Abstract
During animal development, a complex of Par3, Par6 and atypical protein kinase C (aPKC) plays a central role in cell polarisation. The small G protein Cdc42 also functions in cell polarity and has been shown in some cases to act by regulating the Par3 complex. However, it is not yet known whether Cdc42 and the Par3 complex widely function together in development or whether they have independent functions. For example, many studies have implicated Cdc42 in cell migrations, but the Par3 complex has only been little studied, with conflicting results. Here we examine the requirements for CDC-42 and the PAR-3/PAR-6/PKC-3 complex in a range of different developmental events. We found similar requirements in all tissues examined, including polarised growth of vulval precursors and seam cells, migrations of neuroblasts and axons, and the development of the somatic gonad. We also propose a novel role for primordial germ cells in mediating coalescence of the C. elegans gonad. These results indicate that CDC-42 and the PAR-3/PAR-6/aPKC complex function together in diverse cell types.
Keywords: C. elegans, cell migration, polarity, vulva, anchor cell, gonad, Cdc42, Par3, Par6, aPKC
Introduction
A conserved complex of Par3 and Par6 (PDZ domain proteins) and an atypical protein kinase C (aPKC) plays a central role in the establishment and maintenance of cell polarity in animal cells (Macara, 2004). In some systems, this complex has been shown to be activated by the small GTPase Cdc42 (Etienne-Manneville, 2004). In mammalian epithelia Cdc42 and the Par3 complex are required for apical-basal polarity and junction formation (Joberty et al., 2000; Lin et al., 2000). In migratory mammalian cells, Cdc42 and a Par-6/aPKC complex mediate polarisation of the microtubule organising centre towards the leading edge (Etienne-Manneville and Hall, 2001; Solecki et al., 2004). In C. elegans, CDC-42 and the PAR-3/PAR-6/PKC-3 complex regulate polarity in the one celled embryo (Nance, 2005).
Although there are examples where Cdc42 and the Par3 complex are known to function together in cell polarisation, the picture regarding other processes is not clear. For example, Cdc42 has been widely implicated in cell migration, but components of the Par3 complex have been little studied. Furthermore some studies have given conflicting results. Fibroblasts generated from Cdc42 null ES cells show no migration defects (Czuchra et al., 2005), whereas primary fibroblasts from conditional Cdc42 knockout mice show strong defects in wound recruitment and chemotaxis (Yang et al., 2006). Overexpression of mPar6α prevents the migration of glial guided neurons in culture (Solecki et al., 2004). Axon outgrowth, which precedes cell body migration, is also inhibited and might be the cause of the migration defect. Axon outgrowth is also blocked by Par3 complex overexpression in hippocampal neuron cultures (Shi et al., 2003), but evidence from Drosophila mutants suggests that the complex is not required for axon outgrowth or dendrite morphology (Rolls and Doe, 2004). By contrast Par6 and Par3 (Bazooka) are required for the migration of Drosophila border cells (Pinheiro and Montell, 2004), although earlier experiments suggest that Cdc42 is not required for this process (Murphy and Montell, 1996). Thus the role of the Par3 complex in migration and polarised growth, and its relationship to Cdc42 are unclear.
To address this question, we investigated whether CDC-42 and the PAR-3 complex act in the same set of developmental processes in C. elegans. We found that inhibition of CDC-42 or components of the PAR-3/PAR-6/PKC-3 complex causes similar defects in multiple cell types, including somatic gonad precursors, vulval precursors, seam cells and neurons. We suggest that CDC-42 and the PAR-3 complex widely act together in cell migration and polarised cell growth.
Materials and Methods
Strains and worm handling
Standard methods have been used for culturing C. elegans on NGM plates (Lewis and Fleming, 1995). Bristol strain N2 (Brenner, 1974) has been used as wild type throughout. The ehn-3A::GFP reporter (pRA230) contains 3003bp upstream of the ehn-3A translational start and the first two exons of ehn-3A. Genomic sequences were cloned into pPD95.75 using XbaI and PstI sites. pRA230 was injected with pRF4 (Mello et al., 1991) and integrated to generate rdIs2 [ehn-3A::GFP; rol-6(su1006)] V.
Other strains used were bnIs1[pie-1::GFP::pgl-1; unc-119(+)] I, let-23(sy1) II, syIs77[zmp-1::YFP] II, muIs32[mec-7::GFP, lin-15(+)] II, cdc-42(gk388)/mIn1[mIs14 dpy-10(e128)] II, lin-12(n302) III, lin-12(n137) dpy-19(e1259)/lin-12(n137n720) unc-32(e189) III, lin-12(n941) III/ hT2[qIs48] (I; III), jcIs1[ajm-1::GFP; rol-6(su1006)] IV, lin-3(n378) IV, lin-45(n2018) IV, syIs49[zmp-1::GFP; dpy-20(+)] IV, syIs67[zmp-1::CFP; unc-119(+)] V, syIs59[egl-17::CFP] X, zuIs77[par-6::GFP; unc-119(+)], mcIs[let-413::GFP; rol-6(su1006)], arIs51[cdh-3::GFP; dpy-20(+)], syEx[lin-3::GFP; pha-1(+)] , qIs56[lag-2::GFP; unc-119(+)], arIs92[egl-17p::CFP::lacZ; unc-4(+); ttx-3p::GFP] and arIs82[lin-12::GFP; unc-4(+); egl-17p::lac-Z]. For compound microscopy, live specimens were mounted by standard procedures (Sulston and Hodgkin, 1988) on 3% agar pads in 5mM tetramisole in M9. Photomicrographs were generated as described under Immunofluorescence.
RNAi
RNAi was performed by feeding as described previously (Kamath et al., 2001), with minor modifications. Briefly, plates containing NGM agar, 1 mM IPTG and 25μg/ml carbenicillin were inoculated with bacterial cultures grown overnight at 37°C in LB medium supplemented with 50μg/ml ampicillin. Synchronised L4 animals were placed on plates at 15°C for 48 hours for cdc-42, par-3 and pkc-3(RNAi), 24 hours for par-6(RNAi), and 40 hours at 25C for mes-1(RNAi). Longer incubations (72 hours) result in 100% embryonic lethality for cdc-42 or the par-3 complex. After the first incubation adults were transferred to fresh plates and allowed to lay eggs for 24 hours at 20°C before removal. Progeny were incubated at 20°C until they reached the required stage for analysis. Constructs used for RNAi are as previously described (Kamath et al., 2003). These clones are predicted to show no off-target effects (e.g., no primary or secondary off-targets in Wormbase, www.wormbase.org).
Vulval assays
The Muv phenotype was scored under a dissecting microscope. Vulval induction was scored at L4 as previously described (Poulin et al., 2005). 1° fate was scored on the basis of egl-17::CFP expression at the Pn.pxx stage.
Immunofluorescence
Fixation for AJM-1/MH27 and LIN-12::GFP stainings was carried out using a modified version of the Finney-Ruvkun fixation procedure (Shaye and Greenwald, 2002), L1 larval staining and embryo staining was carried out as in Le Bot et al (2003) and LIN-12 and egl-17::CFP stainings as in Hurd and Kemphues (2003). The following antibodies were used: MH27 (Francis and Waterston, 1985), anti-GFP (Molecular Probes or Nacalai Tesque), anti-Pgl-1 (Kawasaki et al., 1998), NE8/4C6.3 (Goh and Bogaert, 1991), and anti-LIN-12 (gift of Stuart Kim). All conjugated secondary antibodies were from Jackson Immunoresearch. Stained worms were mounted in Mowiol (Merck) and viewed with a Zeiss Axioplan 2 microscope. Photomicrographs were obtained using either a Hamamatsu Orca C4742-95 camera and Improvision Openlab software or a Zeiss LSM 500 Meta confocal attachment.
Results
cdc-42 and par-3(RNAi) result in hyperinduction of the vulva
Strong maternal reduction of cdc-42, par-3, par-6 or pkc-3 causes defects in the polarity of the first cell division and embryonic lethality (Gotta et al., 2001; Kemphues et al., 1988; Tabuse et al., 1998; Watts et al., 1996). Components of the PAR-3 complex are expressed widely during later development (Nance, 2005), suggesting additional roles, but such roles show maternal rescue: homozygous loss of function mutants of par-3 and par-6 grow into morphologically normal adults that give rise to 100% dead embryos and a cdc-42 null mutant is viable but homozygous sterile (data not shown) (Kemphues et al., 1988; Watts et al., 1996). Consistent with this, a previous study using RNAi to inhibit zygotic but not maternal par-3 function only identified defects in epithelia that develop in the last larval stage (Aono et al., 2004). In order to more broadly investigate somatic functions of these genes we reduced both their maternal and zygotic activities by carrying out RNAi of mothers for short periods and looked for phenotypes in their surviving progeny (see Methods). We found that RNAi of cdc-42, par-3, par-6 or pkc-3 results in multiple ventral protrusions in adult progeny, characteristic of a Multiple vulva (Muv) phenotype (Table 1). For this paper, we focus on defects induced by RNAi of cdc-42 and use par-3 or par-6 as representative of the PAR-3/PAR-6/PKC-3 complex, as the phenotypes induced by RNAi of par-3, par-6 and pkc-3 are similar.
Table 1.
VPCs are hyperinduced following RNAi of cdc-42, par-3, par-6 or pkc-3
| Treatment | Length of Exposure (hours) |
% animals hypo- induced |
% animals hyper- induced |
Average Induction (Range) |
n | % Muv3 |
n |
|---|---|---|---|---|---|---|---|
| Vector(RNAi) | 48-72 | 0 | 0 | 3.01 (3.0-3.0) | 30 | 0 | 1793 |
| cdc-42(RNAi) | 48-72 | 10 | 47 | 3.41 (2.0-5.0) | 30 | 28 | 1971 |
| par-3(RNAi) | 48-72 | 0 | 10 | 3.11 (3.0-5.0) | 30 | 7 | 188 |
| par-6(RNAi) | 24-48 | 0 | 10 | 3.21 (3.0-6.0) | 30 | 4 | 233 |
| pkc-3(RNAi) | 48-72 | 0 | 13 | 3.11 (3.0-4.5) | 30 | 6 | 151 |
| mes-1(RNAi) | 40-644 | n.d. | n.d. | n.d. n.d. | n.a. | 75 | 206 |
|
| |||||||
| cdc-42(RNAi) 1 AC | 48-72 | 4 | 4 | 3.02 (1.0-4.0) | 55 | n.d. | n.a. |
| cdc-42(RNAi) 2+ AC | 48-72 | 4 | 80 | 4.12 (2.0-6.0) | 45 | n.d. | n.a. |
The number of VPCs induced (from 0-6) was scored by counting vulval cells at L4 Pn.pxx.
The number of VPCs induced (from 0-6) was scored by counting vulval cells at L3 Pn.pxx.
Greater than 3 induced VPCs is hyperinduced and fewer than 3 is hypoinduced. Hypoinduction may be a result of gaps between VPCs or abnormal AC positions.
Muv, multiple protrusions scored using a dissecting microscope.
mes-1(RNAi) incubations were at 25°C.
previously reported by Capowski et al (1991)
AC: Anchor Cell
The C. elegans vulva develops from three of six equipotent vulval precursor cells (VPCs), numbered P3.p-P8.p, induced and patterned by the Ras and Notch signalling pathways (Wang and Sternberg, 2001). A LIN-3/EGF signal produced by the Anchor Cell in the overlying gonad induces vulval fate in the three closest VPCs (P5.p-P7.p) by activating Ras signalling via the EGF receptor LET-23. Lateral signalling between the VPCs, mediated by LIN-12 Notch, results in the central cell (usually P6.p) adopting the primary (1°) vulval fate, while the flanking cells adopt the secondary (2°) fate. The uninduced P3.p, P4.p, and P8.p cells fuse with the surrounding hypodermis after one division, whereas the induced VPCs divide three times to produce 22 adult vulval cells. Counting the number of vulval cells confirmed that RNAi of cdc-42, par-3, par-6 or pkc-3 results in greater than three induced VPCs (Table 1).
Although CDC-42 and the PAR-3 complex play conserved roles in epithelial polarity, the vulval hyperinduction phenotypes observed by their knockdown do not appear to be caused by defects in vulval cell polarity. We found that apical, adherens junction and basal markers are localised to the correct domains in hyperinduced vulval cells of cdc-42(RNAi) and par-3(RNAi) animals (Supplementary Figure 1).
Polarised growth defects of the VPCs and seam cells
Although the apical junctions of individual VPCs are intact, cdc-42 or par-6(RNAi) results in the occurrence of gaps between VPCs during L3 (Figure 1B). VPCs are not born in contact with one another but elongate during L2 to form a continuous array (Liu et al., 2005). The finding of large gaps between VPCs during L3 suggests that CDC-42 and the PAR-3 complex are required for the normal polarised growth of these cells.
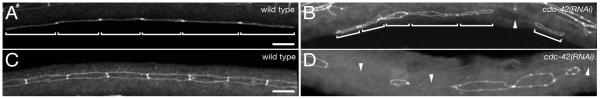
Figure 1. Abnormal VPC and seam cell migrations.
(A,B) AJM-1 at the apical junction of Pn.p stage VPCs (brackets). (A) wild-type: VPCs form a continuous array (n=31). (B) cdc-42(RNAi) animal with a large gap between VPCs (arrowhead); 39% of cdc-42(RNAi) animals (n=28) and 17% of par-6(RNAi) animals (n=30) have such gaps.
(C,D) AJM-1 at the apical junction of L3 stage seam cells. (C) wild-type: seam cells form a continuous array (n=31). (D) cdc-42(RNAi) animal with large gaps (arrowheads) and rounded seam cells; seam cell gaps occur in 86% of cdc-42(RNAi) animals (n=28) and 13% of par-6(RNAi) animals (n=30).
Anterior, left; ventral, down; scale bars, 10μm.
To ask whether a requirement in polarised growth is a general property of these genes, we examined seam cell development. The seam cells divide at the beginning of each larval stage, with the anterior daughters fusing with the hypodermis, while the posterior daughters retain a seam cell fate (Sulston and Horvitz, 1977). The continuous array of seam cells is restored by elongation of the posterior daughters in the anterior/posterior axis. Following RNAi of cdc-42 or par-6, many seam cells do not extend to the neighbouring cell, leading to large gaps between them (Figure 1D). In addition, seam cells have a rounded rather than an elongated morphology consistent with a defect in polarity of growth (Figure 1D). Although apical junctions are continuous, they appear abnormal, with bright spots of AJM-1::GFP at the apical membranes (Figure 1D). Thus CDC-42 and the PAR-3 complex are required for the polarised growth of both VPCs and seam cells and may play a role in regulating the recruitment of junctional components.
Both AC/VU precursors adopt the Anchor Cell fate
We next explored the basis for the Muv phenotype induced by RNAi of cdc-42, par-3, par-6, and pkc-3. Because epistasis analyses showed that CDC-42 acts at or upstream of the LIN-3 signal produced by the Anchor cell (AC) (Supplementary Table 1), we examined AC development.
The AC is derived from one of two equipotent cells, Z1.ppp and Z4.aaa, descendants of the somatic gonad precursors (SGPs), Z1 and Z4 (Kimble and Hirsh, 1979). Z1.ppp and Z4.aaa are brought together by the stereotyped division pattern of the SGPs. Through contact-dependent LIN-12 Notch signalling, either Z1.ppp or Z4.aaa adopts a ventral uterine (VU) fate, while the other forms the AC (Seydoux and Greenwald, 1989). Disruption of this signalling causes both cells to develop as ACs and can result in changes in vulval fate through excess LIN-3 signalling.
We found that instead of the single AC found in wildtype, two ACs were often present following RNAi of cdc-42, par-3, par-6 or pkc-3 (Figure 2, Table 2). This suggests that LIN-12 Notch signalling between AC/VU precursors might be disrupted. CDC-42 does not appear to be required for activity of the LIN-12 Notch pathway downstream of LIN-12, as RNAi of cdc-42 cannot rescue the AC-less phenotype of weak lin-12(gf) mutants (lin-12(n302gf) n=186, lin-12(n137gf)/lin-12(n137n720lf) n=206). This suggests that cdc-42(RNAi) does not cause multiple ACs by downregulating LIN-12 Notch signal transduction.
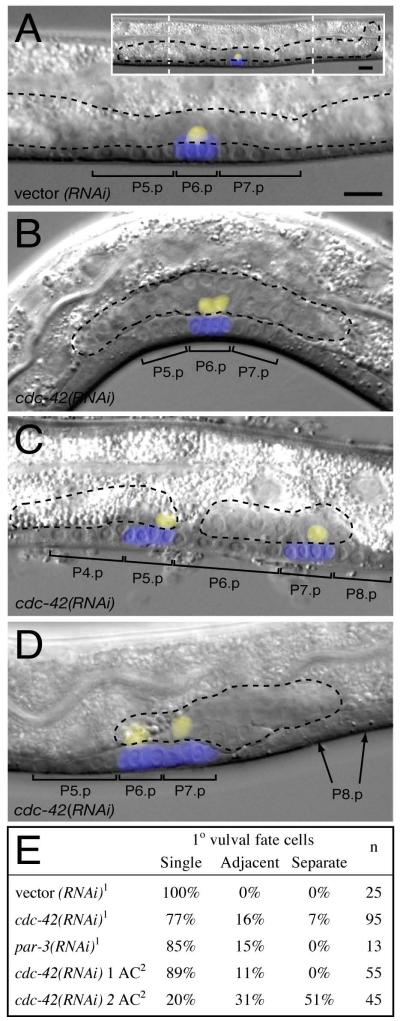
Figure 2. cdc-42(RNAi) results in multiple ACs, adjacent 1° VPC fates and gonad splitting.
Overlays of Nomarski images of L3 Pn.pxx stage animals with projections of zmp-1::YFP expression (yellow) marking the Anchor Cell (AC) and egl-17::CFP expression (blue) marking 1° fate VPCs. Brackets indicate the descendants of induced VPCs and black dotted outlines the morphology of the gonad.
(A) wild-type with extended gonad (inset), a single AC, and three induced VPCs, one of which (P6.p) adopts the 1° fate (marked by egl-17::CFP).
(B-D) cdc-42(RNAi) animals with (B) two touching ACs and a single 1° fate VPC, (C) two ACs in separate gonad fragments and two, non-adjacent, 1° fate VPCs, (D) two separated ACs in an intact, but short, gonad and two adjacent 1° fate VPCs. . VPCs adjacent to 1° fate VPCs sometimes fail to adopt vulval fates (arrows in D), perhaps because of a gap between the VPCs.
Anterior, left; ventral, down; scale bar, 10μm
(E) Table of 1° vulval fate phenotypes
11° fate scored using arIs92[egl-17::CFP::lacZ] (Yoo et al., 2004)
21° fate scored using syIs59[egl-17:CFP] (Inoue et al., 2002)
AC: Anchor Cell
Table 2.
Multiple Anchor Cells induced by RNAi of cdc-42, par-3, par-6 or pkc-3
| Strain | % animals with | n | % 2+ AC animals with |
n | % 2+ separate AC animals with split gonad |
n | |||
|---|---|---|---|---|---|---|---|---|---|
| 1 AC |
2 AC |
>2 AC |
touching ACs |
separate ACs |
|||||
| vector (RNAi)1 | 100 | 0 | 0 | 30 | n.a. | n.a. | n.a. | n.a. | n.a. |
|
cdc- 42(RNAi)1,2 |
55 | 44 | 14 | 100 | 16 | 84 | 15 | 55 | 38 |
| par-3(RNAi) 3 | 85 | 15 | 0 | 26 | 50 | 50 | 4 | n.d. | n.a. |
| par-6(RNAi) 1 | 81 | 19 | 0 | 26 | 0 | 100 | 5 | 40 | 5 |
| pkc-3(RNAi) 1 | 79 | 21 | 0 | 29 | 33 | 67 | 6 | n.d. | n.a. |
|
| |||||||||
| lin-12(n941) 3 | 0 | 78 | 224 | 80 | 100 | 0 | 80 | n.d. | n.a. |
|
lin-12(n941);
cdc-42(RNAi) 3 |
0 | 87 | 134 | 23 | 78 | 22 | 23 | n.d. | n.a. |
|
| |||||||||
| mes-1(RNAi) 1 | 62 | 38 | 0 | 26 | 10 | 90 | 10 | 50 | 8 |
| hnd-1(q740) 1 | 92 | 8 | 0 | 85 | 57 | 43 | 7 | 44 | 9 |
Split gonads have two distinct fragments of gonadal tissue separate from one another.
Anchor Cells (AC) scored using syIs77 [zmp-1::YFP] (Inoue et al., 2002)
AC duplication was confirmed with arIs51[cdh-3::GFP] (Karp and Greenwald, 2003) and syEx[lin-3::GFP] (Wang and Sternberg, 2000) (data not shown)
Anchor Cells scored using syIs49 [zmp-1::GFP] (Wang and Sternberg, 2000)
Additional ACs are derived from the AC/VU precursors (Z1.ppp and Z4.aaa) and occasionally from their sibling cells (Z1.ppa and Z4.aap) (Seydoux et al., 1990)
However, we observed that the two ACs in cdc-42(RNAi) animals were frequently separated from one another (Figure 2C, D; Table 2). This contrasts with the direct contact of multiple ACs in lin-12 mutants (Greenwald et al., 1983), suggesting that RNAi of cdc-42 might cause ACs or their precursors to separate, preventing contact-dependent signalling. Indeed, we found that RNAi of cdc-42 caused separation of the multiple ACs in lin-12 mutants (Table 2).
The additional ACs in cdc-42(RNAi) animals appear to be derived from the normal AC/VU precursors because cdc-42(RNAi) does not increase the number of ACs in a background that already has multiple ACs derived from the normal precursors (lin-12(n941); Table 2). The additional ACs formed in cdc-42(RNAi) animals also occur in the presence of normal distal tip cells (data not shown), whose precursors can instead, in certain mutant backgrounds, give rise to additional ACs (Miskowski et al., 2001).
AC/VU precursors are separated and gonads are fragmented
To further investigate the basis of AC duplication and separation, we examined the development of the somatic gonad in cdc-42(RNAi) and par-3(RNAi) animals. We found that whereas wild-type animals have a single gonad, cdc-42(RNAi) animals often have a gonad split into two separate sections (23%; n=100), each of which usually contains an AC (91%, n=23; Figure 2C). This suggests that the AC/VU precursors might have been physically separated, preventing them from signalling to one another and resulting in the formation of two ACs. Indeed, at the time of signalling between AC/VU precursors (L2), cdc-42(RNAi) and par-3(RNAi) animals showed frequent separation of these precursors (Figure 3B,C; 34%, n=44, 44%, n=16, respectively) whereas separation was never seen in control animals (n=20). AC duplication occurs both in animals with split gonads (47%; n=45) and in those with single, non-split gonads (53%; n=45) suggesting a requirement for CDC-42 and the PAR-3 complex in somatic cell positioning within the gonad. We conclude that RNAi inhibition of CDC-42 or components of the PAR-3 complex causes separation between the AC/VU precursors, resulting in the formation of multiple ACs, presumably by preventing LIN-12 Notch signalling.
Figure 3. AC/VU precursors are separated.
AC/VU precursor cells (filled arrowheads) and their siblings (unfilled arrowheads) at the L2 stage, visualised by LIN-12::GFP expression.
(A) wild-type: AC/VU precursors are in contact.
(B) cdc-42(RNAi) and (C) par-3(RNAi) animals with separated AC/VU precursors.
Anterior, left; ventral, down; scale bar, 10μm
Multiple Anchor Cells and vulval fate changes
To investigate whether the Muv phenotype of cdc-42(RNAi) is due to the presence of multiple ACs, we recovered cdc-42(RNAi) animals with either one or two ACs at L3 and then scored their vulval phenotype at adulthood: 78% (n=27) of 2 AC animals were Muv, compared to only 2% (n=100) of 1 AC animals. Similarly, scoring vulval induction directly showed that 80% (n=45) of 2 AC animals have extra induced VPCs compared with only 4% (n=55) of 1 AC animals (Table 1). These results strongly suggest that the additional ACs induce hyperinduction of the vulva.
In analyzing vulval hyperinduction in cdc-42(RNAi) and par-3(RNAi) animals, we observed unusual patterns of vulval cell fates. In the wild-type and in animals where VPCs are hyperinduced due to excessive Ras signalling, 1° fate VPCs never have 1° fate neighbours but are instead flanked by 2° fate neighbours due to LIN-12 Notch lateral signalling (Wang and Sternberg, 2001). In contrast, 1° fate cells are often adjacent to one another in cdc-42(RNAi) or par-3(RNAi) animals (Figs. 2D,E). These adjacent primary cells both downregulate LIN-12 (Supplementary Figure 2), as do normal 1° cells in wildtype animals (Shaye and Greenwald, 2002).
Adjacent 1° fates usually result from a defect in LIN-12 Notch mediated lateral signalling between VPCs. In a lin-12 null mutant all three induced VPCs adopt the 1° fate (Greenwald et al., 1983). We found that the incidence of adjacent primary fates in cdc-42(RNAi) animals is related to the distance between the ACs. Most cdc-42(RNAi) animals with two touching ACs have only a single 1° fate (71%, n=7, Figure 2B), indicating that the presence of two ACs is not alone sufficient to induce multiple 1° fates. The percentage of these cdc-42(RNAi) animals showing adjacent 1° fates (29%, n=7) is similar to the percentage having gaps between VPCs (39%; n=28). Because LIN-12/Notch signalling is largely contact dependent (Greenwald, 2005), the presence of a gap between VPCs would be expected to prevent lateral signalling and lead to adjacent 1° fates. Gaps between VPCs could also explain the observation of adjacent 1° fates in 11% (n=55) of 1 AC cdc-42(RNAi) animals. Where two ACs are separated by more than one VPC width (21μm, n=55), then most animals display multiple, non-adjacent 1° fates (84%, n=24, Figure 2C).
However, we observed a different phenotype in animals where the two ACs are not touching but are separated by less than the width of a VPC. Most of these animals have adjacent 1° fates (89%, n=9, Figure 2D), a rate more than double that of VPC gaps. This result suggests that when neighbouring VPCs each underlie an AC, the AC signal might override lateral signalling between the VPCs.
Multiple ACs result from disruption of the L1 gonad primordium
To understand the cause of the somatic gonad cell position defects, we examined earlier stages of gonad development. At hatching the gonad primordium is composed of a linear array of four cells, with two SGPs at the poles and two central primordial germ cells (PGCs) (Kimble and Hirsh, 1979). During embryogenesis the SGPs migrate from their anterior birth positions to the PGCs in the posterior region (Sulston et al., 1983).
We found that the L1 gonad primordium was disrupted in 55% (n=49) of cdc-42(RNAi) and 38% (n=60) of par-6(RNAi) animals (4A-E and H). Three different defects were seen: 1) disorganised primordia, with one of the SGPs in a central rather than polar position (Figure 4C, D); 5/5 cdc-42(RNAi) animals of this type developed a single gonad and one had two ACs. 2) separated primordia, with each SGP independently associated with one or more PGCs (Figure 4B); 2/2 cdc-42(RNAi) animals of this type developed split gonads and 2 ACs. 3) SGPs with no associated PGCs (Figure 4H, I); 8/8 cdc-42(RNAi) animals of this type had two ACs. This PGC-less phenotype was the most frequent (Figure 4H) and associated with SGP separation in 77% (n=13) of such animals. cdc-42(RNAi) animals with a normally organised L1 primordium developed intact gonads (18/18) and only a single AC (17/18), suggesting that CDC-42 is no longer required to maintain gonad integrity subsequent to hatching. These results show that cdc-42 and par-6 are required for proper cell positions within the four cell gonad primordium. In addition, reduction of their function leads to the absence of PGCs.
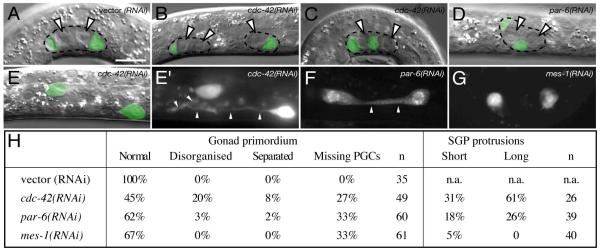
Figure 4. Abnormal SGP positions, gonad splitting and loss of PGCs.
(A-E) SGPs (green) marked by ehn-3A::GFP over Nomarski images of L1 gonad primordia; arrowheads mark PGCs, dotted outlines mark the primordium. (A) wild-type primordium has polar SGPs and central PGCs. (B) cdc-42(RNAi) animal with two separate primordia (C) cdc-42(RNAi) or (D) par-6(RNAi) animals with disorganised single primordia. (E) cdc-42(RNAi) animal with SGPs lacking associated PGCs.
(E’-G) Projections of ehn-3A::GFP expression in SGPs. In (E’) cdc-42(RNAi) or (F) par-6(RNAi) animals lacking PGCs, SGPs extend long processes. (G) in mes-1(RNAi) animals lacking PGCs, processes are absent.
Anterior, left; ventral, down; scale bars, 10μm.
(H) Table of gonad primordium phenotypes.
Loss of PGCs is due to mis-segregation of P-granules during embryogenesis
What is the cause of the striking absence of PGCs at L1 following RNAi of cdc-42 or par-6? During normal development the germ-line P-granules are segregated into the germline precursor P4, which divides to give rise to the PGCs, Z2 and Z3 (Strome and Wood, 1982). In contrast to control animals, where P-granules are found in only the two normal PGCs (n=67, Figure 5A), 55% (n=44) of cdc-42(RNAi) animals contain P-granules in multiple cells that were separated from the SGPs (Figure 5B). These abnormal PGL-1 positive cells coincided with body wall muscles in all cases (n=13, Figure 5B).
Figure 5. Germ cells adopt muscle fates and are not associated with SGPs in cdc-42(RNAi) animals.
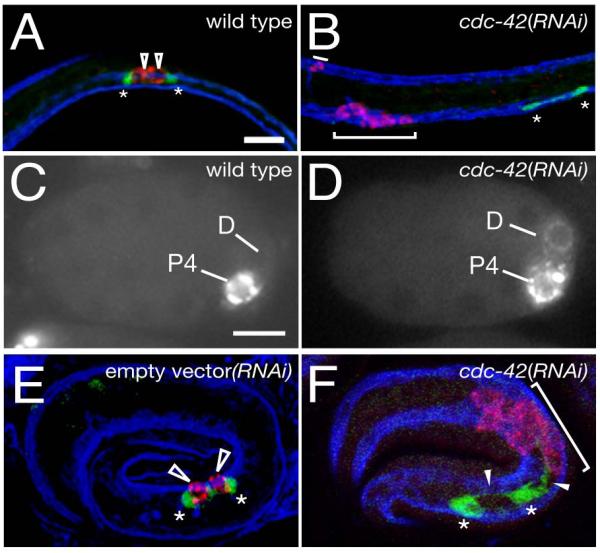
SGPs (ehn-3A::GFP, green), P-granules (anti-PGL-1 (Kawasaki et al., 1998), red in (A,B,E,F), white in (C,D)), and body wall muscles (NE8/4C6.3 marker (Goh and Bogaert, 1991), blue). (A) wild-type L1 gonad primordium with polar SGPs (asterisks) and central PGCs (unfilled arrowheads) (B) cdc-42(RNAi) L1 animal with P-granules (red, brackets) in muscle cells (blue), and SGPs (green) lacking associated PGCs (no P-granule containing cells).
(C) wild-type embryo after the division of P3, the P granule marker pgl-1::GFP (Cheeks et al., 2004) is only in the germline precursor P4. (D) cdc-42(RNAi) embryo with P-granules in both P4 and its sibling D.
(E) wild-type 3-fold embryo; SGPs are associated with PGCs (F) cdc-42(RNAi) embryo; SGPs extend processes that appear to be directed towards the PGL-1 staining cells (filled arrowheads).
Anterior, left; scale bar ,10μm.
The presence of P-granules in body wall muscles is reminiscent of mutations in mes-1, wherein P-granules are mis-segregated into the germline precursor P4 and its muscle precursor sibling D during the division of P3, usually resulting in both cells adopting a D-like muscle fate and the corresponding loss of PGCs (Strome et al., 1995). Studies of the early embryonic roles of CDC-42 and the PAR-3 complex have shown earlier mis-segregation of P-granules between the germline and somatic lineages (Gotta et al., 2001; Kemphues et al., 1988; Tabuse et al., 1998; Watts et al., 1996). Under our RNAi conditions, P-granules also mis-segregate between D and P4 following cdc-42(RNAi) (Figure 5C,D) and are found in body wall muscles later in embryogenesis (Figure 5F).
In cdc-42(RNAi) embryos lacking PGCs, the SGPs are found in the posterior of the embryo but are usually separated from one another (Figure 5F). This separation and lack of association with PGCs presumably leads to the formation of independent gonad primordia, each containing one of the SGPs (but no PGCs).
Gonad development in the absence of PGCs
To ask whether CDC-42 and the PAR complex have roles in gonad development independent of their roles in PGC formation, we compared their RNAi phenotypes to those of mes-1(RNAi) animals, which also lack PGCs but should not disrupt CDC-42 or the PAR-3 complex. Similar to cdc-42(RNAi) and par-6(RNAi) animals, we found that mes-1(RNAi) animals lacking PGCs often have two ACs and split gonads, and become Muv adults (Tables 1 and 2). However, unlike cdc-42(RNAi) or par-6(RNAi) animals, neither abnormal cell positions in single primordia nor separated primordia containing both SGPs and PGCs were seen in mes-1(RNAi) animals. These account for 51% of affected cdc-42(RNAi) and 13% of affected par-6(RNAi) animals. Therefore cdc-42 and par-6 are required for normal SGP cell positions and/or coalescence of the gonad.
We also observed that SGPs in cdc-42(RNAi) and par-6(RNAi) L1 animals extend long protrusions when they are not associated with PGCs (Figure 4E’,F,H). In contrast, protrusions were not seen in mes-1(RNAi) animals lacking PGCs (Figure 4G,H). Abnormal SGP protrusions were also observed in cdc-42(RNAi) embryos lacking PGCs (4/5 embryos after the one-fold stage) (Figure 5F). Interestingly, these protrusions appear to be directed towards the abnormal P-granule containing muscle cells (3/4). These observations suggest that the PGCs might normally provide a cue for positioning the SGPs and that the P-granule containing muscle cells still provide this cue. Since the SGPs in mes-1(RNAi) animals that lack PGCs do not extend long processes, it seems likely that their formation in cdc-42(RNAi) and par-6(RNAi) animals is not the result of loss of the PGCs, but of a separate effect upon cell architecture.
The separation between SGPs observed in the absence of PGCs suggests that PGCs may play an attractive role during normal coalescence of the gonad. However, previous studies that eliminated PGCs by ablating the PGC precursor P4 did not cause SGP separation: the SGPs generated a single small, but normal, somatic gonad (Junkersdorf and Schierenberg, 1992; Sulston et al., 1983). One explanation for the normal coalescence of SGPs following P4 ablation is that the corpse of P4 might provide an attractive cue. Consistent with this possibility, following ablation of P4 we observed that its corpse was clearly associated with the L1 gonad primordium in 5/8 individuals (Supplementary Figure 3). We hypothesise that the PGCs provide an attractive cue that aids in the final steps of gonad coalescence.
Q cell and axon migration defects
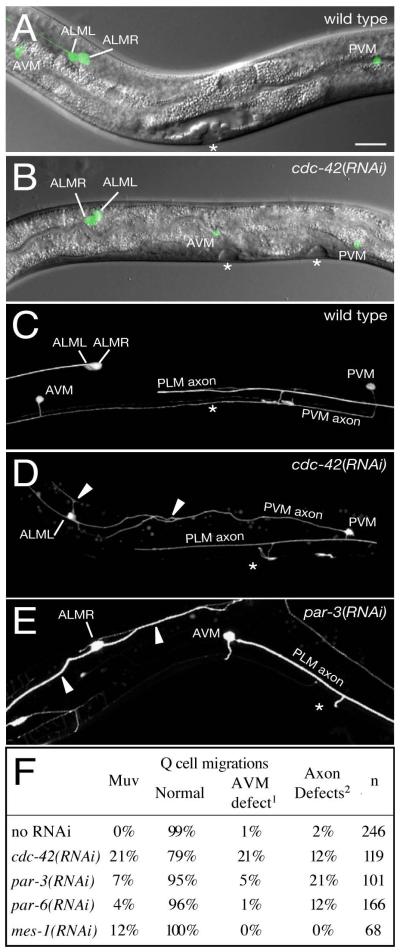
Numerous studies have implicated Cdc42 in regulating cell migrations. Whether or not this cell migration role involves the PAR-3 complex has been little studied. In order to investigate whether CDC-42 and the PAR-3 complex act together during cell migrations in C. elegans, we asked whether RNAi of cdc-42, par-3 or par-6 affected migration of neurons and axons. We used the marker mec-7::GFP (Ch’ng et al., 2003) to visualise the descendants of the migratory Q neuroblasts during L4. In the wild type, QR migrates anteriorly and QL posteriorly during L1. At L4 the descendants of these cells QR.paa (AVM) and QL.paa (PVM) have distinctive anterior and posterior positions respectively and both express mec-7::GFP. In 21% of cdc-42(RNAi) and 5% of par-3(RNAi) animals, AVM failed to complete migration and was posterior to its normal location (Figure 6B,C,E). In addition, 12% of cdc-42(RNAi) and 21% of par-3(RNAi) animals displayed axon migration and/or branching defects in ALM, PLM or A/PVM neurons (Figure 6D,E). We also observed axon migration and/or branching defects of ALM and PLM neurons in 12% of par-6(RNAi) animals (Figure 6E). These phenotypes are unlikely to be a consequence of gonad abnormalities as they are not displayed by mes-1(RNAi) animals (Figure 6F). These results show that CDC-42 and the PAR-3 complex are required for neuroblast and axon migration.
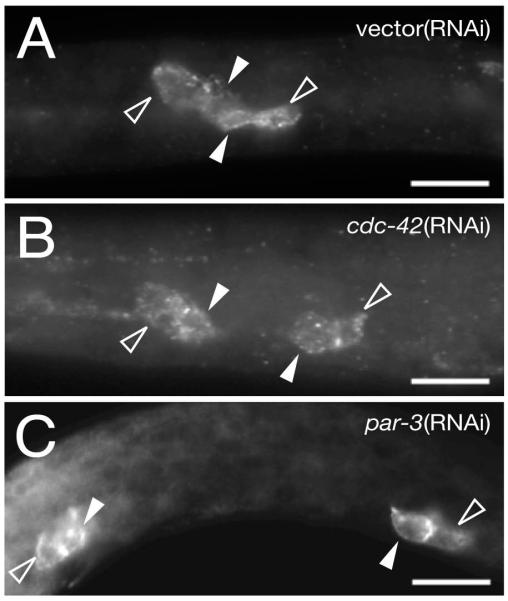
Figure 6. Q cell migrations and axon guidance are disrupted by cdc-42(RNAi).
(A,B) Overlays of Nomarski images of L4 animals with projections of mec-7::GFP expression (green). (A) wild-type; AVM neuron is anterior of the ALM neurons. (B) cdc-42(RNAi) animal with AVM neuron in the mid-body, posterior to ALM.
(C-E) Projections of mec-7::GFP expression in L4 animals marking neurons and axons. In wild type animals the PVM axon migrates ventrally and enters the ventral nerve cord and both PVM and ALM axons are unbranched (C). Some cdc-42(RNAi) (D) and par-3(RNAi) (E) animals show defects of mechanosensory neurons, including dorso-ventral guidance defects of the PVM axon (D), branching defects of both PVM and ALML axons (arrowheads in D) and neuron polarity defects with extended anterior and posterior processes of ALMR (arrowheads in E). Only the left side of the body is shown in (D) and the right side in (E).
Anterior, left; ventral, down; vulvae are marked by asterisks; scale bar, 20μm
(F) Table of Q cell migration and axon guidance defects.
1all affected control and par-6(RNAi) animals had only mild defects, while 7/25 affected cdc-42(RNAi) and 2/5 affected par-3(RNAi) animals had strong defects.2defects include dorso-ventral and anterior-posterior guidance defects, abnormal branching and defasiculation of ALM axons (cdc-42, par-3 or par-6(RNAi)), PLM axons (all treatments) and PVM axons (cdc-42 or par-3(RNAi)) AVM: great grand-daughter (QR.paa) of the anterior migrating QR neuroblast PVM: great grand-daughter (QL.paa) of the posterior migrating QL neuroblast
Discussion
CDC-42 and the conserved PAR-3/PAR-6/aPKC complex were previously shown to function together in establishing cell polarity in a variety of cell types (Etienne-Manneville, 2004; Etienne-Manneville and Hall, 2003b). Here, we show that a range of different polarised growth and cell migration processes show similar requirements for CDC-42 and the PAR-3 complex. We also demonstrate multiple roles for these proteins in formation of the C. elegans gonad. Our results suggest that these proteins are likely to act together widely in cell migration and polarised cell growth in animals.
CDC-42 and the PAR-3/PAR-6/PKC-3 complex act together broadly to regulate cell migration and polarised growth in C. elegans
When cdc-42, par-3 or par-6 are knocked down the migration, polarised growth or positioning of several cell types is perturbed. Abnormal elongation of the VPCs and seam cells results in gaps between cells, and seam cells have a rounded rather than an elongated morphology. In ALM, PLM, and A/PVM neurons, axon migration is abnormal and PVM neuroblast migration is incomplete. Somatic gonad precursors (SGPs) adopt abnormal positions within the gonad primordium or form separated primordia. Thus, for each of these cells types, CDC-42 and the PAR-3 complex function in the same processes.
The phenotypes observed in this study are in the context of partial gene knockdown because complete RNAi inhibition of CDC-42 or members of the PAR-3 complex results in embryonic lethality. Furthermore, the cells assayed retain some polarity. For example, VPCs and seam cells have apical junctions and neurons have axons. Our results do not distinguish whether the observed defects are due to impaired cell polarisation, or are a direct effect on cell migration or polarised growth, or both. We also do not yet know the site of action of these genes. Since RNAi after hatching does not induce the effects we observed here (data not shown), maternal protein is sufficient for these processes. In the future, the use of cell specific constitutively active or dominant negative proteins could address this question. Nevertheless, our results indicate that CDC-42 and the PAR-3 complex function together broadly in a range of different developmental processes.
CDC-42 and the PAR-3/PAR-6/PKC-3 complex together regulate multiple aspects of gonad formation
In many cdc-42(RNAi) or par-6(RNAi) animals, PGCs are missing due to conversion of the germ cell lineage to a muscle fate at the 25-cell stage, a phenotype originally described for mes-1 mutants (Strome et al., 1995). The majority of these animals also show separation of SGPs suggesting that the PGCs are required for coalescence of the SGPs into a single primordium. This partial reliance upon PGCs for coalescence is supported by the fact that the same phenotype is observed in mes-1(RNAi) animals which have a specific defect in PGC specification (Strome et al., 1995). A role for PGCs in attracting SGPs is also suggested by the observation that the abnormal processes generated by cdc-42(RNAi) or par-6(RNAi) SGPs in the absence of PGCs, are usually directed towards the P-granule containing descendants of D and P4.
Our suggestion that PGCs are important for gonad coalescence conflicts with previous studies suggesting that PGCs are dispensable for somatic gonad development: ablation of P4, the precursor of the PGCs, caused no apparent somatic gonad defects (Junkersdorf and Schierenberg, 1992; Sulston et al., 1983). However, in repeating these ablation experiments, we found that the P4 corpse is often incorporated into the gonad primordium, and therefore could be the source of an SGP cue. We propose that the PGCs provide an attractive cue for gonad coalescence.
In addition to those individuals lacking PGCs, cdc-42(RNAi) or par-6(RNAi) also result in animals which retain PGCs but have abnormal SGP positions within the gonad primordium or separate primordia. One possible explanation for these phenotypes is that the SGPs fail to migrate to their normal positions. However we have not detected significant differences in SGP positions during embryogenesis following cdc-42(RNAi) (data not shown). Alternately the primordium might initially form normally but become disrupted by subsequent movement of the SGPs. Whether this defect is a result of abnormal SGP behaviour or abnormal cohesion of the PGCs is an intriguing question for future studies.
CDC-42 and the PAR-3 complex in neuronal development and migration
Cdc42 was first identified in S. cerevisiae by a mutant that failed in polarised growth, arresting as large unbudded cells (Adams et al., 1990; Johnson and Pringle, 1990). Subsequently, it was shown that Cdc42 is also important in polarised cell shape changes in Drosophila epithelia, indicating a conserved function in animals (Eaton et al., 1995). Members of the Par3 complex were identified by their requirement for the polarity of the C. elegans zygote (Gotta et al., 2001; Kemphues et al., 1988; Tabuse et al., 1998; Watts et al., 1996) . Although it is now well-established that Cdc42 and the Par3 complex act together in establishing animal cell polarity, there has been little previous work investigating their shared functions in other processes. We discuss here studies in neuronal and other systems showing roles for members of the Par3 complex or Cdc42 and their possible links and effectors.
Although a role for the Par3 complex in polarised epithelial cell growth has not yet been reported, its requirement in axon formation supports such a function. Overexpression of Par3 or Par6 in mammalian hippocampal neurons in culture inhibits axon formation (Shi et al., 2003). Similarly, overexpression of Par6 in glial-guided neurons inhibits axon formation, causes shorter neurites and impairs migration (Solecki et al., 2004). In contrast, Drosophila interneurons mutant for Par-3 (Baz), Par-6 or aPKC have normal axon development in vivo (Rolls and Doe, 2004). None of these studies investigated involvement of Cdc42, but work in other neuron types in culture has shown a requirement for Cdc42 in neurite outgrowth (Li et al., 2002; Yuan et al., 2003) Cdc42 has also been shown to be involved in axon guidance in Xenopus and Drosophila (Kim et al., 2002; Yuan et al., 2003). In the present study we show that CDC-42 and the PAR-3 complex are both needed for axon guidance and neuronal migration in C. elegans. We did not uncover a role in axon formation, but our RNAi conditions may not have been sufficient to inhibit this process. Our findings, along with data from mammalian systems discussed above, suggest that CDC-42 and the PAR-3 complex act together in regulating neuronal polarity and axon guidance. It may be the case that these functions are also conserved in Drosophila but that low levels of residual maternal protein are sufficient to allow normal development of zygotic mutant neurons.
How might CDC-42 and the PAR-3 complex regulate cell migrations and polarised growth? In mammalian astrocytes, activation of Cdc42 at the leading edge leads to recruitment of a Par-3/Par-6/aPKC complex and reorientation of the MTOC in the direction of migration (Etienne-Manneville and Hall, 2001; Etienne-Manneville and Hall, 2003a). Orientation of the centrosome also plays a role in axon outgrowth and neuronal migrations. In glial guided neurons centrosomal re-orientation precedes axon outgrowth and overexpression of Par6 or aPKC impairs both this centrosome movement and the subsequent axon outgrowth and cell migration (Solecki et al., 2004). In the case of astrocytes, reorientation of the MTOC involves GSK-3β regulation of APC (Etienne-Manneville and Hall, 2003a). As the C. elegans APC homologue APR-1 plays a role in the migration and elongation of hypodermal cells during embryogenesis (Hoier et al., 2000), it is a potential effector of CDC-42 and the PAR-3 complex in regulating cell migration and polarised growth.
Other possible effectors are Rac and Rho GTPases. Rac GTPases regulate cell migration through inducing actin polymerisation at the leading edge (Wittmann and Waterman-Storer, 2001). In mammalian neural cells Cdc42 and Par-3 activate Rac through interaction with Rac GEFs (Chen and Macara, 2005; Nishimura et al., 2005). In C. elegans the three Rac homologues, CED-10, MIG-2 and RAC-2 have partially redundant roles in the migration of a wide-range of cell types (Lundquist et al., 2001). MIG-2, for example, is required for axon guidance and Q cell migrations, with mutants displaying abbreviated migrations of QR and QL (Zipkin et al., 1997). Recently, Norman et al (2005) showed that CDC-42 acts with MIG-2 and CED-10 in the migration of gonadal distal tip cells, supporting a possible role for MIG-2 and CED-10 as CDC-42 effectors. In addition, Cram et al (2006) recently identified cdc-42 as a gene required for distal tip cell migration in an RNAi screen. In contrast to Rac, Rho regulates actin behaviour at the rear of migrating cells and is excluded from the leading edge (Wittmann and Waterman-Storer, 2001). In migrating fibroblasts the Par6/aPKC complex promotes protrusive activity by recruiting the E3 ubiquitin ligase Smurf-1, which locally degrades RhoA (Wang et al., 2003). In C. elegans, the Rho homologue, RHO-1, plays a role in P cell migration (Spencer et al., 2001). Thus CDC-42 and the PAR-3 complex might regulate cell migrations in C. elegans via modulation of Rac and/or Rho activity.
Finally, in addition to regulation of the cytoskeleton, Cdc42 has also been shown to be involved in polarised secretion via the exocyst complex (Zhang et al., 2001). Members of this complex are also potential candidates for mediating elongation of the VPCs and seam cells.
In summary, we have provided evidence that CDC-42 and the PAR-3 complex act together in a range of cell types for polarised cell growth, cell migrations and cell positioning. The challenge for the future will be to link these proteins to their activators and effectors to understand how they together carry out these diverse processes.
Supplementary Material
Acknowledgements
Thanks to Dan Shaye, Neil Hopper, Renaud Legouis, Paul Sternberg, Iva Greenwald, Henrik Korswagen, Jim Priess, Judith Kimble, Susan Strome and Stuart Kim for strains, reagents and advice. We are grateful to Gino Poulin, Monica Gotta, David Rivers, Nathalie Le Bot and Sarah Bray for help and useful discussions and especially to DMR and Pouly for comments on this manuscript. Thanks also to Ravi Kamath and Andy Fraser who made the initial observation of a Muv phenotype in cdc-42(RNAi). Some strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR), and the International C. elegans Gene Knockout Consortium. DW was supported by a Wellcome Trust PhD studentship. LM is supported by a grant from the National Science Foundation (IOB-0617897). JA is a Wellcome Trust Senior Research Fellow (054523).
References
- Adams AE, Johnson DI, Longnecker RM, Sloat BF, Pringle JR. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol. 1990;111:131–42. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono S, Legouis R, Hoose WA, Kemphues KJ. PAR-3 is required for epithelial cell polarity in the distal spermatheca of C. elegans. Development. 2004;131:2865–74. doi: 10.1242/dev.01146. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capowski EE, Martin P, Garvin C, Strome S. Identification of grandchildless loci whose products are required for normal germ-line development in the nematode Caenorhabditis elegans. Genetics. 1991;129:1061–72. doi: 10.1093/genetics/129.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch’ng Q, Williams L, Lie YS, Sym M, Whangbo J, Kenyon C. Identification of genes that regulate a left-right asymmetric neuronal migration in Caenorhabditis elegans. Genetics. 2003;164:1355–67. doi: 10.1093/genetics/164.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeks RJ, Canman JC, Gabriel WN, Meyer N, Strome S, Goldstein B. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr Biol. 2004;14:851–62. doi: 10.1016/j.cub.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol. 2005;7:262–9. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- Cram EJ, Shang H, Schwarzbauer JE. A systematic RNA interference screen reveals a cell migration gene network in C. elegans. J Cell Sci. 2006;119:4811–8. doi: 10.1242/jcs.03274. [DOI] [PubMed] [Google Scholar]
- Czuchra A, Wu X, Meyer H, van Hengel J, Schroeder T, Geffers R, Rottner K, Brakebusch C. Cdc42 is not essential for filopodium formation, directed migration, cell polarization, and mitosis in fibroblastoid cells. Mol Biol Cell. 2005;16:4473–84. doi: 10.1091/mbc.E05-01-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton S, Auvinen P, Luo L, Jan YN, Simons K. CDC42 and Rac1 control different actin-dependent processes in the Drosophila wing disc epithelium. J Cell Biol. 1995;131:151–64. doi: 10.1083/jcb.131.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S. Cdc42--the centre of polarity. J Cell Sci. 2004;117:1291–300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 2001;106:489–98. doi: 10.1016/s0092-8674(01)00471-8. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature. 2003a;421:753–6. doi: 10.1038/nature01423. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Cell polarity: Par6, aPKC and cytoskeletal crosstalk. Curr Opin Cell Biol. 2003b;15:67–72. doi: 10.1016/s0955-0674(02)00005-4. [DOI] [PubMed] [Google Scholar]
- Francis GR, Waterston RH. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J Cell Biol. 1985;101:1532–49. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh PY, Bogaert T. Positioning and maintenance of embryonic body wall muscle attachments in C. elegans requires the mup-1 gene. Development. 1991;111:667–81. doi: 10.1242/dev.111.3.667. [DOI] [PubMed] [Google Scholar]
- Gotta M, Abraham MC, Ahringer J. CDC-42 controls early cell polarity and spindle orientation in C. elegans. Curr Biol. 2001;11:482–8. doi: 10.1016/s0960-9822(01)00142-7. [DOI] [PubMed] [Google Scholar]
- Greenwald I. LIN-12/Notch signaling in C.elegans. In: Community T. C. e. R., editor. WormBook. 2005. doi/10.1895/wormbook.1.7.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald IS, Sternberg PW, Horvitz HR. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell. 1983;34:435–44. doi: 10.1016/0092-8674(83)90377-x. [DOI] [PubMed] [Google Scholar]
- Hoier EF, Mohler WA, Kim SK, Hajnal A. The Caenorhabditis elegans APC-related gene apr-1 is required for epithelial cell migration and Hox gene expression. Genes Dev. 2000;14:874–86. [PMC free article] [PubMed] [Google Scholar]
- Hurd DD, Kemphues KJ. PAR-1 Is Required for Morphogenesis of the Caenorhabditis elegans Vulva. Dev Biol. 2003;253:54–65. doi: 10.1006/dbio.2002.0866. [DOI] [PubMed] [Google Scholar]
- Inoue T, Sherwood DR, Aspock G, Butler JA, Gupta BP, Kirouac M, Wang M, Lee PY, Kramer JM, Hope I, Burglin TR, Sternberg PW. Gene expression markers for Caenorhabditis elegans vulval cells. Mech Dev. 2002;119(Suppl 1):S203–9. doi: 10.1016/s0925-4773(03)00117-5. [DOI] [PubMed] [Google Scholar]
- Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol. 2000;2:531–9. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- Johnson DI, Pringle JR. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111:143–52. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junkersdorf B, Schierenberg E. Embryogenesis in C.elegans after elimination of individual blastonmeres or induced alteration of the cell-division order. Roux’s Archives of Developmental Biology. 1992;202:17–22. doi: 10.1007/BF00364593. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–7. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0002. RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X, Greenwald I. Post-transcriptional regulation of the E/Daughterless ortholog HLH-2, negative feedback, and birth order bias during the AC/VU decision in C. elegans. Genes Dev. 2003;17:3100–11. doi: 10.1101/gad.1160803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I, Shim YH, Kirchner J, Kaminker J, Wood WB, Strome S. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell. 1998;94:635–45. doi: 10.1016/s0092-8674(00)81605-0. [DOI] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–20. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Kim MD, Kolodziej P, Chiba A. Growth cone pathfinding and filopodial dynamics are mediated separately by Cdc42 activation. J Neurosci. 2002;22:1794–806. doi: 10.1523/JNEUROSCI.22-05-01794.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Le Bot N, Tsai MC, Andrews RK, Ahringer J. TAC-1, a regulator of microtubule length in the C. elegans embryo. Curr Biol. 2003;13:1499–505. doi: 10.1016/s0960-9822(03)00577-3. [DOI] [PubMed] [Google Scholar]
- Lewis JA, Fleming JT. Basic Culture Methods. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans: Modern Biological Analysis of an Organism. Academic Press; 1995. [Google Scholar]
- Li X, Saint-Cyr-Proulx E, Aktories K, Lamarche-Vane N. Rac1 and Cdc42 but not RhoA or Rho kinase activities are required for neurite outgrowth induced by the Netrin-1 receptor DCC (deleted in colorectal cancer) in N1E-115 neuroblastoma cells. J Biol Chem. 2002;277:15207–14. doi: 10.1074/jbc.M109913200. [DOI] [PubMed] [Google Scholar]
- Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, Pawson T. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat Cell Biol. 2000;2:540–7. doi: 10.1038/35019582. [DOI] [PubMed] [Google Scholar]
- Liu Z, Fujii T, Nukazuka A, Kurokawa R, Suzuki M, Fujisawa H, Takagi S. C. elegans PlexinA PLX-1 mediates a cell contact-dependent stop signal in vulval precursor cells. Dev Biol. 2005;282:138–51. doi: 10.1016/j.ydbio.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Lundquist EA, Reddien PW, Hartwieg E, Horvitz HR, Bargmann CI. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development. 2001;128:4475–88. doi: 10.1242/dev.128.22.4475. [DOI] [PubMed] [Google Scholar]
- Macara IG. Par proteins: partners in polarization. Curr Biol. 2004;14:R160–2. [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991;10:3959–70. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskowski J, Li Y, Kimble J. The sys-1 gene and sexual dimorphism during gonadogenesis in Caenorhabditis elegans. Dev Biol. 2001;230:61–73. doi: 10.1006/dbio.2000.9998. [DOI] [PubMed] [Google Scholar]
- Murphy AM, Montell DJ. Cell type-specific roles for Cdc42, Rac, and RhoL in Drosophila oogenesis. J Cell Biol. 1996;133:617–30. doi: 10.1083/jcb.133.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J. PAR proteins and the establishment of cell polarity during C. elegans development. Bioessays. 2005;27:126–35. doi: 10.1002/bies.20175. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Yamaguchi T, Kato K, Yoshizawa M, Nabeshima Y, Ohno S, Hoshino M, Kaibuchi K. PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat Cell Biol. 2005;7:270–7. doi: 10.1038/ncb1227. [DOI] [PubMed] [Google Scholar]
- Norman KR, Fazzio RT, Mellem JE, Espelt MV, Strange K, Beckerle MC, Maricq AV. The Rho/Rac-family guanine nucleotide exchange factor VAV-1 regulates rhythmic behaviors in C. elegans. Cell. 2005;123:119–32. doi: 10.1016/j.cell.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Pinheiro EM, Montell DJ. Requirement for Par-6 and Bazooka in Drosophila border cell migration. Development. 2004;131:5243–51. doi: 10.1242/dev.01412. [DOI] [PubMed] [Google Scholar]
- Poulin G, Dong Y, Fraser AG, Hopper NA, Ahringer J. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans. Embo J. 2005;24:2613–23. doi: 10.1038/sj.emboj.7600726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls MM, Doe CQ. Baz, Par-6 and aPKC are not required for axon or dendrite specification in Drosophila. Nat Neurosci. 2004;7:1293–5. doi: 10.1038/nn1346. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Greenwald I. Cell autonomy of lin-12 function in a cell fate decision in C. elegans. Cell. 1989;57:1237–45. doi: 10.1016/0092-8674(89)90060-3. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Schedl T, Greenwald I. Cell-cell interactions prevent a potential inductive interaction between soma and germline in C. elegans. Cell. 1990;61:939–51. doi: 10.1016/0092-8674(90)90060-r. [DOI] [PubMed] [Google Scholar]
- Shaye DD, Greenwald I. Endocytosis-mediated downregulation of LIN-12/Notch upon Ras activation in Caenorhabditis elegans. Nature. 2002;420:686–90. doi: 10.1038/nature01234. [DOI] [PubMed] [Google Scholar]
- Shi SH, Jan LY, Jan YN. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 2003;112:63–75. doi: 10.1016/s0092-8674(02)01249-7. [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci. 2004;7:1195–203. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- Spencer AG, Orita S, Malone CJ, Han M. A RHO GTPase-mediated pathway is required during P cell migration in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2001;98:13132–7. doi: 10.1073/pnas.241504098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Martin P, Schierenberg E, Paulsen J. Transformation of the germ line into muscle in mes-1 mutant embryos of C. elegans. Development. 1995;121:2961–72. doi: 10.1242/dev.121.9.2961. [DOI] [PubMed] [Google Scholar]
- Strome S, Wood WB. Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1982;79:1558–62. doi: 10.1073/pnas.79.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Hodgkin J. Methods. In: Wood WB, Community T. C. e. R., editors. The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–56. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Tabuse Y, Izumi Y, Piano F, Kemphues KJ, Miwa J, Ohno S. Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development. 1998;125:3607–14. doi: 10.1242/dev.125.18.3607. [DOI] [PubMed] [Google Scholar]
- Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–9. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- Wang M, Sternberg PW. Patterning of the C. elegans 1 degrees vulval lineage by RAS and Wnt pathways. Development. 2000;127:5047–58. doi: 10.1242/dev.127.23.5047. [DOI] [PubMed] [Google Scholar]
- Wang M, Sternberg PW. Pattern formation during C. elegans vulval induction. Curr Top Dev Biol. 2001;51:189–220. doi: 10.1016/s0070-2153(01)51006-6. [DOI] [PubMed] [Google Scholar]
- Watts JL, Etemad-Moghadam B, Guo S, Boyd L, Draper BW, Mello CC, Priess JR, Kemphues KJ. par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development. 1996;122:3133–40. doi: 10.1242/dev.122.10.3133. [DOI] [PubMed] [Google Scholar]
- Wittmann T, Waterman-Storer CM. Cell motility: can Rho GTPases and microtubules point the way? J Cell Sci. 2001;114:3795–803. doi: 10.1242/jcs.114.21.3795. [DOI] [PubMed] [Google Scholar]
- Yang L, Wang L, Zheng Y. Gene Targeting of Cdc42 and Cdc42GAP Affirms the Critical Involvement of Cdc42 in Filopodia Induction, Directed Migration, and Proliferation in Primary Mouse Embryonic Fibroblasts. Mol Biol Cell. 2006 doi: 10.1091/mbc.E06-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Bais C, Greenwald I. Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Science. 2004;303:663–6. doi: 10.1126/science.1091639. [DOI] [PubMed] [Google Scholar]
- Yuan XB, Jin M, Xu X, Song YQ, Wu CP, Poo MM, Duan S. Signalling and crosstalk of Rho GTPases in mediating axon guidance. Nat Cell Biol. 2003;5:38–45. doi: 10.1038/ncb895. [DOI] [PubMed] [Google Scholar]
- Zhang X, Bi E, Novick P, Du L, Kozminski KG, Lipschutz JH, Guo W. Cdc42 interacts with the exocyst and regulates polarized secretion. J Biol Chem. 2001;276:46745–50. doi: 10.1074/jbc.M107464200. [DOI] [PubMed] [Google Scholar]
- Zipkin ID, Kindt RM, Kenyon CJ. Role of a new Rho family member in cell migration and axon guidance in C. elegans. Cell. 1997;90:883–94. doi: 10.1016/s0092-8674(00)80353-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.