Abstract
X-linked hypohidrotic ectodermal dysplasia (XHED), an inherited disease recognized in humans, mice, and cattle, is characterized by hypotrichosis, a reduced number or absence of sweat glands, and missing or malformed teeth. In a subset of affected individuals and animals, mutations in the EDA gene (formerly EDI), coding for ectodysplasin, have been found to cause this phenotype. Ectodysplasin is a homotrimeric transmembrane protein with an extracellular TNF-like domain, which has been shown to be involved in the morphogenesis of hair follicles and tooth buds during fetal development. Some human XHED patients also have concurrent immunodeficiency, due to mutations in the NF-κB essential modulator protein (IKBKG; formerly NEMO), which is also encoded on the X chromosome. In a breeding colony of dogs with XHED, immune system defects had been suspected because of frequent pulmonary infections and unexpected deaths resulting from pneumonia. To determine if defects in EDA or IKBKG cause XHED in the dogs, linkage analysis and sequencing experiments were performed. A polymorphic marker near the canine EDA gene showed significant linkage to XHED. The canine EDA gene was sequenced and a nucleotide substitution (G to A) in the splice acceptor site of intron 8 was detected in affected dogs. In the presence of the A residue, a cryptic acceptor site within exon 9 is used, leading to a frame shift and use of a premature stop codon that truncates the translation of both isoforms, EDA-A1 and EDA-A2, resulting in the absence of the TNF-like homology domain, the receptor-binding site of ectodysplasin.
Introduction
In X-linked hypohidrotic ectodermal dysplasia (XHED) in man (Mendelian inheritance in man (MIM) 305100), affected individuals have a developmental disorder characterized by sparse or absent hair, missing and/or malformed teeth, and hypoplastic eccrine glands (Beahrs et al. 1971; Soderholm and Kaitila 1985; Clarke 1987; Clarke et al. 1987; Kere et al. 1996). There is significant morbidity and mortality in affected children as a result of hyperthermia resulting from their inability to sweat, combined with an increased risk of respiratory tract infections (Beahrs et al. 1971; Soderholm and Kaitila 1985; Clarke et al. 1987; Gilgenkrantz et al. 1989). The human X-linked HED phenotype has been shown to be a result of mutations in the ectodysplasin (EDA; formerly EDI) gene (Zonana et al. 1993; Kere et al. 1996; Monreal et al. 1998).
More recently, a clinical syndrome of XHED associated with immune deficiency has been described (HED-ID; MIM 300291) (Zonana et al. 2000; Döffinger et al. 2001; Jain et al. 2001; Kosaki et al. 2001). All of the affected children were male and had phenotypic features of HED. However, they also had dysgammaglobulinemia and severe, recurrent infections with significant morbidity and mortality, but without obvious immunologic defects (Zonana et al. 2000; Döffinger et al. 2001). Because of the similarities to incontinentia pigmenti (IP; MIM 308310), an X-linked disorder affecting the development of skin, teeth, eyes, hair, and the central nervous system, a defect in IKBKG was suspected. Severe loss-of-function defects in IKBKG have been shown to cause a lethal form of IP in male fetuses and in affected carrier females with skewed X-inactivation (Smahi et al. 2000). Most of the mutations responsible for IP result in the loss of the carboxyl terminal portion of the IKBKG protein that is encoded by exons 4–10 of the gene. Therefore, defects that rendered a mildly truncated protein product were hypothesized to result in HED-ID (Zonana et al. 2000). Many mutations causing HED-ID were subsequently found and have all been located in the last exon (exon 10) of the gene for IKBKG (Zonana et al. 2000).
The EDA gene has been shown to produce eight different transcripts, with those producing isoforms EDA-A1 and EDA-A2 being the longest (Bayes et al. 1998). Each of these two isoforms binds to a specific receptor, EDAR and EDA2R (formerly XEDAR), respectively (Yan et al. 2000). EDAR is autosomal while EDA2R, specific for EDA-A2, is X-linked. Therefore, EDA2R can be considered a possible candidate for XHED. However, to date, no mutations in the EDA2R gene have been identified in cases of XHED in humans or other species.
We have established a colony of dogs with XHED for the study of disease mechanisms and therapeutic trials (Casal et al. 1997, 2004). The affected dogs lack all sweat glands and secondary hairs and are completely hairless on their forehead and over the dorsal pelvic area. Most premolars and some incisors are missing and those teeth that are present are mostly conically shaped. As in human XHED, there is increased morbidity and mortality from usually benign and rarely fatal pulmonary infectious diseases among XHED dogs compared with other dogs in the same environment. Most affected dogs have chronic nasal and ocular discharge, often accompanied by corneal ulceration, and a small number of the adult dogs had chronic, treatment-resistant demodecosis (a canine skin parasite) that is associated with a mildly compromised immune system (Caswell et al. 1997). Because of the suspicion of immunodeficiency in the XHED dogs, both EDA and IKBKG were considered as candidate genes for the defect in the XHED dog. We used linkage analysis and cDNA and gene sequencing to identify a mutation in the EDA gene in the XHED dogs.
Materials and methods
Dogs
Blood and tissue samples were obtained from dogs with X-linked ectodermal dysplasia and from and their normal littermates, parents, and grandparents (Casal et al. 1997, 2004). Dogs were raised in the animal colony of the School of Veterinary Medicine, University of Pennsylvania, under National Institutes of Health (NIH) and U.S. Department of Agriculture (USDA) guidelines for the care and use of animals in research.
Linkage analysis
Four polymorphic markers, FH2548, FH3027, FH2584, and F8C, located on the canine X chromosome (CFAX) (Breen et al. 2001) were analyzed in 27 affected dogs and 43 normal relatives. IRDye™ labeled primers (MWG Biotech AG, High Point, NC) were used to produce the polymerase chain reaction (PCR) products that were electrophoresed on a NEN® Global IR2 4200 Sequencer System (Li-cor®, Lhicohi, NE). The genotypes were then determined using SAGA2.1 (Li-cor®) software. Primer sequences and annealing temperatures are available at http://www.fhcrc.org/science/dog_genome/breen2001/breenmaps_data/cfax.html and PCR conditions were described previously (Werner et al. 2004). Linkage analysis was performed using the computer program FASTLINK (Lathrop et al. 1984; Cottingham et al. 1993; Schaffer et al. 1994).
cDNA synthesis and RT-PCR
Total RNA was extracted from skin and kidney from normal and affected dogs using TRIzol reagent (Life Technologies, Grand Island, NY) according to the manufacturer’s protocol. cDNA was synthesized in a 50-μl reaction containing 10 μg total RNA 1 × first-strand synthesis buffer (Invitrogen, Carlsbad, CA), 0.1 mM DTT (Invitrogen), 8 mM dNTPs (Promega, Madison, WI), 0.2 μg/μl BSA (NEB), 8 μM random hexamers (Promega), 8 μM oligo d(T) (Promega), 80 U RNAsin (Promega), and 500 U Superscript II reverse transcriptase (Invitrogen). PCRs were carried out according to the protocols described below, using 1 μl of cDNA as the template for each reaction.
EDA and IKBKG sequence and mutation analysis
Canine EDA and IKBKG gene sequences were retrieved from the 1.5 × poodle genome sequence (Kirkness et al. 2003) and the boxer whole-genome sequences deposited in the Trace Archive (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi) using human and bovine EDA and IKBKG cDNA sequences (GenBank accession Nos. AF040628, AJ300468, AJ300469, AJ278907, AJ271718, and NM174354) as queries. Retrieved sequences were then verified to be from the orthologous gene by BLAST searches (http://www.ncbi.nlm.nih.gov/blast/) of the human genome showing highest homology to the appropriate human gene. PCR primers were designed to amplify cDNA derived from skin and kidney mRNA and to amplify exons from genomicDNA. The sequences of the final PCR primers used to sequence portions of the IKBKG gene and the EDA gene are shown in Tables 1 and 2. PCR reactions containing 50 ng of canine genomic DNA or 1 μl of cDNA reaction as template, 0.2 μMof each primer, 50mMKCl, 10mMTris-HCl (pH 8.3), 3 mM MgCl2, 0.2 mM dNTPs, and 0.5 U Ampli-Taq polymerase (Perkin-Elmer Cetus, Norwalk, CT) were carried out in a volume of 50 μl. Reactions were denatured for 5 min at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at the annealing temperature, and 0.5–2 min at 72°C. For the first five cycles the annealing temperature was reduced by 1°C/cycle, ending at the annealing temperature (Ta) for the remainder of the reaction cycles (Tables 1 and 2). PCR products were purified after gel electrophoresis using the QIAquick gel extraction system (Qiagen, Vaglencia, CA), and were sequenced by the facility at the University of Pennsylvania Abramson Cancer Center.
Table 1.
Primers used to sequence coding regions of exons 9 and 10 of canine IKBKG
| Exon | Primer name | Sequence (5′-3′) | Length (bp) | Ta (°C) | Product (bp) |
|---|---|---|---|---|---|
| 9+10 | i8F | TCC CCT CCG TGC ATA CTT TTG AT | 23 | 63 | 517 |
| u10R | CGA GAG GAG GGC GTG CCA GG | 20 | 69 |
The 5 primer is within intron 8 and the 3 primer within the UTR of exon 10.
Table 2.
Primers used to amplify and sequence all coding exons of canine EDA
| Exon | Primer name | Sequence (5′–3′) | Length (bp) | Ta (°C) | Product (bp) |
|---|---|---|---|---|---|
| 1 | 5′ UTR-F | CCG ATG GCA GGG CAG TAG C | 19 | 61 | 452 |
| i1R | AGT GCG GTT TCT TCG CTT CC | 20 | 59 | ||
| 3 | i2F | ATG GGC TCA GGT TTT AGA CA | 20 | 55 | 421 |
| i3R | AGG GTT GG AGG AGA GAG G | 19 | 56 | ||
| 4 | i3F | GCA GTG TCT TGA GAG TTT CT | 20 | 58 | 165 |
| i4R | TTA GGA AAG ATA TGA CAG CA | 20 | 54 | ||
| 5 | i4F | GGA TAG GGA GTG TGT GTG TAT GTT | 24 | 70 | 471 |
| i5R | CCT GGG AGA CCT GCT TTT CTT ATT | 24 | 70 | ||
| 6 | i5F | GCT GTG AGT GAA CAA CCT | 18 | 56 | 285 |
| i6R | TAA GAG AAG TGA GTG GTG TC | 20 | 51 | ||
| 7 | i6F | CCT AGG CTG TGA TTC TTT G | 19 | 54 | 337 |
| i7R | GAC AGG AGA AAG GGA TCA | 18 | 58 | ||
| 8 | i7F | CCG CTY GAC AAA CAG AAT A | 19 | 53 | 448 |
| i8R | CCT TTC CAC TCC CTC CC | 17 | 55 | ||
| 9 | 8F | TGG AGT GCT CAA TGA CTG G | 19 | 58 | 2386 |
| 3′ UTR-R | AGT CAC TGG GGA ATA AAT AG | 20 | 56 | 524* |
Primers that were designed to lie within intronic sequences are designated by the “i” in front of the primer name.
Amplification product using cDNA.
To determine the sequences of alternatively spliced EDA mRNA molecules produced in affected and carrier dogs, PCR products were cloned using the TOPO TA Cloning Kit (Invitrogen) according to the manufacturer’s specifications. Plasmid DNA was purified by standard phenol-chloroform extraction methods and submitted for sequencing.
Results
Linkage analysis
Five polymorphic markers evenly distributed on the canine X chromosome (CFAX) were used to determine the relative chromosomal location of the canine XHED gene. The EDA gene is located near the centromere of CFAX, whereas IKBKG is located at the distal end of the long arm (http://genome.ucsc.edu). The marker F8C, closest to the IKBKG gene, showed the lowest LODmax score to XHED (0.06), while FH3027 displayed the highest LODmax score (2.59), which is considered statistically significant evidence of linkage because of its location on the X chromosome (Table 3).
Table 3.
Linkage analysis results
| Marker/Gene | Mb position | LODmax score | Recombination fraction |
|---|---|---|---|
| FH2548 | 30.4 | 0.89 | 0.20 |
| FH3027 | 40.9 | 2.59 | 0.06 |
| EDA2R | 54.1 | ||
| EDA | 56.7 | ||
| FH2584 | 103.9 | 1.65 | 0.13 |
| IKBKG | 125.3 | ||
| F8C | 125.9 | 0.06 | 0.38 |
Megabase (Mb) positions of markers and genes on the canine X chromosome based on the first canine genome assembly are indicated. LODmax score values and recombination fraction of polymorphic markers to XHED found in XHED dogs were calculated.
Partial IKBKG sequence
In humans, mutations in exon 10 of the IKBKG gene are found in immunodeficient XHED males, whereas mutations upstream of exon 9 are not compatible with life in males. Therefore, canine IKBKG exons 9–10 were amplified and sequenced (Table 1). There was no difference in the sequences derived from normal and affected dogs.
EDA sequence analysis
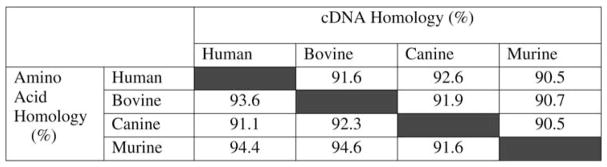
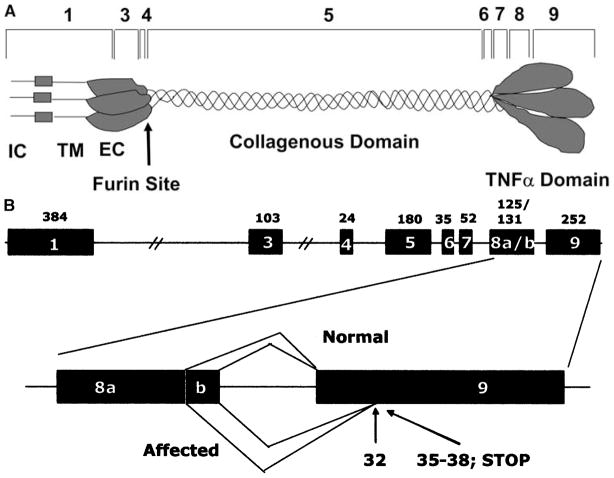
The availability of EDA cDNA sequences from several mammalian species combined with canine genome sequences permitted the design of PCR primers to amplify all of the exons, including splice sites, of the canine EDA gene. All exons were sequenced and the predicted canine EDA cDNA open reading frame (ORF) was compared to human, cattle, and mouse. Interestingly, exons 1 and 3 were shorter by 12 and 3 bp, respectively, in the dog compared to the other species. Thus, the ORF was 1161 nucleotides long for isoform EDA-A1 and 1155 nucleotides long for isoform EDA-A2. [These two isoforms differ by 6 bp (2 amino acids) because of alternative use of splice donor sites at the 3′ of exon 8.] As in cattle and mice, no exon 2 was found. The protein encoded by the deduced mRNA is 386 amino acids long. Comparison of human cDNA to the other three species revealed the highest degree of homology to canine cDNA (Fig. 1). However, when the human protein was compared to that in cattle, mouse, and dog, the murine protein showed the highest degree of homology. Among all three species the cattle and mice shared the most amino acids, and the canine protein was most similar to that in cattle (Fig. 1). Most of the differences in amino acids were found in the N-terminus of the protein, whereas there were 100% identical residues in the protein encoded by the last three exons in all four species. These exons encode the TNF-like domain, the protein’s active site (Fig. 2a).
Fig. 1.
Percent similarity of cDNA and amino acid sequences of EDA from cattle, dogs, mice, and humans. Exons 7–9 are completely homologous among dogs, cattle, mice, and humans (not shown).
Fig. 2.
(A) The homotrimeric protein encoded by EDA. The amino acid sequence expressed by exons 7–9 is 100% homologous in mice, dogs, cattle, and humans. IC: intracellular, TM: transmembrane, EC: extracellular domains. (B) Genomic organization of EDA in dogs and the effect of the mutation. A cryptic splice acceptor site within exon 9 is used in the affected dogs, resulting in a frame shift and a premature stop codon. The numbers above the figure indicate exon sizes and those below indicate the position within the exon.
Canine EDA mutation analysis
Sequencing the EDA exons from an affected dog revealed a point mutation (G → A) in the conserved AG consensus sequence of the splice acceptor site in intron 8. The mutation was confirmed to be present in 25 affected and absent in 25 normal, related dogs. To examine how this single nucleotide change affected splicing, we attempted to sequence the cDNA derived from normal and affected dogs by PCR amplification. However, the PCR products from normal and affected dogs could not be completely sequenced because the alternative splicing that had been shown to occur in other species also occurs in the dog, giving rise to PCR products that differ by 6 bp because of the two different isoforms (exons 8a and 8b). Therefore, the PCR products from normal and affected dogs were cloned and sequenced from the single clones. Sequence analysis of cDNA clones derived from the affected dogs revealed the use of a cryptic splice acceptor site 32 bp from the 5′ end of exon 9, resulting in a frame shift and a premature stop codon 3 bp downstream (Fig. 2b).
Discussion
To establish which of the over 150 different forms of ectodermal dysplasia that have been described to date (Pinheiro and Freire-Maia 1994) was present in our dog model (Casal et al. 1997, 2004), we first determined the X-linked mode of inheritance by performing breeding experiments (Casal et al. 1997). The most common form of X-linked ectodermal dysplasia (XHED) is caused by a defect in EDA (MIM 305100), but there are several other ectodermal dysplasias with the disease-causing mutation in a gene on the X chromosome, such as XHED with immunodeficiency (HED-ID; MIM 300291), BRESHECK syndrome (for the presence of brain anomalies, mental retardation, ectodermal dysplasia, skeletal malformations, Hirschsprung disease, ear and eye anomalies, cleft palate, cryptorchidism, and kidney dysplasia or hypoplasia; MIM 300404), and OLED-AID syndrome (osteopetrosis, lymphedema, anhidrotic ectodermal dysplasia (EDA), and immunodeficiency; MIM 300301).
While our XHED dogs described in this study had classic signs of X-linked ectodermal dysplasia, they had no other signs that would be suggestive of either BRESHECK syndrome or OLEDAID. However, the affected dogs did exhibit clinical signs that could be attributed to an immunodeficiency as in HED-ID (Casal et al. 2003), a clinical syndrome which has been described in children (Zonana et al. 2000; Döffinger et al. 2001; Jain et al. 2001; Kosaki et al. 2001). Defects that render a mildly truncated IKBKG protein product were suspected to result in HED-ID and were subsequently found in the last exon (exon 10) of the IKBKG gene.
EDA2R, the receptor for the EDA-A2 isoform of EDA, also resides on the X chromosome and could potentially be considered as a cause for XHED. However, no naturally occurring mutations causing XHED have been found in the EDA2R gene to date and a recently created EDA2R knockout mouse showed no phenotypic differences compared to wildtype littermates (Newton et al. 2004). In addition, only the EDA-A1 isoform has been shown to be necessary for the development of hair and sweat glands in Tabby mice, a mouse model for XHED (Srivastava et al. 2001). EDA-A1 binds to the EDAR receptor (Elomaa et al. 2001) and not to EDA2R, thus suggesting that mutations in the EDA2R gene are less likely to cause the XHED phenotype. For these reasons, we chose to examine EDA and IKBKG as potential candidates for XHED in our dogs. Linkage analysis placed the XHED locus closer to EDA than to IKBKG. IKBKG was further excluded by determining that the last two exons of canine IKBKG were identical in both normal and affected dogs. Mutations causing HED-ID in humans have been found only in exon 10, and mutations upstream of exon 10 are generally not compatible with life in male fetuses. Thus, EDA became the most likely candidate causing XHED in our dogs.
The normal canine EDA gene has ORFs of 1161 and 1155 bp (EDA-A1 and EDA-A2 isoforms) and contains 8 exons. The exons are conventionally named 1–9, but, as in cattle and mice, exon 2 is missing in the dog. However, unlike humans, cattle, and mice in which the exon lengths are conserved, exons 1 and 3 of the dog gene are shorter by 12 and 3 bp, respectively, than their counterparts. The missing base pairs in exon 1 appear to be in the sequence coding for the protein’s transmembrane domain, and those in exon 3 appear to be in the sequence coding for the extracellular domain but 5′ to the sequence for the putative furin cleavage site. These findings suggest that conservation of these two domains is of less importance and that the differences have no impact on proper function of the protein. On the other hand, the TNF-like domain is 100% conserved at the amino acid level among all species examined, highlighting its importance as the effector portion of the protein, which is cleaved from its anchor in the cell membrane by the furin proteases.
In humans, a number of mutations in EDA have been found that cause XHED. About half of the mutations are missense mutations of which most are located in the sequences coding for (1) the putative transmembrane/extracellular junction domain; (2) the furin cleavage site, where furin-like proteases act to release the active portion of ectodysplasin; (3) the collagenous domain, which is thought to be necessary for trimerization; and (4) in the TNF-like domain, which is the protein’s active domain (Paakkonen et al. 2001; Schneider et al. 2001). These mutations do not alter the overall structure of ectodysplasin but impair the functional domains. The rest of the mutations appear to be randomly placed and usually result in a frame shift with a premature stop codon (Paakkonen et al. 2001; Schneider et al. 2001). In the Tabby mouse, two separate mutations have been found, both in exon 1. In one strain (Ta), a 132-bp in-frame deletion leading to a truncated protein has been described, and in the other strain (Tabby <6J>), a 1-bp deletion was found resulting in a frame shift and premature stop codon. Many similar mutations in exon 1 have been described in humans (Kere et al. 1996; Bayés et al. 1998; Ferguson et al. 1998; Hertz et al. 1998; Paakkonen et al. 2001; Schneider et al. 2001). In black and white German Holstein cattle with XHED, a large genomic deletion that included all of exon 3 caused a frame shift and a premature stop codon resulting in a severely truncated ectodysplasin (Drögemüller et al. 2001). In humans, a single case of XHED with the complete deletion of exon 3 has been described (Bayes et al. 1998); the rest of the cases involving exon 3 are the result of point mutations (Bayés et al. 1998; Monreal et al. 1998; Chen et al. 2001; Paakkonen et al. 2001; Schneider et al. 2001).
The mutation found in canine EDA confirmed its involvement in XHED in our dog model. The canine mutation results in the use of a cryptic splice acceptor site within exon 9, creating a frame shift and a premature protein translation for both the EDA-A1 and EDA-A2 isoforms. Cases of splice site mutations within introns 5, 7, 8, and 9 have been described in humans (Bayes et al, 1998; Schneider et al. 2001) and within intron 8 in red German Holstein cattle (Drögemuller et al. 2002). Interestingly, in cattle with the mutation affecting the splice donor site in intron 8, mRNA studies revealed that both isoforms were affected, even though the splice donor site for EDA-A2 was still intact (Drögemuller et al. 2002). The authors postulated that sequences coding for regulatory elements in the area around the 3′ end of exon 8 were indirectly affected by the mutation. In humans it is not known if the mutation in intron 8 affects both isoforms, although the authors surmised that it would affect only EDA-A1 (Schneider et al. 2001). Clinically, it is unlikely that there would be a difference between individuals lacking both isoforms simultaneously or just EDA-A1, like the DL mouse, which lacks the receptor specific for EDA-A1 (EDAR), is indistinguishable from the Tabby mouse (EDA-A1 and -A2 deficiency). Most importantly, all of these mutations preclude the transcription and subsequent translation of the TNF-like domain, which is vital to the protein’s effect by binding to the respective receptors.
In summary, we have sequenced the canine EDA gene and confirmed that a mutation in this gene causes the XHED phenotype in our dogs. The mutation found in the canine model of XHED prohibits correct splicing of the last exon and is similar to one found in humans and in red Holstein cattle. The results of this study further strengthen the XHED dog as a model to investigate the pathologic basis of disease and provide a large animal model for therapy trials.
Acknowledgments
This work was supported with funds from the National Foundation for Ectodermal Dysplasias and the National Institutes of Health (KO1-AR049817 and P40-RRO2512). The authors thank Dr. Ewan Kirkness and Dr. Claire Fraser for sharing canine genome sequences prior to publication and Dr. Mark Haskins for critical advice.
References
- 1.Bayés M, Hartung A, Ezer S, Pispa J, Thesleff I, et al. The anhidrotic ectodermal dysplasia gene (EDA) undergoes alternative splicing and encodes ectodysplasin-A with deletion mutations in collagenous repeats. Hum Mol Gen. 1998;7:1661–1669. doi: 10.1093/hmg/7.11.1661. [DOI] [PubMed] [Google Scholar]
- 2.Beahrs JO, Lillington GA, Rosan RC, Russin L, Lindgren JA, et al. Anhidrotic ectodermal dysplasia: predisposition to bronchial disease. Ann Intern Med. 1971;74:92–96. doi: 10.7326/0003-4819-74-1-92. [DOI] [PubMed] [Google Scholar]
- 3.Breen M, Jouquand S, Renier C, Mellersh CS, Hitte C, et al. Chromosome-specific single-locus FISH probes allow anchorage of an 1800-marker integrated radiation-hybrid/linkage map of the domestic dog genome to all chromosomes. Genome Res. 2001;11:1784–1795. doi: 10.1101/gr.189401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casal ML, Jezyk PF, Greek JM, Goldschmidt MH, Patterson DF. X-linked ectodermal dysplasia in a German Shepherd dog. J Hered. 1997;88:513–517. doi: 10.1093/oxfordjournals.jhered.a023146. [DOI] [PubMed] [Google Scholar]
- 5.Casal ML, Ryan S, Rhodes JL, Scheidt JL. Immunological aspects of X-linked ectodermal dysplasia: A dog model for the human disease. Am J Humn Genet. 2003;73(Suppl):454. [Google Scholar]
- 6.Casal ML, Mauldin EA, Gaide O, Scheidt JL, Rhodes JL. Canine X-linked ectodermal dysplasia as a model for the human disease: mutational analysis and further characterization of disease. Proceedings of the American Society of Human Genetics; Oct. 2004; Toronto: The American Society of Human Genetics; 2004. p. 485. [Google Scholar]
- 7.Caswell JL, Yager JA, Parker WM, Moore PF. A prospective study of the immunophenotype and temporal changes in the histologic lesions of canine demodicosis. Vet Pathol. 1997;34:279–287. doi: 10.1177/030098589703400403. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Molloy SS, Thomas L, Gambee J, Bachinger HP, et al. Mutations within a furin consensus sequence block proteolytic release of ectodysplasin-A and cause X-linked hypohidrotic ectodermal dysplasia. Proc Natl Acad Sci USA. 2001;98:7218–7223. doi: 10.1073/pnas.131076098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke A. Hypohidrotic ectodermal dysplasia. J Med Genet. 1987;24:659–663. doi: 10.1136/jmg.24.11.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke A, Phillips DI, Brown R, Harper PS. Clinical aspects of X-linked hypohidrotic ectodermal dysplasia. Arch Dis Child. 1987;62:989–996. doi: 10.1136/adc.62.10.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cottingham RW, Jr, Idury RM, Schaffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- 12.Döffinger R, Smahi A, Bessia C, Geissmann F, Feinberg J, et al. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NK-kB signaling. Nat Genet. 2001;27:277–285. doi: 10.1038/85837. [DOI] [PubMed] [Google Scholar]
- 13.Drögemüller C, Distl O, Leeb T. Partial deletion of the bovine ED1 gene causes anhidrotic ectodermal dysplasia in cattle. Genome Res. 2001;11:1699– 1705. doi: 10.1101/gr.182501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drögemüller C, Peters M, Pohlenz J, Distl O, Leeb T. A single point mutation within the ED1 gene disrupts correct splicing at two different splice sites and leads to anhidrotic ectodermal dysplasia in cattle. J Mol Med. 2002;80:319–323. doi: 10.1007/s00109-002-0320-z. [DOI] [PubMed] [Google Scholar]
- 15.Elomaa O, Pulkkinen K, Hannelius U, Mikkola M, Saarialho-Kere U, et al. Ectodysplasin is released by proteolytic shedding and binds to the EDAR protein. Hum Mol Gen. 2001;10:953–962. doi: 10.1093/hmg/10.9.953. [DOI] [PubMed] [Google Scholar]
- 16.Ferguson BM, Thomas NS, Munoz F, Morgan D, Clarke A, et al. Scarcity of mutations detected in families with X-linked hypohidrotic ectodermal dysplasia: diagnostic implications. J Med Genet. 1998;35:112–115. doi: 10.1136/jmg.35.2.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilgenkrantz S, Blanchet-Bardon C, Nazzaro V, Formiga L, Mujica P, et al. Hypohidrotic ectodermal dysplasia. Clinical study of a family of 30 over three generations. Hum Genet. 1989;81:120–122. doi: 10.1007/BF00293886. [DOI] [PubMed] [Google Scholar]
- 18.Hertz JM, Norgaard Hansen K, Juncker I, Kjeldsen M, Gregersen N. A novel missense mutation (402C → T) in exon 1 in the EDA gene in a family with X-linked hypohidrotic ectodermal dysplasia. Clin Genet. 1998;53:205–209. doi: 10.1111/j.1399-0004.1998.tb02678.x. [DOI] [PubMed] [Google Scholar]
- 19.Jain A, Ma CA, Liu S, Brown M, Cohen J, et al. Specific missense mutations in NEMO result in hyper-IgM syndrome with hypohydrotic ectodermal dysplasia. Nat Immunol. 2001;2:223–228. doi: 10.1038/85277. [DOI] [PubMed] [Google Scholar]
- 20.Kere J, Srivastava AK, Montonen O, Zonana J, Thomas N, et al. X-linked anhidrotic (hypohidrotic) ectodermal dysplasia is caused by mutation in a novel transmembrane protein. Nat Genet. 1996;13:409– 416. doi: 10.1038/ng0895-409. [DOI] [PubMed] [Google Scholar]
- 21.Kirkness EF, Bafna V, Halpern AL, Levy S, Remington K, et al. The dog genome: survey sequencing and comparative analysis. Science. 2003;301:1898–1903. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- 22.Kosaki K, Shimasaki N, Fukushima H, Kara M, Ogata T. Female patient showing hypohidrotic ectodermal dysplasia and immunodeficiency (HED-ID) Am J Hum Genet. 2001;69:664–665. doi: 10.1086/323003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lathrop GM, Lalouel JM, Julier C, Ott J. Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA. 1984;81:3443–3446. doi: 10.1073/pnas.81.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monreal AW, Zonana J, Ferguson B. Identification of a new splice form of the EDA1 gene permits detection of nearly all X-linked hypohidrotic ectodermal dysplasia mutations. Am J Hum Genet. 1998;63:380–389. doi: 10.1086/301984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newton K, French DM, Yan M, Frantz GD, Dixit VM. Myodegeneration in EDA-A2 transgenic mice is prevented by XEDAR deficiency. Mol Cell Biol. 2004;24:1608–1613. doi: 10.1128/MCB.24.4.1608-1613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paakkonen K, Cambiaghi S, Novelli G, Ouzts LV, Penttinen M, et al. The mutation spectrum of the EDA gene in X-linked anhidrotic ectodermal dysplasia. Hum Mutat. 2001;17:349. doi: 10.1002/humu.33. [DOI] [PubMed] [Google Scholar]
- 27.Pinheiro M, Freire–Maria N. Ectodermal dysplasias: a clinical classification and a causal review. Am J Med Genet. 1994;53:153–162. doi: 10.1002/ajmg.1320530207. [DOI] [PubMed] [Google Scholar]
- 28.Schaffer AA, Gupta SK, Shriram K, Cottingham RW., Jr Avoiding recomputation in linkage analysis. Hum Hered. 1994;44:225–237. doi: 10.1159/000154222. [DOI] [PubMed] [Google Scholar]
- 29.Schneider P, Street SL, Gaide O, Hertig S, Tardivel A, et al. Mutations leading to X-linked hypohidrotic ectodermal dysplasia affect three major functional domains in the tumor necrosis factor family member ectodysplasin-A. J Biol Chem. 2001;276:18819– 18827. doi: 10.1074/jbc.M101280200. [DOI] [PubMed] [Google Scholar]
- 30.Smahi A, Courtois G, Vabres P, Yamaoka S, Heuertz S, et al. Genomic rearrangement in NEMO impairs NF-kB activation and is a cause of incontinentia pigmenti: The International Incontinentia Pigmenti (IP) Consortium. Nature. 2000;405:466–472. doi: 10.1038/35013114. [DOI] [PubMed] [Google Scholar]
- 31.Soderholm AL, Kaitila I. Expression of X-linked hypohidrotic ectodermal dysplasia in six males and in their mothers. Clin Genet. 1985;28:136–144. doi: 10.1111/j.1399-0004.1985.tb00373.x. [DOI] [PubMed] [Google Scholar]
- 32.Srivastava AK, Durmowicz MC, Hartung AJ, Hudson J, Ouzts LV, et al. Ectodysplasin-A1 is sufficient to rescue both hair: growth and sweat glands in Tabby mice. Hum Mol Genet. 2001;10:2973–2981. doi: 10.1093/hmg/10.26.2973. [DOI] [PubMed] [Google Scholar]
- 33.Werner P, Raducha MG, Shin D, Ostrander EA, Kirkness E, et al. Assignment of 10 canine genes to the canine linkage and comparative maps. Anim Genet. 2004;35:249–251. doi: 10.1111/j.1365-2052.2004.01123.x. [DOI] [PubMed] [Google Scholar]
- 34.Yan M, Wang LC, Hymowitz SG, Schilbach S, Lee J, et al. Two amino acid molecular switch in an epithelial morphogen that regulates binding to two distinct receptors. Science. 2000;290:523–527. doi: 10.1126/science.290.5491.523. [DOI] [PubMed] [Google Scholar]
- 35.Zonana J, Gault J, Davies KJ, Jones M, Browne D, et al. Detection of a molecular deletion at the DXS732 locus in a patient with X-linked hypohidrotic ectodermal dysplasia (EDA), with the identification of a unique junctional fragment. Am J Hum Genet. 1993;52:78–84. [PMC free article] [PubMed] [Google Scholar]
- 36.Zonana J, Elder ME, Schneider LC, Orlow SJ, Moss C, et al. A novel X-linked disorder of immune deficiency and hypohidrotic ectodermal dysplasia is allelic to incontinentia pigmenti and due to mutations in IKK-gamma (NEMO) Am J Hum Genet. 2000;67:1555–1562. doi: 10.1086/316914. [DOI] [PMC free article] [PubMed] [Google Scholar]