Abstract
Endocycles are variant cell cycles comprised of DNA Synthesis (S)- and Gap (G)- phases but lacking mitosis1,2. Such cycles facilitate post-mitotic growth in many invertebrate and plant cells, and are so ubiquitous that they may account for up to half the world’s biomass3,4. DNA replication in endocycling Drosophila cells is triggered by Cyclin E/Cyclin Dependent Kinase 2 (CycE/Cdk2), but this kinase must be inactivated during each G-phase to allow the assembly of pre-Replication Complexes (preRCs) for the next S-phase5,6. How CycE/Cdk2 is periodically silenced to allow re-replication has not been established. Here, using genetic tests in parallel with computational modeling, we show that Drosophila’s endocycles are driven by a molecular oscillator in which the E2F1 transcription factor promotes CycE expression and S-phase initiation, S-phase then activates the CRL4Cdt2 ubiquitin ligase, and this in turn mediates the destruction of E2F17. We propose that it is the transient loss of E2F1 during S-phases that creates the window of low Cdk activity required for preRC formation. In support of this model over-expressed E2F1 accelerated endocycling, whereas a stabilized variant of E2F1 blocked endocycling by de-regulating target genes including CycE, as well as Cdk1 and mitotic Cyclins. Moreover, we find that altering cell growth by changing nutrition or TOR signaling impacts E2F1 translation, thereby making endocycle progression growth-dependent. Many of the regulatory interactions essential to this novel cell cycle oscillator are conserved in animals and plants1,2,8, suggesting that elements of this mechanism act in most growth-dependent cell cycles.
S-phase control in proliferating animal cells depends upon the E3 ubiquitin ligase, APCFzy/Cdc20, which is activated by Cyclin/Cdk1 during mitosis. APCFzy/Cdc20 promotes the degradation of mitotic Cyclins, thereby extinguishing Cdk1 activity following mitosis, and it also promotes the degradation of Geminin (Gem), an inhibitor of the preRC component Cdt1. The combination of low Geminin and low Cyclin/Cdk1 activity during early G1 allows the assembly of preRCs containing origin recognition complex (ORC) proteins, Cdc6, Cdt1/Double-parked (Dup), and MCM2-7 onto replication origins, thus “licensing” the DNA for renewed replication9. Drosophila’s endocycling cells do not express mitotic Cyclin/Cdk1 complexes or APCFzy/Cdc20 10,11, and so this mechanism of S-phase regulation cannot apply to them. These endocycles do however employ CycE/Cdk2 to trigger S-phases12,13 (Fig 1, 2), and they also require the G1-specific APC variant, APCFzr/Cdh1, which mediates cyclic degradation of cell cycle factors including Geminin and Orc110,14. Importantly, while over-expressed CycE/Cdk2 is tolerated in mitotic cell cycles15, it blocks endocycling (Fig 2, 3)5,6. This is likely due to CycE/Cdk2’s ability to suppress APCFzr/Cdh1 and drive Geminin accumulation10,14,16, though CycE may also inhibit preRC formation directly by phosphorylating preRC components. The importance of CycE oscillation for endocycling is underscored by the finding that Archipelago (Ago/Cdc4/Fbw7), which promotes CycE degradation as a component of an SCF ubiquitin ligase, is required for the progression of endocycles, but not mitotic cycles (Fig S1)17. Despite its importance the mechanism controlling CycE/Cdk2 periodicity in the endocycle has remained obscure for over a decade.
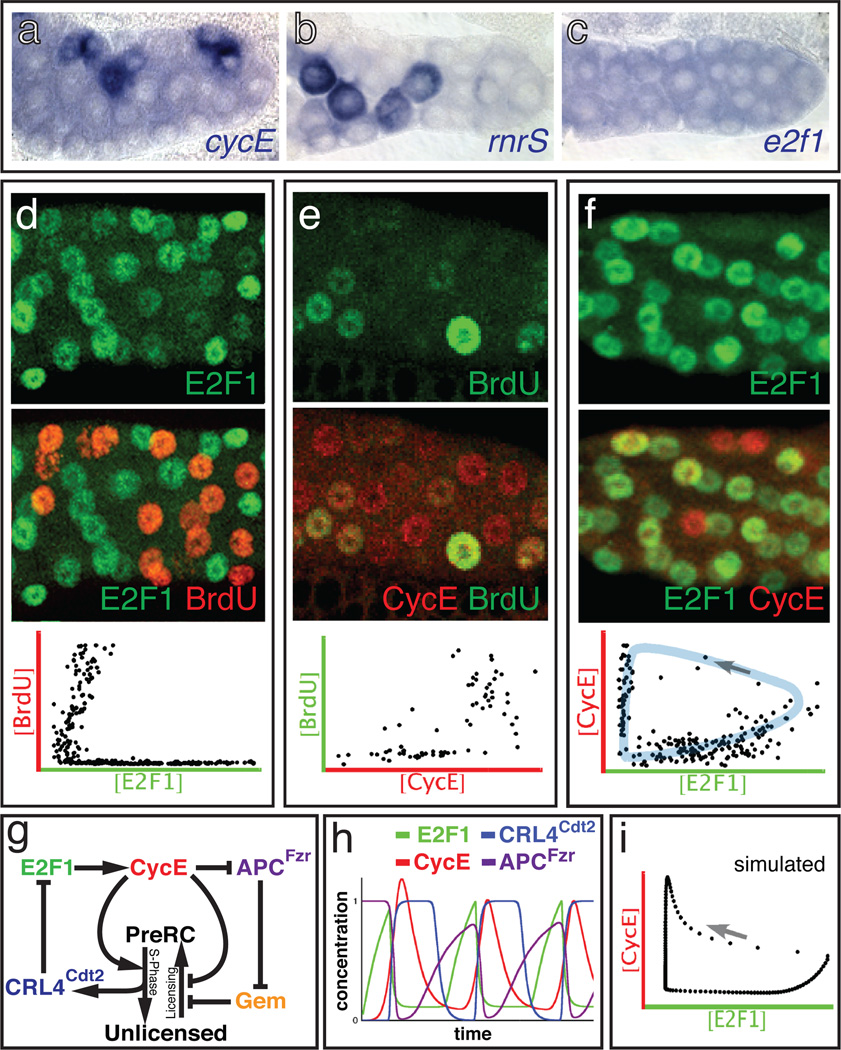
Figure 1. Wildtype salivary gland endocycles.
a-c) In situ hybridization of WT 72h AED glands to the indicated mRNAs. d-f) WT salivary glands at 72h AED double-labeled for: d) E2F1 (green) and BrdU (red); e) CycE (red) and BrdU (green); f) CycE (red) and E2F1 (green). Graphs show nuclear concentrations measured from micrographs of 2–3 glands, in which each dot represents one nucleus. Shaded region (blue) shows trajectory of E2F1/CycE oscillations with an arrow indicating the expected temporal progression. g) Simplified schematic of the computational model. See Fig S4. h) Time plot for WT predicted by the model. i) Nuclear concentrations predicted by the model; arrow represents temporal progression.
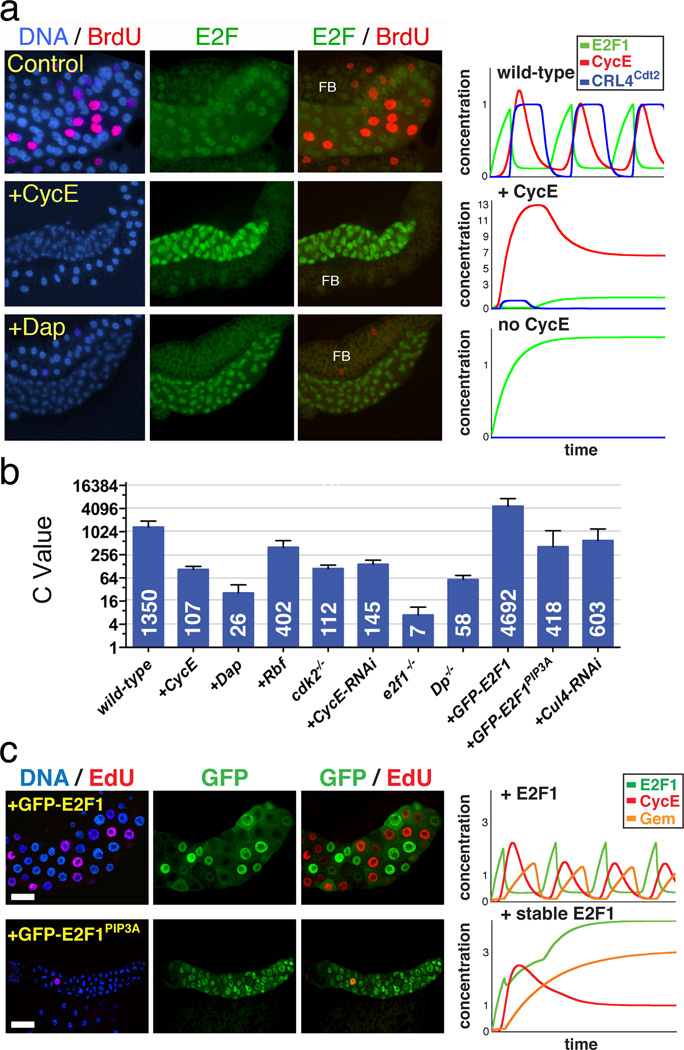
Figure 2. Genetic tests of the endocycle mechanism.
a) Salivary glands (centered) and associated fat body (above or below; FB) from 72h AED larvae expressing the indicated genes under ptc-Gal4/UAS control. ptc-Gal4 expresses in salivary glands but not in fat body. Left column shows DNA (blue) and BrdU (red) incorporated from 71–72h AED. Middle column shows E2F1 (green). Right column shows E2F1 and BrdU. All images had identical exposures and magnifications. Graphs (right) show simulated time plots of E2F1 (green) CycE (red) protein levels and CRL4Cdt2 (Cul4-E3) activity (blue) for each genotype. See Table S1 for parameters. b) Nuclear DNA values from 96h AED glands. For each genotype about 40 nuclei from 6–20 salivary glands were analyzed. Error bars represent standard deviations. ptc-Gal4 drove expression of the UAS-linked transgenes indicated with a “+”. Dp −/− : Dpa2/Df(2R)Exel7124 mutant. E2f1−/− : E2f17172 mutant cells generated by mitotic recombination. cdk2−/− mutant glands were generated as described in methods. c) Salivary glands expressing wild-type GFP-E2F1 (above) or GFP-E2F1PIP3A (below). Layout as in a.
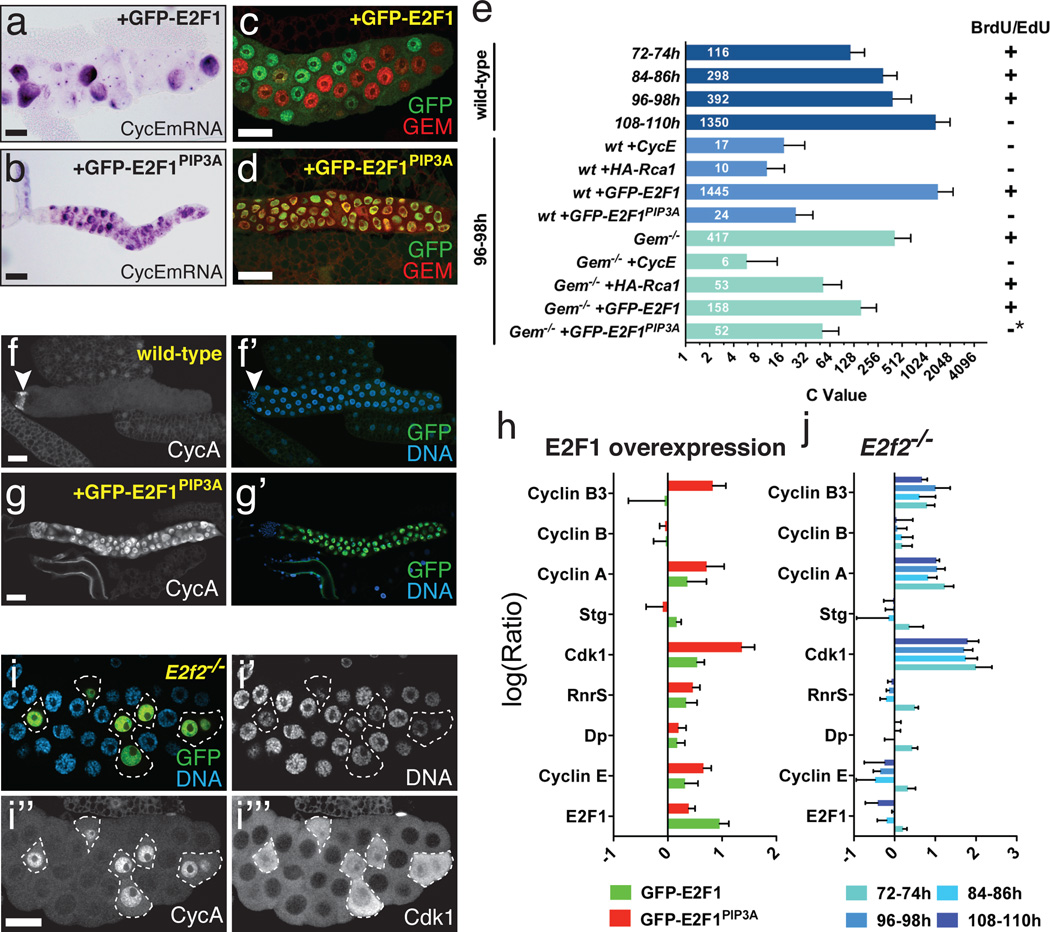
Figure 3. Endocycle arrest by stabilized E2F1.
a-d) Expression of WT GFP-E2F1 with ptc-Gal4 promoted endocycling with cyclic cycE (a) and Gem (c), whereas GFP-E2FPIP3A caused endocycle arrest with uniform cycE (b) and Gem (d) expression. e) C-values per nucleus for the indicated genotypes and timepoints. For each genotype about 40 nuclei from 10–20 salivary glands were analyzed. Error bars represent standard deviations. f-g) CycA expression in WT (f) and glands expressing WT GFP-E2F1 (f) or GFP-E2FPIP3A (g). Arrowhead (f) indicates diploid imaginal ring cells. h) qRT-PCR measurements of the indicated mRNAs, from 72h AED salivary glands expressing GFP-E2F1 (green) or GFP-E2F1PIP3A (red). i) CycA and Cdk1 accumulation in E2f2 mutant cells, generated by MARCM mitotic recombination. GFP in i marks mutant cells (outlined). Cdk1 in i’’’ was detected using anti-PSTAIRE antibody. j) qRT-PCR measurements of the indicated mRNAs, from E2f2 mutant glands at the indicated timepoints. Log10(Ratio)s for h and j are relative to WT controls. Error bars represent standard deviations derived from 3–4 biological replicates.
We addressed this problem in Drosophila’s larval salivary glands, which undergo ~10 asynchronous endocycles from ~7–96 hours after egg deposition (h AED), reaching a final ploidy of ~1350C18. Studies in the fly ovary had suggested that the CycE/Cdk2 inhibitor dacapo (dap) might periodically silence Cdk2 during endocycling19, but our analysis ruled this out for salivary glands (Fig S2)10. Hence we asked whether cyclic CycE/Cdk2 activity might be controlled transcriptionally. CycE transcription is regulated by the E2F1 transcription factor20–22, the accumulation of which is periodic in mitotic Drosophila cells7,23–25 because it is targeted for degradation during S-phase by the PCNA/replication fork-associated E3 ubiquitin ligase CRL4Cdt2 7. In the salivary cells, E2f1 mRNA was ubiquitous (Fig 1c) but E2F1 protein was cyclic, being virtually absent in S-phase nuclei (Fig 1d, 2a). Continuously over-expressed E2F1 proteins were also depleted from S-phase nuclei (Fig 2c, 3c), consistent with periodic degradation. This implied that E2F-dependent transcription might also oscillate. Indeed, the mRNAs encoding CycE and two other E2F targets, RnrS and pcna were periodic (Fig 1, 3, S15, see also13). These mRNAs accumulated when E2F1 was over-expressed (Fig 3a) and were reduced in mutants for Dp, E2F1’s obligate dimerization partner (Fig S15). Thus periodic CycE expression is likely due to periodic activity of its regulator, E2F. CycE protein was also cyclic, being present during a bit of each Gap phase and much of each S-phase (Fig 1e)13,26. Based on these and other results10 we determined that E2F1 accumulates during G-phases and is destroyed upon entry into S-phase, whereas its target CycE rises late in G-phases and persists through most of each S-phase.
These observations suggested that endocycles run using a molecular oscillator in which E2F1 promotes CycE transcription, and then CycE/Cdk2 triggers S-phase and the consequent destruction of E2F1 to reset the cycle (Fig 1g). To evaluate this hypothesis we built a computational model that translated known regulatory interactions into a system of delay differential equations describing the concentrations of E2F1, RBF, CycE, Geminin, and Cdt1/Dup, and the activities of APCFzr/Cdh1 and CRL4Cdt2 (Fig 1g, Supplementary Methods, Fig S4, S5). In this model, when CycE was low Gem was degraded by APCFzr/Cdh1, allowing preRC licensing through Cdt1/Dup. High CycE suppressed APCFzr/Cdh1 activity and allowed Gem accumulation, and also triggered phosphorylation of RBF, S-phase initiation, activation of CRL4Cdt2 and the subsequent degradation of E2F1 and Cdt1/Dup. The model’s behavior depended on unmeasured parameters representing biochemical kinetics (Table S1), but Monte-Carlo searches found numerous parameters sets that simulated actual endocycles (Fig 1h, i). The model robustly produced oscillations of its components despite quantitative parameter variation (Fig S6–9) and did not require exquisitely tuned kinetics to reproduce oscillations like those observed in vivo (Supplementary Discussion).
We tested the computational model by challenging it to reproduce the results of genetic experiments performed in parallel. The model reproduced nearly all observed mutant and gene over-expression phenotypes (Fig 2, S10). Notably, it predicted that increasing E2F1 should accelerate endocycling and lead to hyper-polyploidy, as subsequently observed experimentally (Fig 2, 3, S11). As predicted, we observed increased relative DNA amounts in E2f1+/+ cells generated in an E2f1+/− background, and found that E2f17172 homozygous null mutant cells supported essentially no endocycling (Fig 2b, S13). Thus both loss- and gain-of-function experiments indicated that E2F1 is an essential dose-dependent regulator of endocycle progression.
An important prediction of the computational model was that periodic E2F1 destruction should be essential for endocycling. Drosophila E2F1 is targeted for proteolysis during S-phase via a conserved motif, the PIP box, which binds the replication fork-associated protein, PCNA, and mediates interaction with the CRL4Cdt2 ubiquitin ligase7. Consistent with model predictions, a stabilized but active form of E2F1 lacking the PIP box (GFP-E2F1PIP3A)7 blocked endocycle progression (Fig 2, 3, S11). Likewise RNAi against Cul4, a CRL4Cdt2 component, arrested endocycling (Fig 2b, S11, S12). Levels of E2F1 in cells arrested by GFP-E2F1PIP3A were not higher than in control GFP-E2F1-expressing cells that cycled, suggesting that this arrest was due to inappropriately timed expression of E2F1 rather than its excessive accumulation (Fig 3h, S11). Hence S-phase dependent degradation of E2F1 is essential for endocycling.
One discrepancy between the data and our model was that whereas the model could not readily predict endocycling without E2F (Fig S10), Dp and E2f1 E2f2 mutants support endocycling21,22,26. Our analysis showed that although Dp protein was barely detectable in Dp mutant glands (Fig S15) cells in these mutants nevertheless endocycled slowly and sustained periodic expression of CycE and RnrS, and Geminin oscillation (Fig 2b, S14, S15). One explanation for these apparently discrepant observations is that residual maternal E2F activity persists in these mutants. Consistent with this possibility we found that GFP-E2F1PIP3A was able to block endocycling in Dp mutants (Fig S16). Given this observation, the Dp mutant phenotype cannot be construed as confounding the model (see Supplementary Discussion).
We next asked how stabilized E2F1 arrests endocycling. Consistent with model predictions, cells arrested by GFP-E2F1PIP3A or Cul4-RNAi accumulated CycE and Geminin (Fig 3). In these arrested cells, however, Geminin accumulation occurred following rather than prior to arrest (Fig S12), indicating that it did not initiate the arrest. Interestingly, gem null mutant glands supported rather normal endocycles (Fig S17), but arrest by Rca110,14, an APCFzr/Cdh1 inhibitor, was substantially rescued in the gem mutants (Fig 3e, S18). This demonstrates that the predominant function of APCFzr/Cdh1 in these endocycles is the degradation of Geminin. Importantly, gem mutant cells could be arrested by ectopic CycE10,14 or E2F1PIP3A (Fig 3e, S18, S19). We conclude that while Geminin accumulation might consolidate the arrest caused by excess E2F1, it is neither initiating nor essential for this arrest.
Further investigations revealed that Cyclin A, Cyclin B3 and Cdk1 accumulated in E2F1PIP3A-arrested cells (Fig 3i, S20). These G2/M regulators are not normally expressed in endocycling cells8,10. Large inductions of the mRNAs encoding these factors were observed (Fig 3h), suggesting transcriptional de-repression. Consistent with this notion these factors were also induced in cells mutant for E2f2, Drosophila’s repressor E2F (Fig 3i, j). This suggests that, as in mitotic cells27, excess E2F1 may displace E2F2 and thereby de-repress its targets. In this context E2F2 appears to act as a selectivity factor that represses mitotic targets in endoreplicating cells. Given that Cdk1 is a potent suppressor of PreRCs that can arrest endocycle progression6, its derepression probably contributed to endocycle arrest by E2F1PIP3A.
Altogether our results indicate that periodic E2F1 degradation is necessary for endocycling for three reasons: 1) it creates a window of low CycE/Cdk2 activity; 2) it promotes high APCFzr/Cdh1 activity and thereby suppresses Geminin accumulation; and 3) it allows E2F2 to maintain repression of Cdk1 and its Cyclins. Each of these conditions is required for preRC assembly and endocycle progression. This cell cycle mechanism (Fig 1g, S4) is fundamentally different from that used in mitotic cycles, wherein destruction of the M-phase Cyclins by APCCdc20/Fzy, rather than of E2F1 by the CRL4Cdt2, throws the switch that allows preRC assembly9. Indeed it is noteworthy that the periodic degradation of E2F1 and depletion of CycE are not required for mitotic cell cycles in Drosophila7,12. CRL4Cdt2 is required for endocycling in plants8, suggesting that this element of the endocycle oscillator is conserved.
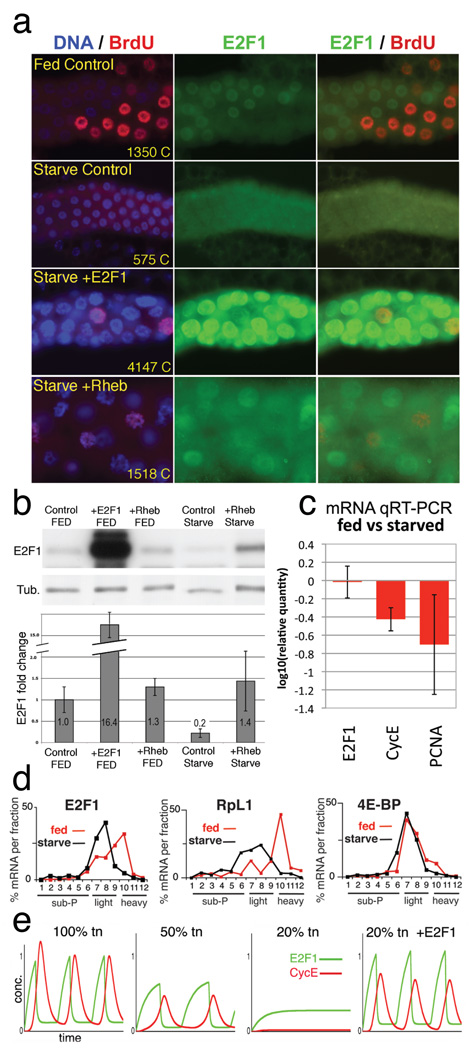
Finally, we asked what factors control E2F production to regulate endocycle rates. Endocycle speed and number can be manipulated by altering cell growth through changes in dietary protein28 or growth-regulatory genes including dMyc1 and Insulin/PI3K/TOR signaling components29. Hence we starved larvae of protein to suppress insulin/TOR signaling, reduce protein synthesis, and block cell growth. Starvation arrested the salivary endocycles within 24h and strongly depleted E2F1 (Fig 4a,b). E2f1 and Dp mRNA levels were not affected, but the E2F targets CycE, pcna, and rnrS were reduced (Fig 4c, not shown). To test whether this was responsible for starvation-induced endocycle arrest we overexpressed E2F1 in the salivary glands of starved animals. Although these glands failed to grow their nuclei incorporated BrdU and accrued ~7-fold more DNA than controls (Fig 4a). Over-expression of Rheb, which activates the Target of Rapamycin (TOR) kinase and increases ribosome biogenesis and cap-dependent translation, also restored cell growth, E2F1 protein, and endocycle progression in starved animals (Fig 4a). Thus E2F1 appears to act as a “growth sensor” that couples rates of endocycle progression to rates of cell growth. A likely mechanism for this, corroborated by modeling (Fig 4e, S8), involves increased translation of E2F1 in rapidly growing cells. Indeed, we found that the association of E2F1 mRNA with polyribosomes was greatly reduced in protein-starved animals (Fig 4d). Translational control of E2F is an attractive mechanism for coupling growth to G1/S progression not only in endocycling cells, but also in growth-dependent mitotic cells with extended G1 periods.
Figure 4. E2F1 is a growth sensor.
a) Salivary glands labeled for DNA (blue), E2F1 (green), and incorporated BrdU (red). Fed Control (WT) was labeled with BrdU at 48h and fixed at 49h. “Starved” animals were transferred to protein-free media at 48h AED, labeled with BrdU at 96h, and fixed at 97h AED. ptc-Gal4 drove expression of UAS-E2F1/DP or UAS-Rheb in the lower two panels. Chromatin (C) values are average nuclear DNA values from 10 glands measured at 120h AED. b) Immunoblot of salivary glands as in a, with quantitation, normalized to tubulin, below. c) mRNA levels from starved and fed control glands, measured by qRT-PCR. d) mRNA levels from 3d protein-starved (black) or fed control (red) whole larvae, quantified from polysome gradient fractions by qRT-PCR. X axis indicates gradient fraction number. e) Computational simulation of starvation by reducing total protein synthesis (tn). In the “20% tn +E2F1” graph, translation of E2F1 was 100% of normal but translation of all other proteins was reduced to 20%. Graphed values (b, c) include standard deviations calculated from 3 independent biological samples.
METHODS SUMMARY
Larvae were raised at 25°C in uncrowded conditions, and salivary glands dissected and analyzed using standard Drosophila genetics and molecular biology methods. DNA quantifications were done using DAPI fluorescence from CCD images. Computational modeling used delay differential equations tracking the concentrations of mRNAs and proteins, and numerically solved in Mathematica 5.2 (Wolfram Research). Full descriptions of experimental and computational methods, genotypes, and reagents is included in the Online Methods section and Supplementary Information.
METHODS
Genetics
To express genes in salivary glands ptc-Gal4, 43B-Gal4 or hey-Gal4 females were crossed to males carrying UAS transgenes. E2f17172/ E2f17172 mutant salivary gland cells were generated by heat shocking hs-Flp; FRT82B E2f17172/FRT82B ub-GPF-nls embryos to 37°C from 2–4h AED. Cdk2 mutant glands were generated using the genotype: F4-Gal4 UAS-GFP/+; Cdk2FRT Cdk23/Cdk22 or F4-Gal4 UAS-GFP/UAS-Flp; Cdk2FRT Cdk23/Cdk22, where Cdk2FRT is a transgene encoding an Flp-excisable Cdk2.
Mutants:
dapg36/CyO, act-GFP32
w; Dpa1 /CyO, act-GFP21
w; Dpa2 /CyO, act-GFP21
w; Df(2R)Exel7124 /CyO(act-GFP) (Bloomington Drosophila Stock Center #7872)
w; FRT80B, e2f17172/TM6B (e2f17172 is described in 34)
w; e2f276Q1, cn, bw/CyO, act-GFP27
w; FRT40A, e2f2C03344, dpov/CyO, act-GFP (Gift from Maxim Frolov, University of Illinois, Chicago/USA)
w; gemininl(2)k14019,c, px, sp/CyO, act-GFP35
w; gemininl(2)k02302,c, px, sp/CyO, act-GFP35
For mutants, we used the strongest alleles available, which in most cases are null alleles. Details on mutant lesions can be found in the cited papers and FlyBase (http://flybase.org/).
Starvation
At 48h or 72h AED larvae were washed with PBS and transferred to PBS+20% sucrose at 25°C, and maintained on this media until 96h or 120h AED, respectively.
DNA quantification
DNA content in nuclei or whole salivary glands was quantified by DAPI fluorescence. Larvae were raised at 25°C to 96h AED, and fixed glands were dissected and stained, using an internal control (ptc-Gal4 UAS-GFPnls) for each sample. Samples were imaged at 10x with a CCD camera (Spot RT or Roper HQ2). Average cytoplasmic intensity was subtracted, and the integrated DAPI intensity was used to measure DNA content for whole glands (Fig 2) or nuclei (Fig 3). All salivary glands had ~the same number of cells (<10% variability). Controls were set to 1350C according to18.
Quantification of nuclear concentrations
Nuclear BrdU, Cyclin E, E2F1, and GFP-E2F1 concentrations as shown in Fig 1(D-F), S3, and S11 were measured from samples stained with DAPI and the indicated antibodies and imaged by confocal microscopy at 20X. We took image stacks (interval size = 0.65μm; optimal overlap under our conditions) with optimized imaging conditions such that the deviation from linearity was <10%. To measure average nuclear concentrations of E2F1 and CycE, we used ImageJ (NIH) and custom software that searched for nuclei by finding ellipsoidal regions that stained brightly for DNA and had the approximate diameter of a nucleus. About half of all nuclei visually overlapped with their neighbors and were not analyzed. To reproducibly set the boundaries for each nucleus, we restricted our analysis to optical sections in which the average nuclear DNA staining was >90% maximal (typically 2–5 sections). Mean intensity in these regions was measured in other channels to determine nuclear concentrations.
BrdU labeling
Embryos were collected on grape-juice/agar plates for 2h and transferred to regular fly-food 24h after egg deposition. At the indicated time points salivary glands were dissected in Drosophila Ringer’s Solution and incubated for 1h at room temperature with 100μg/ml BrdU in Ringer’s Solution. Afterwards, the samples were fixed for 30min in 4% Paraformaldehyde/PBS and subsequently treated for 30min with 2N HCl. BrdU incorporation was detected with a mouse anti-BrdU antibody (Becton Dickinson) diluted 1:20 in 4% NGS/PBS/0.3% Triton-X100 and goat anti-mouse-Alexa Fluor-568 (Invitrogen) as secondary antibody diluted 1:2000 in 4% NGS/PBS/0.3% Triton-X100.
EdU labeling
EdU incorporation was performed analogous to the procedure for BrdU labeling using the Click-It EdU Alexa Fluor-555 imaging kit from Invitrogen.
In situ hybridization
Probes for in situ hybridization were generated with the DIG RNA labeling system (Roche). For in vitro transcription with T7/T3 RNA polymerase the following plasmids were used as template: pT7T3-19U-CycE40; pBLu(2)SKM-RnrS41; pBluSKP-E2F142. Salivary glands were dissected from larvae staged to the indicated time points. Small batches of about 30 larvae were fixed overnight in 8% formaldehyde/PBS, pooled in scintillation vials and stored until usage in ethanol at −80°C. The hybridization procedure was performed according to the protocol developed by the Bier-Lab43. For detection samples were probed with the following antibodies: sheep-anti-DIG-AP (1:500, Roche) or mouse anti-DIG-HRP (1:500, Abcam). BCIP/NBT was used as substrate for the AP reaction according to Tautz & Pfeifle44, while the TSA Alexa Fluor-568 Detection Kit (Invitrogen) was used in combination with HRP.
qRT-PCR
At the indicated time points about 50 salivary glands per genotype were dissected in Drosophila Ringer’s Solution and immediately transferred to the lysis-buffer supplied with the RNAeasy mini kit (Qiagen). Samples were stored at −80°C and then processed with the RNAeasy mini kit (Qiagen) according the manufacturers instructions including the optional on-column DNAseI digestion. 100ng of total RNA were used for cDNA synthesis with the Quantitect Reverse Transcription Kit (Qiagen) or the iScript cDNA synthesis kit (Bio-Rad). qRT-PCR data shown in Figure 3 I-K was acquired on a Light Cycler 480 (Roche) using the indicated UPL assays (Roche) and Light Cycler 480 Probes Master (Roche). Relative expression data presented in Figure 4C was acquired on an iQ5 Instrument (Biorad) using QuantiTect Primer Assays (Qiagen) and the iScript one-step RT-PCR SYBR green kit (Bio- Rad). To ensure statistical significance qRT-PCR was performed in quadruplicates from 3–4 independent samples. Relative expression to GAPDH1 and Actin5c was determined with the ΔΔCT method:
Polysome profiling
Whole larvae were lysed in ice-cold polysome lysis buffer (25mM Tris pH6.8, 10mM MgCl2, 25mM NaCl, 1% Triton-X 100, 0.5% Sodium Deoxycholate, 0.5uM DTT 1ug/ml Cycloheximide, 10ug/ml Heparin, Protease Inhibitor Cocktail (Complete mini, Roche), 2.5 uM PMSF, 5mM Sodium Fluoride, 1mM Sodiumorthovanadate, RNase inhibitor (Ribolock, - Fermentas) using a Dounce Homogenizer. Lysates were then cleared by centrifugation (15,000 rpm, 15 mins, 4C). Equal optical density units (260nM) of cleared lysates were then layered on 15–45% sucrose gradient (prepared in polysome lysis buffer) and centrifuged (37,000 rpm, 2.5 hrs, 4C) in an SW41 Beckman rotor. The gradients were then fractionated using a Brandel BR188 Density Gradient fractionator with continuous OD (254nm) reading and collected into twelve equal fractions. The RNA from each fraction was extracted with Trizol reagent and reversed transcribed using Superscript II (Invitrogen) according to the manufacturers instructions. Quantitative real-time PCR was then performed as described in 45 using a MyIQ PCR machine (BioRad).
Supplementary Material
ACKNOWLEDGEMENTS
Supported by NIH GM51186 to B.A.E., DKFZ, a DAAD fellowship to N.Z., NIGMS 5 P50 GM66050 and NSF MCB0090835 to G.v.D. and K.J.K, DFG LE987/5-1 to C.F.L., CIHR MOP-86622 to S.G., and NIH GM57859 to R.J.D. We thank Yan Liu for help with statistics.
Footnotes
AUTHOR CONTRIBUTIONS
The E2F1-based oscillator was conceived by B.A.E.. N.Z. developed the framework for licensing control and E2F2-mediated repression of mitotic genes. K.J.K. did most of the computational modeling, which was initiated by G.v.D.. Initial experiments were done by V.T., who, with help from B.W. and K.J.K., contributed Fig 1d-f, 2a-b, 4a-b and S13. N.Z. carried out much of the later experimental work with help from M.v.S, and contributed Fig 2b-c, 3c-j, 4c, S1, S3, S11, S12 and S14–20. S.T.S and R.J.D. contributed the GFP-E2F1PIP3A transgenics and controls. M.J.B. contributed Fig 1A-C, 3A-B and S15 G-H. S.N. and S.G. contributed Fig 4D. C.R. and C.F.L. contributed Fig S2 and the cdk2−/− data in Fig 2B. B.A.E. directed the project and wrote the manuscript.
REFERENCES
- 1.Edgar BA, Orr-Weaver TL. Endoreplication cell cycles: more for less. Cell. 2001;105:297–306. doi: 10.1016/s0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- 2.Lilly MA, Duronio RJ. New insights into cell cycle control from the Drosophila endocycle. Oncogene. 2005;24:2765–2775. doi: 10.1038/sj.onc.1208610. [DOI] [PubMed] [Google Scholar]
- 3.Sugimoto-Shirasu K, Roberts K. "Big it up": endoreduplication and cell-size control in plants. Curr Opin Plant Biol. 2003;6:544–553. doi: 10.1016/j.pbi.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Follette PJ, Duronio RJ, O'Farrell PH. Fluctuations in cyclin E levels are required for multiple rounds of endocycle S phase in Drosophila. Current Biology. 1998;8:235–238. doi: 10.1016/s0960-9822(98)70089-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss A, Herzig A, Jacobs H, Lehner CF. Continuous Cyclin E expression inhibits progression through endoreduplication cycles in Drosophila. Current Biology. 1998;8:239–242. doi: 10.1016/s0960-9822(98)70090-9. [DOI] [PubMed] [Google Scholar]
- 7.Shibutani ST, et al. Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev Cell. 2008;15:890–900. doi: 10.1016/j.devcel.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roodbarkelari F, et al. Cullin 4-ring finger-ligase plays a key role in the control of endoreplication cycles in Arabidopsis trichomes. Proc Natl Acad Sci U S A. 107:15275–15280. doi: 10.1073/pnas.1006941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14:R778–R786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 10.Zielke N, Querings S, Rottig C, Lehner C, Sprenger F. The anaphase-promoting complex/cyclosome (APC/C) is required for rereplication control in endoreplication cycles. Genes Dev. 2008;22:1690–1703. doi: 10.1101/gad.469108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maqbool SB, et al. Dampened activity of E2F1-DP and Myb-MuvB transcription factors in Drosophila endocycling cells. J Cell Sci. 2010;123:4095–4106. doi: 10.1242/jcs.064519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knoblich JA, et al. Cyclin E controls S-phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994;77:107–120. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 13.Lilly MA, Spradling AC. The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes & Development. 1996;10:2514–2526. doi: 10.1101/gad.10.19.2514. [DOI] [PubMed] [Google Scholar]
- 14.Narbonne-Reveau K, et al. APC/CFzr/Cdh1 promotes cell cycle progression during the Drosophila endocycle. Development. 2008;135:1451–1461. doi: 10.1242/dev.016295. [DOI] [PubMed] [Google Scholar]
- 15.Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- 16.Sigrist SJ, Lehner CF. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell. 1997;90:671–681. doi: 10.1016/s0092-8674(00)80528-0. [DOI] [PubMed] [Google Scholar]
- 17.Shcherbata HR, Althauser C, Findley SD, Ruohola-Baker H. The mitotic-to-endocycle switch in Drosophila follicle cells is executed by Notch-dependent regulation of G1/S, G2/M and M/G1 cell-cycle transitions. Development. 2004;131:3169–3181. doi: 10.1242/dev.01172. [DOI] [PubMed] [Google Scholar]
- 18.Hammond MP, Laird CD. Control of DNA replication and spatial distribution of defined DNA sequences in salivary gland cells of Drosophila melanogaster. Chromosoma. 1985;91:279–286. doi: 10.1007/BF00328223. [DOI] [PubMed] [Google Scholar]
- 19.Hong A, et al. The cyclin-dependent kinase inhibitor Dacapo promotes replication licensing during Drosophila endocycles. Embo J. 2007;26:2071–2082. doi: 10.1038/sj.emboj.7601648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duronio RJ, O'Farrell PH. Developmental control of the G1 to S transition in Drosophila: cyclin E is a limiting downstream target of E2F. Genes and Development. 1995;9:1456–1468. doi: 10.1101/gad.9.12.1456. [DOI] [PubMed] [Google Scholar]
- 21.Royzman I, Whittaker AJ, Orr-Weaver TL. Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcriptional program. Genes and Development. 1997;11:1999–2011. doi: 10.1101/gad.11.15.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duronio RJ, Bonnette PC, O'Farrell PH. Mutations of the Drosophila dDP, dE2F, and cyclin E genes reveal distinct roles for the E2F-DP transcription factor and cyclin E during the S-phase transition. Molecular and Cellular Biology. 1998;18:141–151. doi: 10.1128/mcb.18.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asano M, Nevins JR, Wharton RP. Ectopic E2F expression induces S-phase and apoptosis in Drosophila imaginal discs. Genes and Development. 1996;10:1422–1432. doi: 10.1101/gad.10.11.1422. [DOI] [PubMed] [Google Scholar]
- 24.Reis T, Edgar BA. Negative regulation of dE2F1 by cyclin-dependent kinases controls cell cycle timing. Cell. 2004;117:253–264. doi: 10.1016/s0092-8674(04)00247-8. [DOI] [PubMed] [Google Scholar]
- 25.Heriche JK, Ang D, Bier E, O'Farrell PH. Involvement of an SCFSlmb complex in timely elimination of E2F upon initiation of DNA replication in Drosophila. BMC Genet. 2003;4:9. doi: 10.1186/1471-2156-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weng L, Zhu C, Xu J, Du W. Critical role of active repression by E2F and Rb proteins in endoreplication during Drosophila development. Embo J. 2003;22:3865–3875. doi: 10.1093/emboj/cdg373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frolov MV, et al. Functional antagonism between E2F family members. Genes & Development. 2001;15:2146–2160. doi: 10.1101/gad.903901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Britton JS, Edgar BA. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;125:2149–2158. doi: 10.1242/dev.125.11.2149. [DOI] [PubMed] [Google Scholar]
- 29.Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell. 2002;2:239–249. doi: 10.1016/s1534-5807(02)00117-x. [DOI] [PubMed] [Google Scholar]
- 30.Moberg KH, Mukherjee A, Veraksa A, Artavanis-Tsakonas S, Hariharan IK. The Drosophila F box protein archipelago regulates dMyc protein levels in vivo. Curr Biol. 2004;14:965–974. doi: 10.1016/j.cub.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 31.Lane ME, et al. Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell. 1996;87:1225–1235. doi: 10.1016/s0092-8674(00)81818-8. [DOI] [PubMed] [Google Scholar]
- 32.Lane ME, et al. A screen for modifiers of cyclin E function in Drosophila melanogaster identifies Cdk2 mutations, revealing the insignificance of putative phosphorylation sites in Cdk2. Genetics. 2000;155:233–244. doi: 10.1093/genetics/155.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frolov MV, Moon NS, Dyson NJ. dDP is needed for normal cell proliferation. Mol Cell Biol. 2005;25:3027–3039. doi: 10.1128/MCB.25.8.3027-3039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duronio RJ, O'Farrell PH, Xie J-E, Brook A, Dyson N. The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes and Development. 1995;9:1445–1455. doi: 10.1101/gad.9.12.1445. [DOI] [PubMed] [Google Scholar]
- 35.Quinn LM, Herr A, McGarry TJ, Richardson H. The Drosophila Geminin homolog: roles for Geminin in limiting DNA replication, in anaphase and in neurogenesis. Genes Dev. 2001;15:2741–2754. doi: 10.1101/gad.916201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Speicher SA, Thomas U, Hinz U, Knust E. The Serrate locus of Drosophila and its role in morphogenesis of the wing imaginal discs: control of cell proliferation. Development. 1994;120:535–544. doi: 10.1242/dev.120.3.535. [DOI] [PubMed] [Google Scholar]
- 37.Xin S, Weng L, Xu J, Du W. The role of RBF in developmentally regulated cell proliferation in the eye disc and in Cyclin D/Cdk4 induced cellular growth. Development. 2002;129:1345–1356. doi: 10.1242/dev.129.6.1345. [DOI] [PubMed] [Google Scholar]
- 38.Saucedo LJ, et al. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- 39.Grosskortenhaus R, Sprenger F. Rca1 inhibits APC-Cdh1(Fzr) and is required to prevent cyclin degradation in G2. Dev Cell. 2002;2:29–40. doi: 10.1016/s1534-5807(01)00104-6. [DOI] [PubMed] [Google Scholar]
- 40.Richardson HE, O'Keefe LV, Reed SI, Saint R. A Drosophila G1-specific cyclin E homolog exhibits different modes of expression during embryogenesis. Development. 1993;119:673–690. doi: 10.1242/dev.119.3.673. [DOI] [PubMed] [Google Scholar]
- 41.Duronio RJ, O'Farrell P. Developmental control of a G1-S transcriptional program in Drosophila. Development. 1994;120:1503–1515. doi: 10.1242/dev.120.6.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dynlacht BD, Brook A, Dembski M, Yenush L, Dyson N. DNA-binding and trans-activation properties of Drosophila E2F and DP proteins. Proceedings of the National Academy of Sciences USA. 1994;91:6359–6363. doi: 10.1073/pnas.91.14.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kosman D, et al. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- 44.Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 45.Van Gilst MR, Hadjivassiliou H, Yamamoto KR. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc Natl Acad Sci U S A. 2005;102:13496–13501. doi: 10.1073/pnas.0506234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.