) gel (c) Bands corresponding to intron retention (IR) and use of de novo acceptor and donor splice sites (ASS and DSS), as well as normally spliced mRNA (N) could be seen and are described graphically (d) Other bands were presumed to be heteroduplexes of these products, as gel purified bands produced mixed sequence. The products (shown in (c)) were sequenced in both directions using intronic-specific primers, which demonstrated that the variant sequence was utilised as both an ASS and DSS as well as resulting in intron retention (e).
COL2A1 intron 48 c.3435+18C>A; c.3435+79A>T; c.3435+83C>G
These variants were found in three different cases of Stickler syndrome.
The +18C>A change was seen in a mother and daughter, the vitreous phenotype was undetermined because of previous surgery and young age. The +79A>T change was seen in a family with variable vitreous architecture most of who had the hypoplastic phenotype but one of which had the membranous anomaly typical of COL2A1-associated Stickler syndrome. Finally, the +83C>G change was seen in a family with the hypoplastic vitreous phenotype.
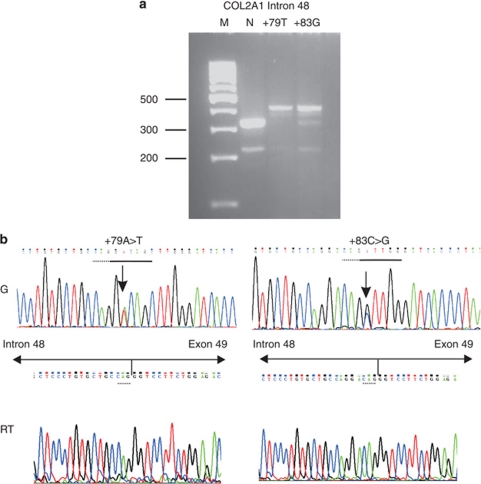
Alamut predictions are shown in Table 1. The +79 and +83 variants were predicted to create different de novo DSS close to each other. All three variants were analysed using minigenes consisting of exons 47–49. In contrast to the normal and +18C>A variant that spliced the exons normally, the +79 and +83 variants inserted part of intron 48 consistent with use of the de novo donor splice sites predicted by Alamut (Figure 4).
Figure 4.
Minigene analysis for COL2A1 c.3435+79A>T and +83C>G. RT-PCR products from minigene experiments using the normal allele (N) and two variant alleles (+79T and +83G) are shown in (a). Genomic sequences (G) and those from the RT-PCRs (RT) for each of the variants are shown in (b). The de novo DSS created by each variant are indicated in the genomic sequence by the dashed and solid lines, indicating the ‘exonic' and ‘intronic' region of the DSS. The arrows indicate the cDNA sequence corresponding to intron 48 and exon 49. Standard size markers (M) are indicated in bp.
COL11A1 intron 35 c.2755+5G>A
This mutation was previously reported by us,11 in a family with the beaded vitreous phenotype, but without any functional analysis. Because the gene structure did not allow exon 35 to be analysed within both of its neighbouring exons, (exon 36 is >3 kb downstream) it was cloned into the splicing reporter USR13 described above. Exons 33–35 of COL11A1 are also very close together. For this reason, all three COL11A1 exons 33, 34 and 35 were cloned into USR13, and analysed by RT-PCR and sequencing using a primer sited within COL11A1 exon 33. In the normal allele there was correct splicing of all three COL11A1 exons. However, in the mutant allele there was a misspliced product. Here, instead of using the mutant donor splice site in COL11A1 intron 35, a cryptic donor splice site present within COL2A1 intron 44 of USR13 was used instead (Figure 1b). Clearly this site could not be utilised in the natural mutant allele, and so we expect that the usual exon skipping-mechanism involving exon 35 would be the outcome in vivo (see discussion).
COL11A1 intron 43 c.3385-6T>G
This mutation was seen in a patient with type 2 Stickler syndrome and the beaded vitreous phenotype.11 No other potentially pathogenic mutation was seen, and Alamut predicted that it disrupted the normal ASS of exon 44 (Table 1) and created a possible de novo ASS. The effect of the variant was analysed using the patient's own cultured dermal fibroblasts. RT-PCR demonstrated skipping of exon 44. This removed 54 bp, consistent with the commonly observed mechanism of dominant negative mutations in type 2 Stickler syndrome. Although Alamut had also predicted a de novo ASS, there was no indication in the sequence chromatogram that this was utilised (sequence data not shown).
Discussion
Identification of a pathogenic mutation in a clinically affected individual allows testing and counselling of other at risk family members, without the need for expert clinical examination. This is particularly important for disorders where phenotypic variability can sometimes make identifying clinically affected individuals difficult. In type 1 Stickler syndrome this is important, as there is a high risk of retinal detachment that can occur at any age, and young children often present late to the ophthalmologist. The availability of prophylactic cryotherapy to reduce the risk of retinal detachment22 in type 1 Stickler syndrome makes the identification of at-risk-children especially important. The presence of a characteristic vitreous membrane11, 13 in Stickler syndrome is a strong indicator that the causative mutation is in COL2A1; however, in around 5% of these cases a mutation cannot be found.10, 11 The suspicion is that a large proportion of these 5% are deep intronic mutations, not easily detected by exon sequencing.
We have previously postulated that modification of the phenotype in Stickler syndrome can potentially result from variation in missplicing events that lead either to haploinsufficiency or a dominant negative effect.17 Two results here demonstrate how some mutations can produce these variable effects on pre-mRNA splicing, which can potentially affect the resulting phenotype. First, the COL2A1 Intron 11 c.762+3G>C mutation did not result in utilisation of a cryptic splice site, instead it produced a mixture of exon skipping and normal splicing. This is similar to a case of mosaicism where a child with Kniest dysplasia had a mutation that caused exon skipping, but the mosaic mother had Stickler syndrome.23 The c.762+3G>C differs in that, instead of some cells being normal, all cells contain the mutant sequence but the mutant pre-mRNA can be processed normally, therefore increasing the normal:mutant ratio and resulting in Stickler syndrome instead of Kniest dysplasia. Second, the COL2A1 intron 12 c.817-9G>A mutation results in two different types of transcript, one leading to nonsense-mediated decay and the other to a dominant negative effect. This will potentially modify the phenotype depending upon the ratio of each isoform expressed by different tissues, with the dominant negative, exon skipping, producing a more severe effect. We also noted that those mutations, which created de novo splice sites in addition to the normal splice sites, resulted in a more variable vitreous phenotype, with some family members having a hypoplastic vitreous instead of the more common membranous phenotype. This may reflect the possibility that both normal and mutant transcripts can be processed from those alleles.
Reports from diagnostic laboratories may be used during decision processes regarding clinical prophylactic or therapeutic treatment and family planning. It is therefore critical that accurate information regarding the nature of sequence variants detected during screening is reported. Because of the uncertain pathogenicity of many intronic variants, these are most likely to be classified as ‘of unknown clinical significance', even if they appear unique to the family. Two tools are now becoming more commonly utilised in the analysis of these variants. First, the availability of in silico analysis packages has allowed the diagnostic laboratories to quickly examine sequence variants for possible pathogenic effects on both the protein and RNA. However, in silico predictions may not be a true reflection of the in vivo situation. Less widely used are functional studies that assess the effect of the variants on splicing. The pros and cons of the various methods available have been discussed by Baralle et al.6 Briefly, minigenes allow expression of the variants in cell lines that may reflect more accurately, than illegitimate transcripts, the tissues in which the mRNA is naturally processed. Of the two types of minigenes, it is probable that those that clone the variant within the context of its natural surrounding exons, are more likely to reflect the true effect on splicing, than hybrid minigenes. The advantage of using cells from the patient is that they may more accurately reflect the true in vivo situation, especially if cells that express the gene are available. The minigene used here for the positive control, COL2A1 intron 23 +135 mutation replicated the effect seen using illegitimate transcripts.10 Similarly, previous studies using a minigene for a silent mutation in COL2A111 were confirmed by another laboratory12 using illegitimate transcripts, indicating that various functional strategies are valid. Here, we have used both in silico analysis and different functional studies to investigate intronic variants detected during routine gene sequencing, and identified those that affect pre-mRNA splicing in ex vivo experiments. The majority of functional assays involved minigenes, but a hybrid minigene and RNA isolated from a patient's cells were also utilised. The use of the hybrid minigene highlighted the problem with this type of construct. Although the assay demonstrated missplicing, owing to the altered donor splice site, a cryptic donor splice site within the hybrid part of the construct was used instead, whereas previous investigations,9, 11, 18 using patient-derived cultured-dermal fibroblasts, have consistently demonstrated exon skipping for this type of mutation in type 2 Stickler syndrome. COL11A1 mRNA/cDNA is easily amplified from these types of cells indicating some significant transcription of this gene under culture conditions. Exon skipping was also observed here for the COL11A1 intron 43 c.3385-6T>G mutation using mRNA from cultured cells, and exon skipping is most likely to be the result of the COL11A1 intron 35+5G>A mutation in vivo.
In COL2A1, four deep intronic mutations have now been located in just two introns, namely 23 and 48. This may in part be because of the relatively poor natural splice sites that define these introns. For instance, using the SpliceSiteFinder-like scoring system (Alamut version 2.0, Interactive Biosoftware), the ASS of intron 23 is rated at 76/100 compared with an average of 88/100 for constitutive COL2A1 exons. In addition, the donor splice site of intron 23 (AGAgttaag) is so poor compared with the consensus sequence (CAGgtaagt) that despite being the natural splice site it is only recognised by one (human splice finder) of the four different splice site prediction tools within Alamut, with a score of 64/100. Similarly, the DSS of intron 48 is rated as 74/100, in comparison, the de novo DSS sites created by the mutations in intron 48 have scores of 75/100 and 84/100. Other deep intronic mutations may go undetected, as some introns are not as comprehensively sequenced owing to their large size and the location of primers used for amplification.
In addition to the deep intronic mutations, variants in positions of −9, −6, +3 and +5 were also identified as having an effect on mRNA processing. Functional studies on three COL2A1 variants, c.1222-98A>G, c.2356-62C>A and c.3435+18C>A, did not demonstrate missplicing. However, this does not mean that they are necessarily benign changes, but only that the particular minigene constructs and cell line utilised did not demonstrate any effect upon mRNA processing. Two of these variants were predicted by Alamut to alter possible binding sites for splicing factors, namely SC35, SRp40 and SRp55 (Table 1). Because of the degenerate nature of sequences that bind these factors many such ESEs/ISEs are identified by Alamut. Therefore, it is possible that such predictions are not as significant as those that create cryptic splice sites.
The emergence of next generation sequencing for use in diagnostic laboratories24 will have a number of consequences in the characterisation of unclassified variants. First, as the numbers of complete genomes that have been sequenced increases, then so will the knowledge about which sequence variants are rare polymorphisms within a population and which may be unique mutations, specific to a particular family or genetic disorder. Second, there is likely to be a greater number of these unclassified variants identified. Strategies open to the diagnostic laboratories are the sequencing of whole exomes, but with the analysis of only certain genes of interest.25 However, this will still limit the amount of intronic sequence screened. The likelihood is that eventually complete genes will be able to be sequenced either as part of a complete genome capture or as long range amplification of selected genes, with subsequent comparison with a reference sequence and known variants. Complete gene sequencing will also resolve how frequent these deep intronic mutations are in Stickler syndrome. In type 1 Stickler syndrome, this is likely to be between 2 and 5% as this represents those deep intronic variants that we have already characterised and the number of patients in which mutations cannot be found by exon sequencing. Tools such as the in silico analysis and functional studies used here will be critical in evaluating a variant's pathogenicity.
Acknowledgments
We thank all the staff in the Molecular Genetics Department at Addenbrooke's Hospital for their contribution to this work.
The authors are commissioned by the Advisory Group for National Specialised Services (http://www.specialisedservices.nhs.uk) and the Department of Health to provide a national diagnostic service for Stickler syndrome.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Patthy L. Modular assembly of genes and the evolution of new functions. Genetica. 2003;118:217–231. [PubMed] [Google Scholar]
- Liu M, Grigoriev A. Protein domains correlate strongly with exons in multiple eukaryotic genomes-evidence of exon shuffling. Trends in Genet. 2004;20:399–403. doi: 10.1016/j.tig.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- Boue S, Letunic I, Bork P. Alternative splicing and evolution. Bioessays. 2003;25:1031–1034. doi: 10.1002/bies.10371. [DOI] [PubMed] [Google Scholar]
- Wang G-S, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- Baralle D, Lucassen A, Buratti E. Missed threads. EMBO R. 2009;10:810–816. doi: 10.1038/embor.2009.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad NN, Ala-Kokko L, Knowlton RG, et al. Stop codon in the procollagen II gene (COL2A1) in a family with the Stickler syndrome (arthro-ophthalmopathy) Proc Natl Acad Sci USA. 1991;88:6624–6627. doi: 10.1073/pnas.88.15.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AJ, Yates JRW, Williams R, et al. A family with Stickler syndrome type 2 has a mutation in the COL11A1 gene resulting in the substitution of glycine 97 by valine in α1(XI) collagen. Hum Mol Genet. 1996;5:1339–1343. doi: 10.1093/hmg/5.9.1339. [DOI] [PubMed] [Google Scholar]
- Annunen S, Korkko J, Czarny M, et al. Splicing mutations of 54-bp exons in the COL11A1 gene cause Marshall syndrome, but other mutations cause overlapping Marshall/Stickler phenotypes. Am J Hum Genet. 1999;65:974–983. doi: 10.1086/302585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AJ, Laidlaw M, Whittaker J, et al. High efficiency of mutation detection in type 1 stickler syndrome using a two-stage approach: vitreoretinal assessment coupled with exon sequencing for screening COL2A1. Hum Mut. 2006;27:696–704. doi: 10.1002/humu.20347. [DOI] [PubMed] [Google Scholar]
- Richards AJ, McNinch A, Martin H, et al. Stickler syndrome and the vitreous phenotype: mutations in COL2A1 and COL11A1. Hum Mut. 2010;31:E1461–E1471. doi: 10.1002/humu.21257. [DOI] [PubMed] [Google Scholar]
- Hoornaert KP, Vereecke I, Dewinter C, et al. Stickler syndrome caused by COL2A1 mutations: genotype-phenotype correlation in a series of 100 patients. Eur J Hum Genet. 2010;18:872–880. doi: 10.1038/ejhg.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead MP, Yates JRW. Clinical and Molecular Genetics of Stickler syndrome. J Med Genet. 1999;36:353–359. [PMC free article] [PubMed] [Google Scholar]
- Carroll C, Papaioannou D, Rees A, et al. The clinical effectiveness and safety of prophylactic retinal interventions to reduce the risk of retinal detachment and subsequent vision loss in adults and children with Stickler syndrome: a systematic review. Health Technol Assess. 2011;15:1–62. doi: 10.3310/hta15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi H, Ikeda T, Osada H, et al. Identification of the COL2A1 mutation in patients with type I Stickler syndrome using RNA from freshly isolated peripheral white blood cells. Genet Test Mol Biomarkers. 2011;15:231–237. doi: 10.1089/gtmb.2010.0138. [DOI] [PubMed] [Google Scholar]
- Spranger J, Winterpacht A, Zabel B. Kniest dysplasia: Dr. W. Kniest, his patient, the molecular defect. Am J Med Genet. 1997;69:79–84. doi: 10.1002/(sici)1096-8628(19970303)69:1<79::aid-ajmg15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Richards AJ, Snead MP. The influence of pre-mRNA splicing on phenotypic modification in Stickler's syndrome and other type II collagenopathies. Eye. 2008;22:1243–1250. doi: 10.1038/eye.2008.34. [DOI] [PubMed] [Google Scholar]
- Martin S, Richards AJ, Yates JRW, et al. Stickler syndrome: further mutations in COL11A1 and evidence for additional locus heterogeneity. Eur J Hum Genet. 1999;7:807–814. doi: 10.1038/sj.ejhg.5200377. [DOI] [PubMed] [Google Scholar]
- Gaildrat P, Killian A, Martins A, et al. Use of splicing reporter minigene assay to evaluate the effect on splicing of unclassified genetic variants. Methods Mol Biol. 2010;653:249–257. doi: 10.1007/978-1-60761-759-4_15. [DOI] [PubMed] [Google Scholar]
- Limb GA, Salt TE, Munro PM, et al. In vitro characterization of a spontaneously immortalized human Müller cell line (MIO-M1) Invest Ophthalmol Vis Sci. 2002;43:864–869. [PubMed] [Google Scholar]
- Richards AJ, Meredith S, Poulson A, et al. A novel mutation of COL2A1 resulting in dominantly inherited rhegmatogenous retinal detachment. Invest Ophthalmol Vis Sci. 2005;46:663–668. doi: 10.1167/iovs.04-1017. [DOI] [PubMed] [Google Scholar]
- Ang A, Poulson AV, Goodburn SF, et al. Retinal detachment and prophylaxis in type 1 Stickler syndrome. Ophthalmology. 2008;115:164–168. doi: 10.1016/j.ophtha.2007.03.059. [DOI] [PubMed] [Google Scholar]
- Winterpacht A, Hilbert M, Schwarze U, et al. Kniest and Stickler dysplasia phenotypes caused by collagen type II gene (COL2A1) defect. Nat Genet. 1993;3:323–326. doi: 10.1038/ng0493-323. [DOI] [PubMed] [Google Scholar]
- Voelkerding KV, Dames SA, Durtschi JD. Next-generation sequencing: from basic research to diagnostics. Clin Chem. 2009;55:641–658. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]
- Kuhlenbaumer G, Hullmann J, Appenzeller S. Novel genomic techniques open new avenues in the analysis of monogenic disorders. Hum Mut. 2011;32:144–151. doi: 10.1002/humu.21400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.