Abstract
We studied the genomic instability and methylation status of the mismatch-repair (MMR) genes hMLH1 and hMSH2, and the imprinted genes H19/IGF2, in fetuses with neural tube defects (NTDs) to explore the pathogenesis of the disease. Microsatellite instability (MSI) was observed in 23 of 50 NTD patients. Five NTD patients showed high-degree MSI (MSI-H) and 18 showed low-degree MSI (MSI-L). The frequencies of mutated microsatellite loci were 3/50 (6%) for BatT-25, 10/50 (20%) for Bat-26, 3/50 (6%) for Bat34C4, 6/50 (12%) for D2S123, 4/50 (8%) for D2S119, and 3/50 (6%) for D3S1611. The promoter regions of the hMLH1 and hMSH2 genes were unmethylated in NTD patients, as determined by methylation-specific PCR. The hMLH1 and hMSH2 promoter methylation patterns, the methylation levels of H19 DMR1, and IGF2 DMR0 were detected by bisulfite sequencing PCR, sub-cloning, and sequencing. The hMSH2 promoter sequence was unmethylated, and the hMLH1 promoter showed a specific methylation pattern at two CpG sites. The methylation levels of H19 DMR1 in the NTD and control groups are 73.3%±15.9 and 58.3%±11.2, respectively. The methylation level of the NTD group was higher than that of the control group (Student's t-test, P<0.05). There is no significant difference in IGF2 DMR0 methylation level between the two groups. All of the results presented here suggest that genomic instability, the MMR system, and hyper-methylation of the H19 DMR1 may be correlated with the occurrence of NTDs.
Keywords: neural tube defects, microsatellite instability, mismatch repair, methylation, IGF2/H19
Introduction
Neural tube defects (NTDs), including anencephaly and spina bifida, are a common group of central nervous system anomalies affecting 0.5–2 fetuses per 1000 pregnancies worldwide.1 Shanxi Province in China has the highest reported incidence of NTDs in the world. A population-based study conducted in 2003 reported a prevalence of 138.7 per 10 000 births in four counties within Shanxi Province with elevated NTD risks.2 Children with severe birth defects have a 15-fold increased risk of death during their first year of life, with 9–10% dying during this period.3 NTDs are multifactorial disorders that may be ascribed to genetic or environmental factors, including maternal nutritional factors, infection, physical, and chemical factors, or combinations of these factors.4 Among the environmental factors related to NTDs, low maternal folate status and vitamin B12 appear to be risk factors. Many studies have now conclusively proved the beneficial effects of folic acid supplementation in preventing NTDs.5, 6 These findings suggest that folate intake is a key determinant of susceptibility to NTDs. As a methyl donor, folic acid is involved in DNA synthesis, DNA methylation, and other physiological processes. In addition, folate deficiency increases uracil mis-incorporation into DNA in mice, which could lead to genomic instability.7 Genomic instability and changes in DNA methylation status may, therefore, have significant effects on the development of NTDs.
DNA mismatch repair (MMR) is an evolutionarily conserved process that corrects mismatches generated during DNA replication and escape proofreading. MMR proteins also participate in many other DNA transactions, such that inactivation of MMR can have wide-ranging biological consequences, which can be either beneficial or detrimental.8 Microsatellite instability (MSI) is one signal of genomic instability and reflects the status of the MMR genes. MSI has been found in hereditary non-polyposis colorectal cancer (HNPCC), sporadic colorectal, endometrial, pancreatic, breast, and gastric cancers.9 Changes in microsatellite sequence may affect functions related to gene expression, such as transcription rate, RNA stability, splicing efficiency, and RNA–protein interactions.10, 11 DNA methylation is an important component of epigenetics and the regulation of gene expression. Incorrect development of DNA methylation patterns can lead to embryonic lethality and developmental malformations.12 Promoter region methylation has recently been demonstrated to be an important mechanism of gene inactivation in cancer. MSI has been associated with hyper-methylation of the hMLH1 promoter region in sporadic colorectal cancer, gastric carcinoma, and endometrial carcinomas.13
Genomic imprinting regulates the expression of a group of genes that are monoallelically expressed in a parent-of-origin-specific manner. Allele-specific DNA methylation occurs at differentially methylated regions (DMRs) of these genes. IGF2 and H19 belong to the same cluster of imprinted genes, but are expressed differently, depending on whether they are carried by a chromosome of maternal or paternal origin. The most important regulating area of IGF2 and H19 is the sixth CCCTC-binding factor (CTCF)-binding site in the DMRs upstream from H19. The paternal CTCF allele is methylated, whereas the maternal allele is unmethylated in normal tissues. An enhancer-competition model describes how the IGF2 and H19 promoters compete on the same chromosome for a shared enhancer. Access of the unmethylated maternal IGF2 allele to the enhancer is blocked by the H19 DMRs because of the insulator activity of CTCF binding to the unmethylated H19 DMRs. As methylation is a key regulator of imprinted gene expression, methylation disturbances at DMRs have been associated with many congenital growth disorders.14 Research has shown that the methylation patterns of imprinted genes are sensitive to malnutrition in early embryonic development.15
Despite the increased understanding of birth defects, the mechanism underlying the formation of NTDs remains unclear. Because folate deficiency, genomic instability, DNA methylation, and MMR are highly correlated, this study explores the role in NTD-associated gene regulation played by genomic instability, MMR, and epigenetic alterations.
Materials and methods
Samples
Stillborn NTD case subjects were obtained from Shanxi Province of China. Pathologic diagnosis of NTDs was completed by experienced pathologists according to the International Classification of Disease. Fifty NTD samples (brain tissue) were retained for analysis, including 27 subjects with spina bifida, 17 subjects with anencephalus, 5 subjects with encephaloceles, and 1 subject with sacrococcygeal defect. Fifty control subjects, who were aborted for non-medical reasons, were also obtained from the Shanxi Province and selected according to sex, gestational week, and tissue. Any fetuses showing pathologic malformation or family history or chromosomal disorder were excluded from the control group. Routine prenatal checkup and questionnaire and the autopsy report were completed for all NTD case and control subjects. The study was approved by the local ethics committee, and written informed consent was obtained from all parents. Information relating to all of the subjects used in the study is provided in Table 1.
Table 1. Characteristic of NTD and control group.
| Characteristic | Case | Control | Pa |
|---|---|---|---|
| n | 50 | 50 | |
| Age (weeks) | 0.966 | ||
| <20 | 7 | 8 | |
| 20–24 | 26 | 25 | |
| 25–29 | 14 | 13 | |
| ≥30 | 3 | 4 | |
| Sex | 0.680 | ||
| Female | 32 | 30 | |
| Male | 18 | 20 |
Calculated by χ2-test.
Microsatellite analysis
A panel of six microsatellite markers was selected, consisting of three mononucleotide repeats (Bat25, Bat26, and Bat34C4), and three dinucleotide repeats (D2S123, D2S119, and D3S1611). Genomic DNA was extracted from samples using the standard phenol/chloroform method. Then, genomic DNA was amplified by PCR (primers were obtained from http://www.gdb.org/). PCR products were denatured, electrophoresed on 8% denaturing polyacrylamide gels, and visualized by silver staining. MSI was determined by the presence of clearly visualized altered allelic shifts in the PCR products derived from the NTD tissue specimens, when compared with the corresponding control DNA.16 NTDs with instability in two or more markers are characterized as MSI-H (high-degree MSI), whereas NTDs with instability in one marker are defined as MSI-L (low-degree MSI), and NTDs in which none of the markers show MSI are defined as MSS.17 GeneScan was also used to analyze the MSI status of the Bat25 and Bat26 markers in NTDs. Genomic DNA was amplified by PCR using 6-FAM and HEX-labeled primers. The GeneScan analysis software GeneMarker V1.75 was used to analyze MSI status.
MSP and BSP, sub-cloning, and sequencing
Methylation-specific PCR (MSP) was used to investigate the methylation pattern of the MMR genes hMLH1 and hMSH2 (GenBank accession numbers U83845 and U23824). The target regions were a 180-bp fragment upstream from exon-1 of hMLH1 (−271 to −92 bp, containing 11 CpG sites) and a 143-bp fragment upstream of exon-1 from hMSH2 (−42 to +100 bp, containing 14 CpG sites). Owing to degradation of genomic DNA and the limited availability of sample sources, 22 NTD samples (13 MSI-positive NTD samples, 9 MSS NTD samples) and seven control samples were selected for this analysis. Genomic DNA from NTD and control samples was treated with sodium bisulfite and purified by using the Wizard DNA Clean-Up System, and then MSP18 was used to analyze the methylation status of the hMLH1 and hMSH2 promoters. Human placental gDNA either unmodified or methylated in vitro with SssI methylase was used as validation controls. For each PCR, 25 m MgCl2, 25 m dNTP, 20 μ forward and reverse primers, 1.5 U of platinum Taq, and 10 × platinum Taq buffer were used in a 25-μl total reaction volume. The PCR cycles were as follows: pre-denaturation at 95 °C for 5 min, denaturation at 95 °C for 45 s, annealing at Tm of the primer for 1 min, and extension at 72 °C for 1 min, followed by 40 cycles, followed by a final extension at 72 °C for 5 min. PCR products were run on 1.2% agarose gels and visualized after staining with ethidium bromide. Those same samples were detected by BSP (BSP), sub-cloning, and sequencing to analyze methylation patterns at the hMLH1, hMSH2 promoter. The bisulfite-treated DNA was amplified by a nested PCR. The PCR conditions were similar as those described above except for the primers. The primer sequences and Tm are provided in Supplementary Table S1; the MSP primers were as outlined previously;19, 20 whereas the nested primers were designed using Methyl Primer Express v1.0 (ABI). About five colonies for each allele were performed by automated sequencing using M13 primers. We used QUantification tool for Methylation Analysis (QUMA) (http://quma.cdb.riken.jp/) as the analysis software to evaluate the samples' methylation status.21
A total of 17 NTD samples were detected by BSP, sub-cloning, and sequencing for analysis of the methylation status of the H19 DMR1 and IGF2 DMR0 (GenBank accession numbers AF125183 and Y13633). The target regions were a 201-bp region of the human H19 DMR1 containing 18 CpG sites and one CTCF-binding site, which was 5 kb upstream from H19 and known to be the key site of methylation, and the IGF2 DMR0 within intron-2 of IGF2 and methylated on the maternal allele. These target loci were amplified by nested PCR. PCR was performed under the similar reaction conditions as those described above except for the primers. The primer sequences and Tm are provided in Supplementary Table S2; the primers of H19 DMR1 and IGF2 DMR0 were as previously outlined;22, 23 whereas the nested primers were designed with Methyl Primer Express v1.0 (ABI). After that, the PCR products were cloned into the pGEM-T vector. About 10 colonies for each allele were performed by automated sequencing using M13 primers. The methylation pattern of each allele was analyzed with the online software QUMA.
Results
Presence of MSI associated with NTDs
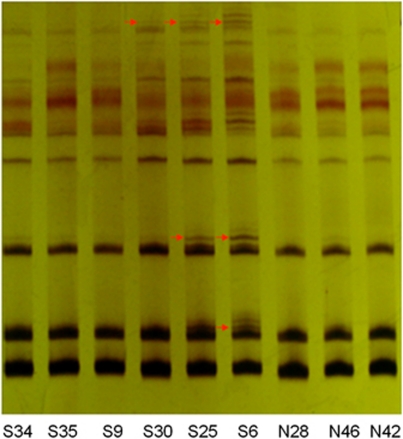
We evaluated six microsatellite markers to investigate genomic instability in NTD tissues. MSI was observed in 23 of the 50 NTD samples using PCR–urea denaturing PAGE (see Table 2) and GeneScan. The total positive rate of MSI in the NTD group was 23/50 (46%). In the MSI subset, the frequencies of mutated microsatellite loci were 3/50 (6%) for Bat-25, 10/50 (20%) for Bat-26, 3/50 (6%) for Bat34C4 3, 6/50 (12%) for D2S123, 4/50 (8%) for D2S119, and 3/50 (6%) for D3S1611. Five samples showed MSI on more than one site, which defined them as MSI-H; they all showed spina bifida. Furthermore, a total of 18 samples were defined as MSI-L, with MSI at one site only. Figure 1 depicts a representative analysis of the MSIs.
Table 2. Summary of MSI assays.
| Number | NTDs | Bat25 | Bat26 | Bat34C4 | D2S123 | D2S119 | D3S1611 |
|---|---|---|---|---|---|---|---|
| 1 | S2 | + | |||||
| 2 | S3 | + | |||||
| 3 | S6 | + | |||||
| 4 | S11 | + | |||||
| 5 | S12 | + | |||||
| 6 | S14 | + | + | ||||
| 7 | S19 | + | |||||
| 8 | S21 | + | |||||
| 9 | S22 | + | |||||
| 10 | S25 | + | |||||
| 11 | S26 | + | |||||
| 12 | S30 | + | + | + | |||
| 13 | S31 | + | |||||
| 14 | S33 | + | |||||
| 15 | S34 | + | + | ||||
| 16 | S40 | + | |||||
| 17 | S42 | + | |||||
| 18 | S43 | + | |||||
| 19 | S44 | + | |||||
| 20 | S45 | + | |||||
| 21 | S47 | + | + | ||||
| 22 | S48 | + | + | ||||
| 23 | S50 | + |
Abbreviation: MSI, microsatellite instability.
Bat25, Bat26, Bat34C4, D2S123, D2S119, D3S1611: microsatellite sites.
S, NTDs group; +, MSI-positive.
Figure 1.
MSI in NTDs. Electrophoretic analysis of urea denaturing PAGE of Bat25. The red arrows indicate abnormal electrophoresis bands. S, NTDs; N, control.
The methylation patterns of the hMLH1 and hMSH2 promoters
To date, little research has been conducted to investigate the relationship between the methylation patterns of the hMLH1 and hMSH2 promoters and the occurrence of NTDs. In our study, we used MSP to analyze the methylation status of the hMLH1 and hMSH2 promoter sequences in the two groups. However, we did not detect significantly more methylation of the hMLH1 and hMSH2 promoters in the NTD samples than in the control samples. Then 22 NTD samples (13 MSI-positive NTD samples, 9 MSS NTD samples) and seven non-NTD samples were detected by BSP, sub-cloning, and sequencing. The hMSH2 promoter sequence was not methylated, in agreement with the MSP analysis. The hMLH1 promoter showed two specific methylation patterns at ‘4,5 CpG' sites (upstream to downstream from the 4,5th CpG sites in the target region) in some NTD samples, but no significant difference was seen in the methylation patterns between the NTD and control groups (see Table 3).
Table 3. Methylation status of the hMLH1 and hMSH2 promoter regions.
| hMLH1 promoter | hMSH2 promoter | ||||
|---|---|---|---|---|---|
| Group | Unmethylated | Methylated 4,5 CpG sites | Unmethylated | Methylated sites | Total |
| Control | 2 | 5 | 7 | 0 | 7 |
| NTDs (MSI−a) | 2 | 7 | 9 | 0 | 9 |
| NTDs (MSI+a) | 3 | 10 | 13 | 0 | 13 |
MSI+, MSI-positive NTDs; MSI−, microsatellite stability NTDs.
The methylation patterns of IGF2 and H19
The human IGF2 harbors two DMRs, a DMR0 that is located at the P0 promoter and an analog DMR2 at exon-9. As the H19 DMR1 and the IGF2 DMR0 are primary regulators of H19 and IGF2 gene expression, we therefore analyzed their methylation status by BSP, sub-cloning, and sequencing to determine whether DNA methylation changes were associated with NTDs.
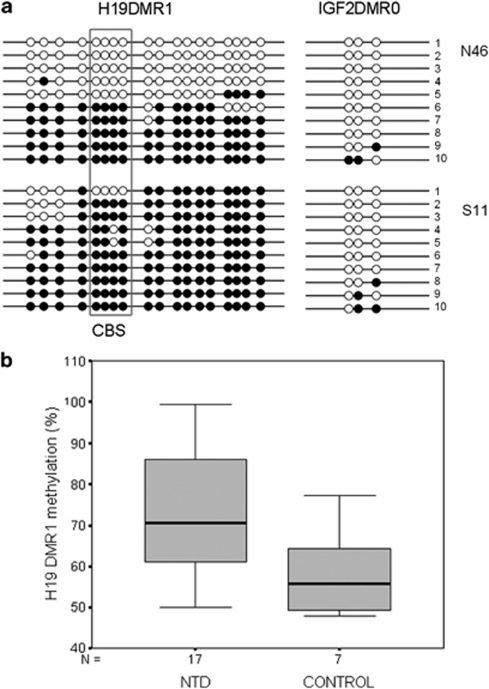
A total of 17 NTD samples were analyzed by BSP, sub-cloning, and sequencing for analysis of the methylation status of the H19 DMR1 and IGF2 DMR0. The methylation levels of H19 DMR1 in the NTD and control groups are 73.3%±15.9 and 58.3%±11.2, respectively (see Figure 2). The methylation level of NTD group was higher than that of control group (Student's t-test, P<0.05). The normal pattern observed at the H19 DMR1 and the IGF2 DMR0 is approximately 50% methylation.24 We defined normal methylation pattern as 45–60% in our study, meaning that methylation levels higher than 60% or lower than 45% were considered to represent aberrant methylation. Of the 17 NTD patients, 14 showed hyper-methylation of DMR1 (14/17), and two samples showed hyper-methylation out of seven control samples (2/7). Figure 2a shows a representative result of S11 and N46. Among the 14 NTD patients who showed hyper-methylation of DMR1 (>60%), eight showed MSI (2 patients were MSI-H and 6 patients were MSI-L; 8/14); in the three NTD patients who did not show hyper-methylation of DMR1 (45–60%), one NTD patient showed MSI (all of them were MSI-L, 1/3). The incidence of MSI (especially MSI-H) tended to be higher in NTD patients with hyper-methylation of DMR1 than in NTD patients with no DMR1 hyper-methylation. The methylation status of the H19 DMR1 in NTD samples appeared to vary substantially, as 6 of 17 patients had methylation levels in excess of 80%, whereas the control group methylation status changed very little, with most levels at 45–60%.
Figure 2.
The methylation levels of H19 DMR1 and IGF2 DMR0 in the NTD and control groups. (a) Methylation status analysis of the H19 DMR1 and the IGF2 DMR0 by BSP, sub-cloning, and sequencing in two samples. Each line represents a separate clone. CpG sites marked by a box indicate the CTCF-binding site. The methylation pattern of each allele was analyzed using the online soft QUMA. The methylation levels of H19 DMR1 in N46 and S11 are 49.9% and 90.0%, respectively. •, methylated CpG sites; ○, unmethylated CpG sites; N46, control; S11, NTDs. (b) Box plot showing mean methylation levels between NTD and control samples for H19 DMR1 methylation analysis by BSP, sub-cloning, and sequencing. The bottom and top of the box are the 25th and 75th percentile (the lower and upper quartiles, respectively), and the band near the middle of the box is the 50th percentile (the median). The ends of the whiskers represent 5th and 95th percentile. The methylation level of NTD group was higher than that of the control group (Student's t-test, P<0.05).
IGF2 DMR0 methylation level was detected in NTD and control samples by BSP, sub-cloning, and sequencing. The methylation level was approximately 10% in both groups. There is no significant difference between the two groups. The results did not show that the IGF2 DMR0 was associated with the hyper-methylation status of the H19 DMR1 in NTD samples.
Discussion
NTDs are caused by a failure in neural tube closure during prenatal development. However, the pathogenesis of the NTDs remains unclear. In this study, we have explored the role of genomic instability and epigenetic alterations in the gene regulation associated with NTDs. Tissue samples from patients were collected from Shanxi Province in China, which has been identified as a region with a high occurrence of NTDs.25
MSI is one marker of genomic instability, reflecting the status of the MMR genes. MSI presents in many kinds of tumors, especially in HNPCC. In this study, we assessed the MSI of NTD samples. In total, 23 of the 50 NTD samples were MSI-positive, for a positive rate of 46%. Of these samples, five were MSI-H. The remaining 18 samples were MSI-L, with single-site MSI. Genomic instability clearly existed in the NTD samples, suggesting a defect in MMR function. Importantly, Bat25, Bat26, and Bat34C4 are located within intron-16 of the c-kit oncogene, intron-5 of the hMSH2, and the 3′-untranslated region of exon-11 of P53,26, 27 respectively. D2S123 and D2S119 surround hMSH2 and hMLH1, whereas D3S1611 is located within hMLH1.28, 29 Length changes in these microsatellites might serve as expression-regulating factors, affecting the activity of these genes.
Our previous research suggested that NTD samples had low folic acid and vitamin B12 content, and the fact that the whole genome of NTD samples showed a low global methylation status indicated that a one-carbon metabolism disorder may affect the DNA methylation in NTD samples. We investigated the methylation status of the hMLH1 and hMSH2 genes to determine whether defects in the MMR pathway were associated with human NTDs. hMLH1 and hMSH2, the major active components of the MMR pathway, not only participate in DNA MMR, but also function in cell apoptosis and cell-cycle regulation.30 Mutations to and aberrant methylation of hMLH1 and hMSH2 have been proposed as contributing factors for some cancers.31 MSI is observed in approximately 15% of colorectal, gastric, and endometrial cancers, and, in lower frequencies, in a minority of other tumors, where it is associated with hyper-methylation of the promoter region of hMLH1.32 In our study, the hMSH2 promoter region was unmethylated in the NTD samples. Conversely, the hMLH1 promoter region showed a specific methylation pattern in the ‘4,5 CpG' sites in some NTD patients, but no significant difference was seen between the two groups. Nevertheless, we cannot exclude the existence of other CpG methylation sites that may regulate hMLH1 and hMSH2 gene expression, or defects in other MMR genes that may also lead to MSI. The relations between MMR and NTDs still need further exploration.
The H19 and IGF2 genes share enhancers located downstream from H19.33 In both mice and humans, H19 and IGF2 are broadly expressed during embryonic development and are postnatally downregulated in most tissues. H19 encodes a fully processed 2.3-kb non-coding RNA and was initially implicated as a tumor suppressor. IGF2 encodes a protein involved in promoting embryonic and placental growth, and development.34 Disturbances of methylation in the H19 germline DMR are known to lead to dysregulation of H19/IGF2 imprinting, causing intrauterine growth retardation (Silver–Russell syndrome) and overgrowth (Beckwith–Wiedemann syndrome),35 respectively. Biallelic expression of IGF2 has been reported in a number of cancer types. Methylation changes of the H19 DMR have been associated with loss of imprinting (LOI) of IGF2. The aberrant methylation at this site correlates with LOI in Wilms' tumors, bladder cancer, and colon cancer, as well as chronic myelogenous leukemia.36 Researchers have studied the methylation changes at the H19/IGF2 locus in congenital growth disorders and cancer. They conclude that mechanisms of imprinting loss in neoplastic and non-neoplastic cells may be different.37 Wang et al38 researched the relation between hypo-methylation of long interspersed nucleotide elements (LINE-1) and risk of neural tube defects, and found that methylation the levels of genomic DNA and LINE-1 decreased significantly in the neural tissue of NTD samples. Hypo-methylation of LINE-1 is thought to be connected to genomic instability and endogenous DNA double-strand breaks in some cancers. In the current study, we found MSI present in NTD samples; thus, decreased genomic stability is likely to contribute to the development of NTDs.
For the purpose of researching the relationship between imprinted genes and NTDs, we selected the H19 DMR1 and IGF2 DMR0 as the objective regions to conduct BSP, sub-cloning, and sequencing. We found the H19 DMR1 methylation level of NTD group was significantly higher than that of the control group (Student's t-test, P<0.05), whereas the IGF2 DMR0 showed similar methylation level in both groups, no significant difference was seen. These results indicated that the mechanisms that cause imprinting defects at the IGF2/H19 locus in NTDs may be different from the enhancer-competition model. The methylation status of IGF2 DMR0 may be independent of H19 DMR1. Studies showed that some DMRs are marked by allelic covalent modifications on histones as well39 (ie, histone methylation and acetylation), and that DNA methylation is not the sole determinant of imprints. One study indicates that IGF2 DMR0 hypo-methylation occurs as an acquired tissue-specific somatic event, rather than a constitutive innate epimutation in tumor tissues.40 It should be important to further explore the mechanisms that regulate the imprinting of the IGF2/H19 in the NTDs.
As hyper-methylation of DMR1 and genomic instability co-existed in NTD samples, they may correlate with the occurrence of NTDs. The methylation status of the H19 DMR1 in the NTD samples appeared to vary substantially, as 6 of 17 patients had methylation levels in excess of 80%, whereas the control group methylation status changed very little, with most levels at 45–60%. This result implied that the H19 DMR1 hyper-methylation status may be a marker for DNA methylation disorders.
The closing of the neural tube is a complex event that can be influenced by both genes and the environment. The interactions between genetic and environmental factors are now being untangled. In our study, MSI was observed in some NTD patients, which suggests that genomic instability and MMR defects occur in fetuses with NTDs. The hyper-methylation status of the H19 DMR1 indicated that abnormal expression of H19 may affect the occurrence of NTDs. Methylation abnormalities may, therefore, be pathological factors in NTDs. Genomic instability, the MMR pathway, and the imprinting genes IGF2/H19 are likely to exert combined effects on the pathogenesis of NTDs. Future investigation is needed for a better understanding of the mechanisms underlying the occurrence of NTDs.
Acknowledgments
This work was supported by the National Basic Research Program of China ‘973' (2007CB511903), the National Natural Science Foundation of China (30671156), and Beijing Natural Science Foundation (5072014).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Greene ND, Stanier P, Copp AJ. Genetics of human neural tube defects. Hum Mol Genet. 2009;18:113–129. doi: 10.1093/hmg/ddp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZW, Ren AG, Zhang L, et al. Prevalence of major external birth defects in high and low risk areas in China, 2003. Zhonghua Liu Xing Bing Xue Za Zhi. 2005;26:252–257. [PubMed] [Google Scholar]
- Malcoe LH, Shaw GM, Lammer EJ, Herman AA. The effect of congenital anomalies on mortality risk in white and black infants. Am J Public Health. 1999;89:887–892. doi: 10.2105/ajph.89.6.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp AJ, Greene ND. Genetics and development of neural tube defects. J Pathol. 2010;220:217–230. doi: 10.1002/path.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MRC Vitamin Study Research Group Prevention of neural tube defects: results of the Medical Research Council Vitamin Study. MRC Vitamin Study Research Group. Lancet. 1991;338:131–137. [PubMed] [Google Scholar]
- Grosse SD, Collins JS. Folic acid supplementation and neural tube defect recurrence prevention. Birth Defects Res. 2007;79:737–742. doi: 10.1002/bdra.20394. [DOI] [PubMed] [Google Scholar]
- Burren KA, Savery D, Massa V, et al. Gene–environment interactions in the causation of neural tube defects: folate deficiency increases susceptibility conferred by loss of Pax3 function. Hum Mol Genet. 2008;17:3675–3685. doi: 10.1093/hmg/ddn262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- Li YC, Korol AB, Fahima T, Nevo E. Microsatellites within genes: structure, function, and evolution. Mol Biol Evol. 2004;21:991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- Hui J, Hung LH, Heiner M, et al. Intronic CA-repeat and CA-rich elements: a new class of regulators of mammalian alternative splicing. EMBO J. 2005;24:1988–1998. doi: 10.1038/sj.emboj.7600677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay JA, Williams EA, Mathers JC. Folate and DNA methylation during in utero development and aging. Biochem Soc Trans. 2004;32 (Part 6:1006–1007. doi: 10.1042/BST0321006. [DOI] [PubMed] [Google Scholar]
- Salvesen HB, MacDonald N, Ryan A, et al. Methylation of hMLH1 in a population-based series of endometrial carcinomas. Clin Cancer Res. 2000;6:3607–3613. [PubMed] [Google Scholar]
- Riccio A, Sparago A, Verde G, et al. Inherited and sporadic epimutations at the IGF2-H19 locus in Beckwith–Wiedemann syndrome and Wilms' tumor. Endocr Dev. 2009;14:1–9. doi: 10.1159/000207461. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20:63–68. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Cao Z, Song JH, Kim CJ, et al. Genetic and epigenetic analysis of the VHL gene in gastric cancers. Acta Oncol. 2008;47:1551–1556. doi: 10.1080/02841860802001459. [DOI] [PubMed] [Google Scholar]
- Loukola A, Eklin K, Laiho P, et al. Microsatellite marker analysis in screening for hereditary nonpolyposis colorectal cancer (HNPCC) Cancer Res. 2001;61:4545–4549. [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Rashid A, Lee JH, Kim SG, Hamilton SR, Wu TT. Frequent CpG island methylation in serrated adenomas of the colorectum. Am J Pathol. 2003;162:815–822. doi: 10.1016/S0002-9440(10)63878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008;36 (Web Server issue:W170–W175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil SG, Riksen NP, Boers GH, Smulders Y, Blom HJ. DNA methylation status is not impaired in treated cystathionine beta-synthase (CBS) deficient patients. Mol Genet Metab. 2007;91:55–60. doi: 10.1016/j.ymgme.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62:6442–6446. [PubMed] [Google Scholar]
- Sparago A, Cerrato F, Vernucci M. Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith–Wiedemann syndrome. Nat Genet. 2004;36:958–960. doi: 10.1038/ng1410. [DOI] [PubMed] [Google Scholar]
- Gu X, Lin L, Zheng X, et al. High prevalence of NTDs in Shanxi province: a combined epidemiological approach. Birth Defects Res A Clin Mol Teratol. 2007;79:702–707. doi: 10.1002/bdra.20397. [DOI] [PubMed] [Google Scholar]
- Deschoolmeester V, Baay M, Wuyts W, et al. Detection of microsatellite instability in colorectal cancer using an alternative multiplex assay of quasi-monomorphic mononucleotide markers. J Mol Diagn. 2008;10:154–159. doi: 10.2353/jmoldx.2008.070087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana-Pereira M, Almeida I, Sousa S, et al. Analysis of microsatellite instability in medulloblastoma. Neuro Oncol. 2009;11:458–467. doi: 10.1215/15228517-2008-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Del-Campo A, Salinas-Sánchez AS, Sánchez-Sánchez F, et al. Implications of mismatch repair genes hMLH1 and hMSH2 in patients with sporadic renal cell carcinoma. BJU Int. 2008;102:504–509. doi: 10.1111/j.1464-410X.2008.07581.x. [DOI] [PubMed] [Google Scholar]
- Stuckless S, Parfrey PS, Woods MO, et al. The phenotypic expression of three MSH2 mutations in large Newfoundland families with Lynch syndrome. Fam Cancer. 2007;6:1–12. doi: 10.1007/s10689-006-0014-8. [DOI] [PubMed] [Google Scholar]
- Köster F, Schröer A, Fischer D, Greweldinger T, Diedrich K, Friedrich M. Correlation of DNA mismatch repair protein hMSH2 immunohistochemistry with p53 and apoptosis in cervical carcinoma. Anticancer Res. 2007;27:63–68. [PubMed] [Google Scholar]
- Gazzoli I, Loda M, Garber J, Syngal S, Kolodner RD. A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res. 2002;62:3925–3928. [PubMed] [Google Scholar]
- Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis. 2008;29:673–680. doi: 10.1093/carcin/bgm228. [DOI] [PubMed] [Google Scholar]
- Demars J, Shmela ME, Rossignol S, et al. Analysis of the IGF2/H19 imprinting control region uncovers new genetic defects, including mutations of OCT-binding sequences, in patients with 11p15 fetal growth disorders. Hum Mol Genet. 2010;19:803–814. doi: 10.1093/hmg/ddp549. [DOI] [PubMed] [Google Scholar]
- Chao W, D′Amore PA. IGF2: epigenetic regulation and role in development and disease. Cytokine Growth Factor Rev. 2008;19:111–120. doi: 10.1016/j.cytogfr.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LE, Fernandes K, McEvoy TG, et al. Epigenetic change in IGF2R is associated with fetal overgrowth after sheep embryo culture. Nat Genet. 2001;27:153–154. doi: 10.1038/84769. [DOI] [PubMed] [Google Scholar]
- Byun HM, Wong HL, Birnstein EA, et al. Examination of IGF2 and H19 loss of imprinting in bladder cancer. Cancer Res. 2007;67:10753–10758. doi: 10.1158/0008-5472.CAN-07-0329. [DOI] [PubMed] [Google Scholar]
- Murrell A, Ito Y, Verde G, et al. Distinct methylation changes at the IGF2-H19 locus in congenital growth disorders and cancer. PLoS One. 2008;3:e1849. doi: 10.1371/journal.pone.0001849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang F, Guan J, et al. Relation between hypomethylation of long interspersed nucleotide elements. Am J Clin Nutr. 2010;91:1359–1367. doi: 10.3945/ajcn.2009.28858. [DOI] [PubMed] [Google Scholar]
- Delaval K, Feil R. Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev. 2004;14:188–195. doi: 10.1016/j.gde.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Ito Y, Koessler T, Ibrahim AE, et al. Somatically acquired hypomethylation of IGF2 in breast and colorectal cancer. Hum Mol Genet. 2008;17:2633–2643. doi: 10.1093/hmg/ddn163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.