Abstract
Family history-based risk assessment (FHRA) is a genetic tool for identifying those at risk of disease. Genome-wide association studies have shown that single nucleotide polymorphisms (SNP) are statistically associated with low- to moderate-level risks of diseases. There has been limited study of complementarity for these two assessment methods. We sought to compare cancer risk categorizations from FHRA and from Navigenics Personal Genome Screening (PGS). We compared FHRA with PGS for breast (22 females), prostate (22 males), and colon cancer (44 males and females) assessed by kappa (κ) statistic. We also assessed each participant's hereditary risk based on clinical criteria and/or gene-test results. Both FHRA and PGS placed 59%, 68% and 44% of participants into the same risk categories for breast, prostate, and colon cancer, respectively. Overall, however, there was little concordance in FHRA versus PGS for all three cancer risks (κ<0.2). FHRA assigned 22 with hereditary risk compared with PGS, which identified one as high risk (P<0.0001). We assessed nine with hereditary colorectal cancer risk, five with germline mutations, but none were classified as PGS high risk (P=0.0001). FHRA and PGS may be complementary tools for cancer risk assessment. However, evaluation of family history remains the standard to evaluate an individual's cancer risk until further research.
Keywords: breast cancer, colon cancer, Navigenics, prostate cancer, risk assessment
Introduction
It is believed that the two largest contributors to premature mortality are behavior (40%) and genetics (35%).1, 2 When direct-to-consumer personal genome screening (PGS) rapidly appeared on an almost worldwide basis, subsets of consumers, healthcare providers and policy decision makers at all levels believed and continue to believe that it and standard clinical evaluation for heritable risk of disease are equivalent, a phenomenon we term the problem of misattributed equivalence.3 Importantly, physicians, especially in the primary-care setting, are in the first line of contact when patients have questions about any medical and seemingly medical issues. Direct-to-consumer PGS is no exception.4 With increasing access and decreasing price point for this type of risk assessment for common diseases such as cancer, healthcare providers and consumers are questioning how this type of profiling can be integrated into mainstream medicine, and how this type of genetic risk assessment compares with the standard of family health history-based risk assessment (FHRA).5
Of all common disorders, scientific knowledge regarding genetic predisposition to cancer is amongst the most advanced. An average of 10% (range 1–30%) of all cancers can be attributed to high penetrance genetic predisposition.6 Indeed, an average of 5–10% of all diseases have a high penetrance genetic cause. Many more have a genetic component. Currently, obtaining a medical and family history is the gold standard in the initial assessment for the presence of a heritable cancer syndrome. It is well documented and acknowledged that the family history-based risk assessment is one of the most effective tools for predicting what diseases an individual may be at risk for developing.7 FHRA is the current standard genetic tool used to identify those at moderate and high risks of disease predisposition, and is particularly useful for cancer risk assessment. These will yield the clinical ‘red flags' suggesting heritable cancer: early age of onset, multi-focal disease, bilateral disease in paired organs, associated cancers (eg, colon and endometrial, breast and ovarian) and familial clustering. Based on medical history and family history, a genetic differential diagnosis is formulated and genetic testing offered based on the clinical differential diagnosis. This model of Mendelian genetic testing focuses only on one or a few genes, which are known to be strongly associated with the differential diagnosis list. If a cancer-predisposition gene mutation is identified, the individual is at high risk (sometimes close to 100% likelihood) of developing the associated cancer(s). Importantly, identifying a predisposing gene mutation leads to genotype-informed personalization of medical management, such as organ-specific surveillance and/or prophylactic surgery, to effect the earliest cancer diagnosis and prevention.8, 9 This also allows for genotype-specific predictive testing of as yet unaffected family members with mutation-positive relatives offered similar tailored clinical care.
Genome-wide association studies are case–control studies that study hundreds to thousands of individuals to search for genetic markers, typically single nucleotide polymorphisms (SNPs) that are statistically found to be more common in individuals with disease over controls. They are common in the general population and may or may not have a known functional consequence. Genome-wide association studies typically provide a relative risk of the likelihood beyond some population mean that an individual with a particular SNP genotype will have disease, which significantly differs from the highly penetrant, deleterious mutations identified via Mendelian genetic testing. With vast amounts of genome-wide association data available, several for-profit companies worldwide have developed testing services to provide clients with predictions about their risks for a broad spectrum of medical and non-medical conditions and traits, ranging from ancestry grouping to earwax type to prostate cancer risk. Many of these companies provide services direct to consumer.
To date, there has been limited study of the overlap between these two methods of risk assessment for common diseases. We, therefore, sought to compare FHRA with SNP-based risk assessment, as available from Navigenics, for risks of three common cancers, those of the breast, prostate and colorectum.
Materials and methods
Research participants
All research participants signed informed consent in accordance with the Cleveland Clinic Institutional Review Board for Human Subjects' Protection.
Males and females aged 18 years or older were enrolled from the cancer genetics clinical service in the Center for Personalized Genetic Healthcare of the Cleveland Clinic Genomic Medicine Institute if they met the following eligibility criteria: a personal or family history of breast, prostate or colorectal cancer. When a research participant from this clinic was enrolled, if he/she had a spouse/partner, the spouse was offered enrollment irrespective of personal or family history of cancer. We selected a target minimum sample size of 40 to be powered (at P>0.9) to detect 5% prevalence of events (eg, a risk status). We also selected sample size by examining if an outcome (ie, the call of risk level) would be correlated between PGS and family history-based cancer risk assessment at least at 0.30. In this situation, a sample size of 30 would yield sufficient power (P>0.8, alpha=0.05, two-tail). Thus, between these two statistical considerations, we aimed for a sample size of 40.
Cancer risk assessment
Upon study enrollment, a personal medical history and at least a three-generation family cancer history was obtained from each subject in a manner consistent with a cancer genetics clinic. A cancer family history-based risk assessment, taking into account both personal medical history and family cancer history, was performed, and when indicated, clinical testing for the appropriate predisposition gene was offered in the setting of genetic counseling. Based on personal and family cancer history, each subject was classified as general population, moderate, or high risk (Table 1). The Surveillance Epidemiology and End Results general population lifetime risk was utilized for family history-based risk assessment was 12.08%, 5.20%, and 15.9% for breast, colorectal, and prostate cancer, respectively (http://www.seer.cancer.gov). Subsequently, based on personal and family cancer history or the presence of a germline mutation in a cancer-predisposition gene in the participant or in his/her family member, each participant is additionally classified as at hereditary risk or not. A single genetic counselor (BH) and a single cancer-genetics physician (CE) reviewed all risk assessments.
Table 1. Cancer Risk Assessment Comparing Family History-Based Risk Assessment (FHRA) to personal genome screening (PGS).
| Risk class | PGS low | PGS moderate | PGS high | Total |
|---|---|---|---|---|
| (a) Breast cancer (κ=0.12) | ||||
| FHRA GenPop | 12 | 1 | 0 | 13 |
| FHRA moderate | 1 | 0 | 0 | 1 |
| FHRA high | 6 | 1 | 1 | 9a |
| 19 | 2 | 1 | 22 | |
| (b) Prostate cancer (κ=0.14) | ||||
| FHRA GenPop | 15 | 2 | 2 | 19 |
| FHRA moderate | 0 | 1 | 0 | 1 |
| FHRA high | 2 | 0 | 0 | 2 |
| 17 | 3 | 2 | 22 | |
| (c) Colorectal cancer (κ=−0.05) | ||||
| FHRA GenPop | 18 | 12 | 0 | 30 |
| FHRA moderate | 5 | 2 | 0 | 7 |
| FHRA high | 4 | 3 | 0 | 7 |
| 27 | 17 | 0 | 44 | |
Note that there is one research participant who was classed as high risk by FHRA, where PGS results for breast cancer risk were not available.
First, breast cancer family history-based risk assessment was based on three sets of established clinical criteria, the Scheuner criteria, which classes into general population, moderate and high risk based on the genetic ‘red flags' (Supplementary Table 1), the Gail Model, and the Claus model.10, 11, 12 Thereafter, our family history-based risk cancer assessment utilized the following clinical criteria to assign hereditary risk: US National Comprehensive Cancer Center Hereditary Breast/Ovarian Cancer v.1.2.2009 diagnostic criteria (www.nccn.org); International Cowden Consortium Clinical Diagnostic Criteria 2000;13 criteria for classic Li–Fraumeni syndrome or relaxed criteria for Li–Fraumeni-like syndrome,14, 15, 16 for hereditary diffuse gastric cancer syndrome, which carries a lobular breast carcinoma risk,17 and for the clinical diagnosis of Peutz–Jegers syndrome18 (Supplementary Table 2). Individuals or families meeting diagnostic criteria for any of the heritable breast cancer syndromes are classified as hereditary risk. Subjects or families who have a known germline mutation in a breast cancer-predisposition gene are classified as hereditary risk.
Prostate cancer risk assessment was based on the Scheuner criteria, which, like breast cancer, classifies individuals as general population, moderate and high risk (Supplementary Table 1). Familial prostate cancer (hereditary) risk was based on the Hopkins criteria (Supplementary Table 2).19
Colorectal cancer risk assessment was based on the Scheuner criteria, which classifies individuals as general population, moderate and high risk (Supplementary Table 1). Participants' medical and family histories were then re-examined for evidence of hereditary risk. Hereditary risk was assigned if the subject or family met the Amsterdam II or Revised Bethesda Guidelines for Lynch syndrome;20, 21 or met accepted criteria for familial adenomatous polyposis or attenuated FAP,22, 23 MYH-associated polyposis,24 juvenile polyposis syndrome,25 or Peutz–Jeghers syndrome18 (Supplementary Table 2). Research participants or family members who carry a known germline mutation in a colorectal cancer-predisposition gene were classified as hereditary risk.
In addition to their personal and family cancer histories, the subjects were offered Navigenics PGS. Each subject donated a saliva sample. All samples were unlinked from personal identifiers, before shipment to Navigenics. Only the SNP-based risk profiles for breast, prostate and colorectal cancer were reviewed by the study genetic counselor.
Statistical analysis
The concordance amongst the three categories (general population, moderate and high risk) for each of breast, prostate and colorectal cancer risk was compared between family history-based risk assessment and Navigenics PGS using the kappa (κ) statistic. Subjects classified as hereditary risk for any of the three cancers were compared with the PGS high-risk classification using Fisher two-tailed exact test, with P<0.05 considered significantly different.
Results
Demographic data
A total of 44 participants (22 female, 22 male) were enrolled prospectively. Of the 44, 22 visited a cancer genetics clinic and the other 22 were their spouses (see Materials and methods). There were 42 White individuals, one Asian, and one Pacific Islander. Overall, mean age at enrollment was 55.2 years (18–84 years). Amongst the 22 women who underwent breast cancer risk assessment, 7 had a personal history of breast cancer with the mean age at diagnosis 48 years (33–62 years). The lifetime risk of breast cancer for these subjects, based on family history, ranged 6.4–87%, whereas the range based on PGS was 9–27%. Of the 22 men who underwent prostate cancer risk assessment, 2 had a personal history of prostate cancer, with the mean age at diagnosis of 62 years (59–65 years). The lifetime risk of prostate cancer based on family history assessment ranged 15.9–31.8% with a similar range based on PGS, 10–31%. All 44 participants received family cancer history-based risk assessment and PGS for colorectal cancer, and of those, 4 had a personal history of colorectal cancer diagnosed at a mean age of 51.8 years (29–74 years) and 11 had a personal history of adenomatous polyps (mean 52.8 years, 28–74 years). Family history-based assessments estimated a lifetime risk of colorectal cancer for these subjects in the range of 5.2–80%, whereas the PGS lifetime risk was significantly lower, 3.6–9%.
Family history-based risk assessment compared with PGS risk assessment for three common cancers
Between the time the first individual enrolled in this study and the time of analysis (about 1 year), Navigenics altered a subset of SNPs and/or statistical algorithm used for risk assessment for all three cancers. For each cancer type, we first compared the concordance between the first and second Navigenics PGS risk assessments and found that there was little difference between the two versions in classifying our 44 subjects into their three risk categories (κ=0.7–1.0). For purposes of comparing family history-based risk assessment and PGS, therefore, we chose to use the first PGS risk assessment.
Both family history-based cancer risk assessment and PGS placed 59%, 68% and 44% of participants into the same risk categories for breast, prostate, and colon cancer, respectively (Table 1). Overall, however, the concordance between FHRA and PGS risk assessments for breast (κ=0.12), prostate (κ=0.14) and colorectal (κ=−0.05) cancer was low (Table 1). For example, family history-based assessments classified nine participants as high risk for breast cancer, whereas only one of these was classified as high risk by PGS (Table 1a). PGS predicted a high risk for prostate cancer for two participants, who were assessed to be at general population risk by family history evaluation (Table 1b). There were 17 individuals with a moderate colorectal cancer risk on PGS, whereas family history-based assessments classified 12 as general population risk and 3 at high risk (Table 1c). Similarly, family history-based risk assessments classed seven research participants as high risk for colorectal cancer, whereas none were noted as high risk by PGS.
Using family histories, we assessed nine individuals as having a hereditary risk of breast cancer, three of whom harbored germline mutations in BRCA1/2 defining hereditary breast–ovarian cancer syndrome, and a fourth with a family history suspicious for hereditary breast–ovarian cancer syndrome. Of these nine, only one was classified as high risk by PGS (P<0.0001). Family history-based assessment classified nine participants with hereditary risk for colorectal cancer, five of whom met syndromic clinical criteria, whereas the remaining four had family histories meeting hereditary colorectal cancer criteria. Amongst the nine individuals, five were found to have germline mutations in MLH1 (N=2), MSH2 (N=1), MYH (bi-allelic mutations, N=1), SMAD4 (N=1), defining Lynch syndrome, MYH-associated polyposis and juvenile polyposis syndrome, respectively. In contrast, none of these nine were classified by PGS as high risk (P=0.0001). None of the three individuals classified by family history risk assessment as having a hereditary prostate cancer risk were assessed as high risk by PGS (P=0.1).
Discussion
Our comparison of family history-based and Personal Genome Scanning for breast, prostate and colorectal cancer risk assessments suggest little concordance although there is some partial overlap (illustrated by Figure 1). However, it is not haphazard concordance or haphazard discordance. The most striking observation is the almost complete discordance between family history-based and PGS risk assessments for the hereditary cancer risks, whereby PGS is unable to pick up individuals at hereditary or highest risks of common cancers. This raises the ethical concern that without a formal family history risk assessment, consumers who may be at high risk of hereditary disease, could be falsely reassured by the results of PGS. From a clinical point of view, this observation suggests that when a consumer brings a low-risk PGS cancer profile to his/her primary-care physician, the physician should always take a family health history, including that for cancer, for risk assessment for potential heritable diseases.26 On the other hand, our observation may suggest further investigation to determine the utility, validity and actionability of SNP-based risk assessment after hereditary risk for disease, in this case, cancer, is rigorously excluded.
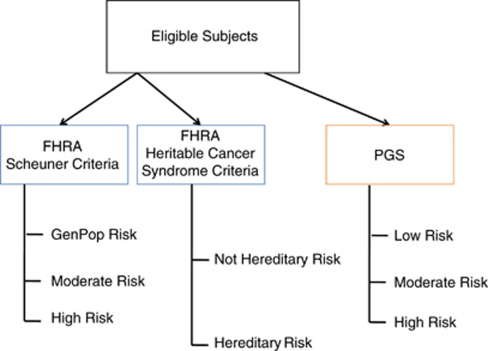
Figure 1.
Cancer risk assessment study schema. Cancer risk assessment schema for personal and FHRA based on the clinical red flags suggesting genetic risk (Scheuner criteria; Supplementary Table 1) for the three common cancers and FHRA based on clinical criteria for hereditary cancer risk (see Supplementary Table 2 also); and PGS of SNP-associated cancer risks.
Judicious and targeted gene testing for Mendelian cancer-predisposition genes is accepted as beneficial after clinical risk assessment based on personal medical and family health history. In general, when an individual or family is deemed high or moderate risk by family history-based risk assessment, targeted gene testing in the setting of genetic counseling acts as a powerful molecular diagnostic test.27 Furthermore, the types of cancers in a family of an individual presenting with, for example, breast cancer at the age of 45 years will hone the differential diagnosis as to which of the eight high-penetrance breast cancer-susceptibility genes to analyze.27 Because of genotype–phenotype and clinical outcomes research over many years, we know the gene-specific organ-specific cancer risks and ages of maximal risk, and so, surveillance or prophylaxis can be effectively instituted. Additionally, once a family-specific mutation is found, predictive testing of as yet unaffected family members is 100% accurate. Although we know the relative risk or increased lifetime risk of a specific cancer(s) conferred by a germline mutation in a Mendelian gene, such as BRCA1 (up to 80% lifetime risk of breast cancer and up to 40% lifetime risk of ovarian cancer compared with 13% and 1%, respectively, in the general population), we do not know which particular 80% or which particular 40% of mutation carriers are the ones who will develop these component cancers. It was hoped that these common SNPs would help to pinpoint who precisely, on an individual basis, are destined to develop component cancers. Multi-center, multi-national studies recently showed that such SNPs either did not modify the risk of breast cancer in BRCA1/2 mutation carriers or only modestly (eg, RR 1.1–1.3).28, 29 Xu et al analyzed the utility of the 14 SNPs associated with prostate cancer risk.30 By themselves, the SNPs were not particularly helpful for identifying men who would go on to develop prostate cancer, as we have noted in our present study. However, the SNPs had value in specific situations. Xu et al examined the utility of these SNPs in men with borderline PSA levels, using the SNPs in conjunction with PSA levels to deliver an SNP-adjusted PSA score to help in the biopsy decision. The investigators also examined the utility of combining SNP risks. Without a family history of prostate cancer, they found that the presence of all 14 SNPs raised the risk of a white individual developing prostate cancer between the ages of 55 and 72 years from 13% (the general population risk among whites) to 20%.30 In the presence of a family history of prostate cancer, however, having eight or more of these SNPs raised the risk of developing prostate cancer between the ages of 55 and 72 years from 13 to 25%. If these data could be replicated independently, then having >8 SNPs delivered incremental risks beyond 25% and would be actionable.
This study has several important strengths. These data were obtained in a prospective manner, accruing consecutive eligible research participants from an academic cancer genetics clinical practice, but also enrolling their corresponding spouses/partners, the latter of whom would represent the population-at-large. Similarly, both sets of data, family health history-based and SNP-based, were obtained on the same series of participants. Although SNP-based testing is not intended for populations at high risk of hereditary disease, such as those attending a cancer genetics clinic, we believe our study population represents the spectrum of consumers who may elect to pursue direct-to-consumer genetic testing.
Notably, this is the first formal comparison between current practices and personal genome scanning using cancer risk assessment as the model, thus addressing in an evidence-based manner, the issue of misattributed equivalence. Whereas our study focuses on three of the most common cancers, we suspect, given the available data, that our observations may be generalizable to other common diseases as well. A limitation of our study includes the relatively small sample size (N=44). Nonetheless, it would be ideal if similar studies and larger independent studies could be performed for various cancers and other diseases.
Two of the proposed goals for Healthy People 2020 are the identification of as many people as possible who are at genetic risk of breast or colorectal cancer, so that they can be directed to proper care including genetic counseling, appropriate clinical surveillance and personalized prevention strategies (www.healthypeople.gov/hp2020/Objectives/ TopicArea.aspx?id=23&TopicArea=Genomics). It may seem obvious that taking a family health history would be the simplest, most cost-effective manner of obtaining these goals. However, it is also well known that taking complete, accurate family health histories occurs in <30% of all healthcare encounters. Therefore, when novel technologies are developed, the healthcare community considers whether such new methods can replace (because it is simpler, less expensive, etc) or complement the existing gold standard (Figure 1). This is no different when SNP-based risk assessment becomes possible and becomes available direct-to-consumer; stakeholders are becoming fascinated and are wondering about the concordance or complementarity of SNP-based PGS to current standard family and medical history-based assessment of genetic risk of disease (Figure 1). Here, we have used assessing genetic risk of three common cancers as a model. If PGS and FHRA were 100% or almost completely concordant (Figure 1c), then a serious consideration of utilizing only one method over the other based on practical considerations and best endpoints would be reasonable. If there was absolute discordance (Figure 1a), then there are two hypothetical conclusions: both PGS and family history-based risk assessment might have roles, or either PGS or family history-based assessment should be discarded. Finally, if there was some partial overlap, then PGS and FHRA may have different albeit useful roles in identifying genetic risk of disease. For healthcare policy makers and other hospital/medical center decision makers, it would be important for them to recognize the problem of misattributed equivalence.3
All in all, our data demonstrate that family medical history-based risk assessment and PGS risk assessments for breast, prostate and colorectal cancers are not concordant, but may suggest that together both tools could be complementary tools for cancer risk assessment. In this situation, FHRA to exclude hereditary and highest risks for cancer should be carried out first, followed by PGS, if borne out by further investigation. It is important to emphasize that evaluation of family health history is still the standard in current clinical care, and this should be used to clinically evaluate an individual's risk of developing cancer and other diseases until further research is done to prove that PGS should be discarded, modified or can be used to predict an individual's risk in some manner and/or can be integrated with family-based health risk assessment to increase sensitivity.
Acknowledgments
Navigenics performed Personal Genomic Screening on a research basis without charge to the investigators or the research participants. CE is the Sondra J and Stephen R Hardis Endowed Chair of Cancer Genomic Medicine at the Cleveland Clinic and is an American Cancer Society Clinical Research Professor, generously funded, in part, by the FM Kirby Foundation.
Web Resources
DHHS Healthy People 2010 Genomics, www.healthypeople.gov/hp2020/Objectives/ TopicArea.aspx?id=23&TopicArea=Genomics.
National Comprehensive Cancer Network (NCCN), http://www.nccn.org/.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Sorensen TI, Nielsen GG, Andersen PK, Teasdalte TW. Genetic and environmental influences on premature mortality in adult adoptees. N Engl J Med. 1988;318:727–732. doi: 10.1056/NEJM198803243181202. [DOI] [PubMed] [Google Scholar]
- Petersen L, Nielsen GG, Andersen PK, Sorensen TI. Case-control study of genetic and environmental influences of premature death of adult adoptees. Genet Epidemiol. 2002;23:123–132. doi: 10.1002/gepi.1122. [DOI] [PubMed] [Google Scholar]
- Eng C, Sharp RR. Bioethical and clinical dilemmas of direct-to-consumer genomic testing: the problem of misattributed equivalance. Science Transl Med. 2010;2:1–5. doi: 10.1126/scitranslmed.3000214. [DOI] [PubMed] [Google Scholar]
- McGuire AL, Burke W. An unwelcome side effect of direct-to-consumer personal genome testing: raiding the medical commons. JAMA. 2008;300:2669–2671. doi: 10.1001/jama.2008.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon PW, Scheuner MT, Peterson-Oehlke KL, Gwinn M, Faucett A, Khoury MJ. Can family history be used as a tool for public health and preventive medicine. Genet Med. 2002;4:304–310. doi: 10.1097/00125817-200207000-00009. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Holm N, Verkasalo P, et al. Environmental and heritable factors in the causation of cancer. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- Guttmacher AE, Collins FS, Carmona RH. The family history – more important than ever. N Engl J Med. 2004;351:2333–2336. doi: 10.1056/NEJMsb042979. [DOI] [PubMed] [Google Scholar]
- Kloos RT, Eng C, Evans DB, et al. Medullary thyroid carcinoma: Management Guidelines of the American Thyroid Association. Thyroid. 2009;19:565–612. doi: 10.1089/thy.2008.0403. [DOI] [PubMed] [Google Scholar]
- Zbuk K, Eng C. Cancer phenomics: RET and PTEN as illustrative models. Nature Rev Cancer. 2007;7:35–45. doi: 10.1038/nrc2037. [DOI] [PubMed] [Google Scholar]
- Scheuner MT, Wang SJ, Raffel LJ, Larabell SK, Rotter JI. Family history: a comprehensive genetic risk assessment method for the chornic conditions of adulthood. Am J Med Genet. 1997;71:315–324. doi: 10.1002/(sici)1096-8628(19970822)71:3<315::aid-ajmg12>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Gail MH, Brinton LA, Byar DP, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81:1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- Claus EB, Risch N, Thompson WB. The calculation of breast cancer risk for women with a first degree family history of ovarian cancer. Breast Cancer Res Treat. 1993;28:115–120. doi: 10.1007/BF00666424. [DOI] [PubMed] [Google Scholar]
- Eng C. Will the real Cowden syndrome please stand up: revised diagnostic criteria. J Med Genet. 2000;37:828–830. doi: 10.1136/jmg.37.11.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FP, Fraumeni JF., Jr Collaborative interdisciplinary studies of p53 and other predisposing genes in Li-Fraumeni syndrome. Cancer Epidemiol Biomark Prevent. 1994;3:715–717. [PubMed] [Google Scholar]
- Varley JM, McGown G, Thorncroft M, et al. Germ-line mutations of TP53 in Li-Fraumeni families: an extended study of 39 families. Cancer Res. 1997;57:3245–3252. [PubMed] [Google Scholar]
- Eeles RA. Germline mutations in the TP53 gene. Cancer Surv. 1995;25:101–124. [PubMed] [Google Scholar]
- Brooks-Wilson AR, Kaurah P, Suriano G, et al. Germline E-cadherin mutations in hereditary diffuse gastric cancer: assessment of 42 new families and review of genetic screening criteria. J Med Genet. 2004;41:508–517. doi: 10.1136/jmg.2004.018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardello FM, Walsh SB, Hamiton SR, et al. Increased risk of cancer in Peutz-Jeghers syndrome. N Engl J Med. 1987;316:1511–1514. doi: 10.1056/NEJM198706113162404. [DOI] [PubMed] [Google Scholar]
- Carter BS, Beaty TH, Steinberg GD, Childs B, Walsh PC. Mendelian inheritance in familial prostate cancer. Proc Natl Acad Sci USA. 1992;89:3367–3371. doi: 10.1073/pnas.89.8.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasen HF, Watson P, Mecklin J-P, Lynch HT. New clinical criteria for hereditary non-polyposis colon cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative Group on HNPCC. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda guidelines for hereditary nonpolyposis colon cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, Hes FJ, Nagengast FM, et al. Germline mutations in APC and MUTYH are responsible for the majority of families with attenuated familial adenomatous polyposis. Clin Genet. 2007;71:427–433. doi: 10.1111/j.1399-0004.2007.00766.x. [DOI] [PubMed] [Google Scholar]
- APC-associated polyposis conditions 2009 . http://www.ncbi.nlm.nih.gov/books/NBK1345/ . (Accessed 24 August 2009).
- Sieber OM, Lipton L, Crabtree M, et al. Multiple colorectal adenomas, classic adenomatous polyposis and germ-line mutations in MYH. N Engl J Med. 2003;348:791–799. doi: 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- Jass JR, Williams CB, Bussey HJR, Morson BC. Juvenile polyposis–a precancerous condition. Histopathology. 1988;13:619–630. doi: 10.1111/j.1365-2559.1988.tb02093.x. [DOI] [PubMed] [Google Scholar]
- Edelman E, Eng C. A practical guide to the interpretation and clinical application of personal genomic screening. Br Med J. 2009;339:b4253. doi: 10.1136/bmj.b4253. [DOI] [PubMed] [Google Scholar]
- Eng C. Mendelian genetics of rare-and not so rare-cancers. Ann NY Acad Sci. 2010;1214:70–82. doi: 10.1111/j.1749-6632.2010.05789.x. [DOI] [PubMed] [Google Scholar]
- Osorio A, Milne RL, Pita G, et al. Evaluation of a candidate breast cancer associated SNP in ERCC4 as a risk modifier in BRCA1 and BRCA2 mutation carriers. Results from the Consortium of Investigators of Modifiers of BRCA1/BRCA2 (CIMBA) Br J Cancer. 2009;101:2048–2054. doi: 10.1038/sj.bjc.6605416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou AC, Spurdle AB, Sinilnikova OM, et al. Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am J Hum Genet. 2008;82:937–948. doi: 10.1016/j.ajhg.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Sun J, Kader AK, et al. Estimation of absolute risk for prostate cancer using genetic markers and family history. Prostate. 2009;69:1562–1572. doi: 10.1002/pros.21002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.