Abstract
Population stratification occurs when a study population is comprised of several sub-populations, and can result in increased false positive findings in genomewide-association studies. Recently published work shows that sub-population-specific positive assortative mating at the genotypic level results in population stratification. We show that if the allele frequency of a single nucleotide polymorphism responsible for a trait varies between sub-populations and there is no dominance variance, then the heritability of the trait increases, primarily due to an increase in the additive genetic variance of the trait.
Keywords: population stratification, heritability, genomewide-association studies
Introduction
The phenotype of an individual is affected by both genetic and environmental factors. One of the major challenges of the Human Genome Project is identifying the genetic causes underlying phenotypic variation of quantitative traits (Hill et al., 2008). Fisher (1918) unified biometrical and Mendelian genetics by demonstrating theoretically the observed resemblance between relatives due to genetic factors. The heritability (‘narrow-sense heritability') of a trait is the ratio of the additive genetic variance (VA.Hardy–Weinberg equilibrium (HWE)) to the total variance of the trait (VP), where the total variance of the trait is the sum of the genetic variance and the environmental variance of the trait. Therefore, the heritability of a trait lies between 0 and 1. The heritability of a trait is a population-specific parameter, and can vary over time from generation to generation based on the change in the frequency of an allele that changes the value of the trait due to drift, selection, mutation and migration or due to changes in environment or other non-genetic effects. Because heritability is population specific, there is variability in the estimates of heritability obtained from different studies for the same trait. For example, estimates of heritability for the psychiatric disorders schizophrenia and bipolar disorder range from 80 to 90% (Crow, 2011).
The heritability of a trait increases with phenotypic assortative mating (Falconer and Mackay, 1996, pp 175–176). Sebro et al. (2010) showed that a predictable phenomenon is seen in the presence of population stratification—an increase in the number of unions between spouse pairs with the same genotypes compared with that expected assuming random mating. Sebro et al. (2010) also showed that this increase is directly proportional to the variance of the genotype frequency between sub-populations when the same genotypes comprise the mating types. Sub-population or ancestry-related positive assortative mating (Risch et al., 2009; Sebro et al., 2010) results in population stratification, and is seen at all loci where the allele frequency differs between sub-populations.
Positive assortative mating increases the heritability of a trait. However, assortative mating occurs at the phenotypic level, where mates select each other based on a physical or observable trait. The positive assortative mating noted within a stratified population is at the genetic level (genetic homogamy). No calculations exist in the literature that show how positive assortative mating at the genetic level affects the heritability of a trait. We use two methods for calculating the mating type frequencies in the presence of population stratification (Sebro and Rogus, 2010; Sebro et al., 2010), and show theoretically how the heritability of a trait changes with population stratification using single-locus and two-locus models.
Materials and methods
Single-locus model
Consider the case where the population is comprised of G sub-populations, where G, as well as the members of each sub-population is unknown. We assume that there is random mating within each sub-population, but no mating between sub-populations, and no selection from generation to generation. We further assume that generations are distinct and do not overlap, and that there is no change in the number of individuals in each sub-population across generations. Consider a single biallelic marker or single nucleotide polymorphism (SNP) with alleles A and B. Let the frequency of the A allele in the total population be p, and the frequency of the B allele be 1−p=q. Let a represent the average phenotypic value of individuals with the AA genotype, d represent the average phenotypic value of individuals with the AB genotype and −a represent the average phenotypic value of individuals with the BB genotype. We use the method described by Sebro et al. (2010) (Method 1) and the method described by Sebro and Rogus (2010) (Method 2) for calculating the frequencies of the mating types in the presence of population stratification.
If we assume symmetry between the genotypes of spouse pairs (AA × AB≡AB × AA), then there are only 6 unique mating types. The frequency of each of the 6 mating types can be calculated using Method 1 which is based on the variance of the allele A frequency between sub-populations, Var(pi), and the variance of the AA genotype frequency between sub-populations, Var(p2i) and the variance of the BB genotype frequency between sub-populations, Var(q2i). Alternatively, the frequency of each of the six mating types can be calculated using Method 2, which is based on the average allele A frequency in the population, p; the variance (second central moment) of the frequency of the A allele between sub-populations, Var(pi); the third and fourth central moments of the frequency of the A allele between sub-populations.
We then use these mating type frequencies to calculate the covariance between the following relatives: monozygous (MZ) twins, siblings and dizygotic twins, parent-offspring, half-siblings, grandparent-offspring, avuncular pairs, first-cousin pairs, and unrelated individuals. These results are shown in Table 1. Recent data show that most of the genetic variance of complex traits are due to the additive genetic variance and not the dominance genetic variance, which is negligible (d∼0) (Hill et al., 2008). We recalculate the covariances between the relative pairs assuming no dominance variance (d=0) in Table 2. Our calculations assume that the genetic variability in a quantitative trait is caused by a single SNP in a single gene, and that the SNP allele frequency varies between sub-populations.
Table 1. Comparison of the covariance between relatives assuming HWE compared with that calculated in the presence of population stratification.
| HWE | Population stratification Model 1 | Population stratification Model 2 | |
|---|---|---|---|
| Monozygotic twins | VA.HWE+VD.HWE | VA.HWE+VD.HWE+2Var(pi)a2+ad × (4 × Var(pi) −8 × p × Var(pi))+d2(−2Var(pi)+8pVar(pi) −8p2Var(pi)−4Var(p2i)) | VA.HWE+VD.HWE+2Fpq((a+d(p−q))2+2Fpq(−2d2(p−q)2 −4ad(p−q)−2d2Fpq)) |
| Sib-pairs/dizygotic twins | ½ VA. HWE+¼ × VD.HWE | ½ VA.HWE+¼ × VD.HWE+3a2Var(pi)+ad(−4pVar(pi) +2Var(pi)+Var(p2i)−Var(q2i))+d2(2Var(pi)(−1+3p−3p2 +1/2Var(pi)−4Var(pi)2) | ½ VA.HWE+¼ × VD.HWE+3a2Fpq+ad(Fpq(4p−2)+4ϕ3)+ d2(2Fp2q2(1−2F)+ϕ3(−2+4p)+ϕ4) |
| Parent-offspring | ½ × VA.HWE | ½ VA.HWE+3a2Var(pi)+ad(−4p Var(pi)+2Var(pi)+ Var(p2i)−Var(q2i))+d2(1+8pq+4Var(pi)) | ½ VA.HWE+3a2Fpq+ad(Fpq(4p−2)+4ϕ3)+d2(Fpq(−1+8pq−4Fpq)) |
| Grandparent-offspring | ¼ × VA.HWE | ¼ × VA.HWE+7/2a2Var(pi)+ad(Var(pi)−2pVar(pi) +3/2Var(p2i)−3/2Var(q2i))+d2(Var(pi) × (−5/2−12p +12p2+2Var(pi)+Var(p2i)+Var(q2i)) | ¼ × VA.HWE+a2(7/2Fpq)+ad(−9Fpq+14Fp2q+4Fpq2+6ϕ3) +d2(3/2Fpq−12Fp2q+12Fp3q−8Fp2q2−4ϕ3+8pϕ3+2ϕ4) |
| Avuncular pairs | ¼ × VA.HWE | ¼ × VA.HWE+7/2a2Var(pi)+ad(Var(pi)−2pVar(pi)+ 3/2Var(p2i)−3/2Var(q2i))+d2(Var(pi) × (−5/2−12p +12p2+2Var(pi)+Var(p2i)+Var(q2i)) | ¼ × VA.HWE+a2(7/2Fpq)+ad(−9Fpq+14Fp2q+4Fpq2+6ϕ3) +d2(3/2Fpq−12Fp2q+12Fp3q-8Fp2q2−4ϕ3+8pϕ3+2ϕ4) |
| Half-siblings | ¼ × VA.HWE | ¼ × VA.HWE+7/2a2Var(pi)+ad(Var(pi)−2pVar(pi) +3/2Var(p2i)−3/2Var(q2i))+d2(Var(pi) × (−5/2−12p+12p2+2Var(pi)+Var(p2i)+Var(q2i)) | ¼ × VA.HWE+a2(7/2Fpq)+ad(−9Fpq+14Fp2q+4Fpq2+6ϕ3) +d2(3/2Fpq−12Fp2q+12Fp3q−8Fp2q2−4ϕ3+8pϕ3+2ϕ4) |
| First-cousins | 1/8 × VA.HWE | 1/8 × VA.HWE+15/4a2Var(pi)+ad(−13/2Var(pi) +13pVar(pi))+d2(Var(pi) × (−13/4+2p−2p2+3Var(pi)) +3/2Var(p2i)+3/2Var(q2i)) | 1/8 × VA.HWE+a2(15/4Fpq)+ad(−13/2Fpq+13Fp2q)+d2 (11/4Fpq−10Fp2q+10Fp3q+6(p−q)ϕ3+3ϕ4) |
| Unrelated individuals | 0 | 4a2Var(pi)+ad(2Var(p2i)−2Var(q2i))+d2(−4Var(pi) +2Var(p2i)+2Var(q2i)) | 4a2Fpq+ad(Fpq(16p−8)+8ϕ3)+d2(Fpq(4−16pq−4Fpq)+ (16p−8)ϕ3+4ϕ4) |
Abbreviation: HWE, Hardy–Weinberg equilibrium.
VA.HWE=2pq(a+d(p−q))2.
VD.HWE=4d2p2q2.
φ3=third central moment of the frequency of the A allele between subpopulations.
φ4=fourth central moment of the frequency of the A allele between subpopulations.
Var(p2i)=variance of the frequency of the AA genotype between subpopulations.
Var(q2i)=variance of the frequency of the BB genotype between subpopulations.
Table 2. Comparison of the covariance between relatives assuming HWE compared with that calculated in the presence of population stratification, when there is no dominance variance (d=0).
| HWE | Population stratification Model 1 | Population stratification Model 2 | |
|---|---|---|---|
| Monozygotic twins | VA.HWE | VA.HWE+2a2Var(pi) | VA.HWE+2Fpqa2 |
| Sib-pairs/dizygotic twins | ½ VA.HWE | ½ VA.HWE+3a2Var(pi) | ½ VA.HWE+3a2Fpq |
| Parent-offspring | ½ × VA.HWE | ½ VA.HWE+3a2Var(pi) | ½ VA.HWE+3a2Fpq |
| Grandparent-offspring | ¼ × VA.HWE | ¼ × VA.HWE+7/2a2Var(pi) | ¼ × VA.HWE+a2(7/2Fpq) |
| Avuncular pairs | ¼ × VA.HWE | ¼ × VA.HWE+7/2a2Var(pi) | ¼ × VA.HWE+a2(7/2Fpq) |
| Half-siblings | ¼ × VA.HWE | ¼ × VA.HWE+7/2a2Var(pi) | ¼ × VA.HWE+a2(7/2Fpq) |
| First-cousins | 1/8 × VA.HWE | 1/8 × VA.HWE+15/4a2Var(pi) | 1/8 × VA.HWE+a2(15/4Fpq) |
| Unrelated individuals | 0 | 4a2Var(pi) | 4a2Fpq |
Abbreviation: HWE, Hardy–Weinberg equilibrium.
VA.HWE=2pqa2.
VD.HWE=4d2p2q2.
Var(pi)=variance of the frequency of the A allele between subpopulations.
Theoretical plots of the correlation between different relative pairs in the presence of population stratification are generated, and the values obtained are compared with those calculated assuming HWE. The mean trait value in a population in HWE, μHWE, is equal to a(p−q)−2dpq, and the variance of the trait in a population in HWE, VP.HWE is 2pq[a+d(p−q)]2+4d2p2q2. Population stratification leads to excess homozygotes and a deficiency of heterozygotes, a phenomenon known as the Wahlund Effect. The change in expected genotype frequencies in a stratified population compared with that expected in a population in HWE affects the mean and variance of the trait. The mean trait value in a stratified population, μSTRAT, is equal to a(p−q)−2d(1−F)pq, where F is Wright's coefficient of inbreeding, and the variance of the trait in the stratified population, VP.STRAT is 2pq[a+d(p−q)]2+4d2p2q2+2Fpq{[a+d(p−q)]2+2Fpq[−2d2(p−q)2−4ad(p−q)−2d2Fpq]}.
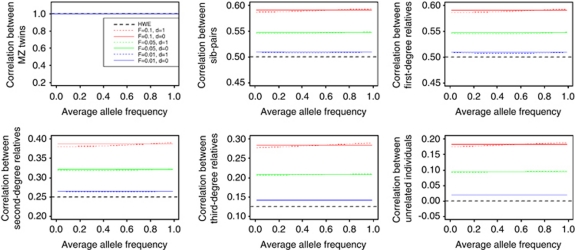
To assess the impact of a, d and F on the correlation between relative pairs, seven models are considered: a=10, d=0, F=0 (HWE model with no dominance variance); and six models, each with varying levels of population stratification and dominance variance: a=10, d=0, F=0.01; a=10, d=1, F=0.01; a=10, d=0, F=0.05; a=10, d=1, F=0.05; a=10, d=0, F=0.1 and a=10, d=1, F=0.1. These results are shown in Figure 1.
Figure 1.
Correlation between relative pairs assuming HWE compared to that calculated assuming population stratification using a single-locus model.
Two-locus model
Complex disease traits are polygenic and the genetic variants discovered so far have small effect sizes. To evaluate the impact of population stratification on heritability in a polygenic model, we consider a two-locus model, where the variance of the trait is affected by two loci.
There are 9 possible genotypes and 81 possible mating types in a two-locus model with biallelic SNPs at each locus. If we assume symmetry of the mating types, then there are 45 possible mating types.
Wright's coefficient of inbreeding, F is used as a measure of the correlation between uniting gametes at a single locus; however, two other parameters are needed to adequately describe the mating frequencies in a stratified population at a single locus (Sebro et al., 2010). When two loci are considered, an additional measure of correlation must be accounted for—the correlation between genes at different loci in the same gamete because of population stratification. This correlation between genes at different loci in the same gamete can be measured using ñ(correlation), which is equal to D/√[p(1−p)r(1−r)], where D is the linkage disequilibrium induced between the loci due to population stratification.
Consider a two-locus model with alleles A and a at the first locus and alleles B and b at the second locus. The frequency of the A allele is p and the frequency of the B allele is r. We assume that both loci are in complete linkage equilibrium within sub-population (D=0). The degree of population stratification at the first locus is F1 and the linkage disequilibrium induced by population stratification is D. If we restrict the model to the situation where the stratified population comprising two equally sized sub-populations, then only p, r, F1 and D are required for calculation of the 81 mating types. This simplification allows us to assess the impact of F1 and D on heritability. We allow for stratification at both loci, and consider only the case where the degree of stratification at the first locus, F1 is greater than that calculated at the second locus.
Let the phenotypic value of the AABB, AABb, AAbb, AaBB, AaBb, Aabb, aaBB, aaBb and aabb genotypes be α1, α2, α3, α4, α5, α6, α7, α8, and α9 respectively. The mean and variance of the trait value differ from that calculated when assuming HWE because of the change in the genotype frequency distribution. The joint genotype probability distribution for MZ twins, parent-offspring, sib-pairs/dizygotic twins and unrelated individuals are used together with the phenotypic value for each genotype to calculate the covariance between these relative pairs. The correlation for each relative pair was calculated for a simple additive model with no interaction, where α1=a1+a2, α2=a1+d2, α3=a1−a2, α4=d1+a2, α5=d1+d2, α6=d1−a2, α7=−a1+a2, α8=−a1+d2 and α9=−a1−a2 respectively.
Eight models are considered: a1=5, a2=5, d1=0, d2=0, F1=0.001, D=0.00001 (small F1, small D, with no dominance variance); a1=5, a2=5, d1=0, d2=0, F1=0.001, D=0.00005 (small F1, larger D, with no dominance variance); a1=5, a2=5, d1 =0.5, d2=0.5, F1=0.001, D=0.00001 (small F1, small D, with dominance variance); a1=5, a2=5, d1=0.5, d2=0.5, F1=0.001, D=0.00005 (small F1, larger D, with dominance variance); a1=5, a2=5, d1=0, d2=0, F1=0.01, D=0.0001 (large F1, small D, with no dominance variance); a1=5, a2=5, d1=0, d2=0, F1=0.01, D=0.0005 (large F1, larger D, with no dominance variance); a1=5, a2=5, d1=0.5, d2=0.5, F1=0.01, D=0.0001 (large F1, small D, with dominance variance); a1=5, a2=5, d1=0.5, d2=0.5, F1=0.01, D=0.0005 (large F1, larger D, with dominance variance). Values of F1 and D were arbitrarily chosen to reflect a mild degree of population stratification, mild linkage disequilibrium between loci, and are values that could be seen in practice.
If there is gene–gene interaction (epistasis), then four additional variances are required for parameterization of the two-locus model—the additive-additive variance Iaa, the additive-dominance variance Iad, the dominance-additive variance Ida and the dominance-dominance variance Idd. However, in our model, we assume no epistasis, so that Iaa=Iad=Ida=Idd=0.
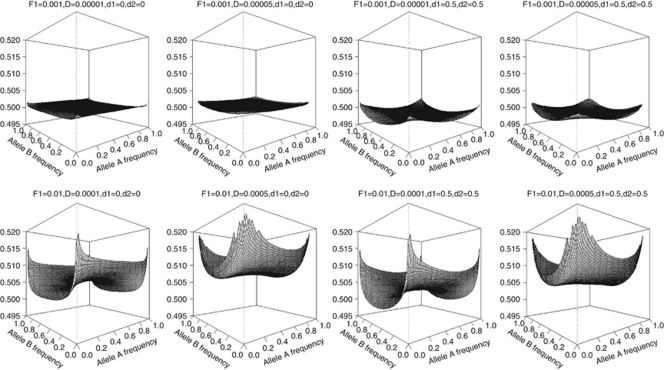
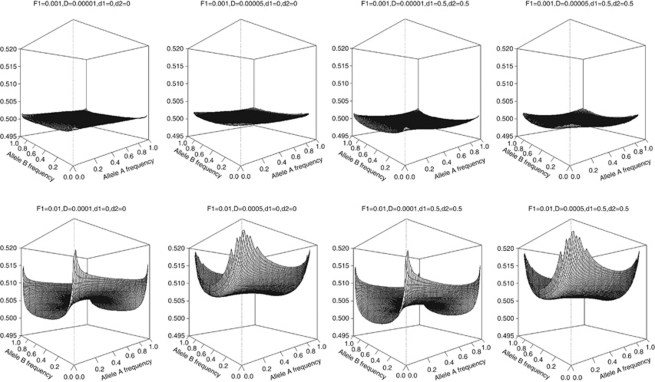
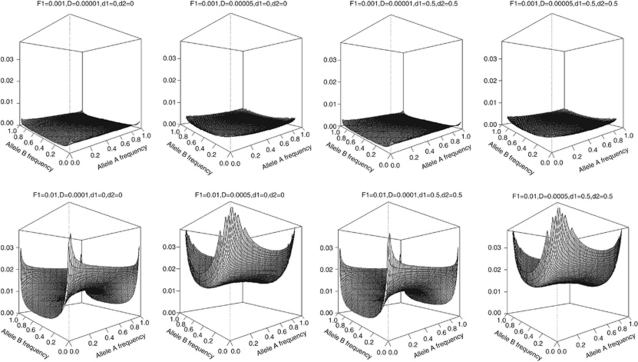
The theoretical correlation between first-degree relatives (parent-offspring pairs), sib-pairs and unrelated individuals are shown in Figures 2,3,4, respectively.
Figure 2.
Genetic correlation (z-axis) between parent-offspring pairs in the presence of population stratification using a two-locus model.
Figure 3.
Genetic correlation (z-axis) between sib-pairs in the presence of population stratification using a two-locus model.
Figure 4.
Genetic correlation (z-axis) between unrelated individuals in the presence of population stratification using a two-locus model.
Results
Although there is no genetic correlation between spouses (random mating) within sub-populations, when the entire stratified population is considered, there is a significant positive genetic correlation between spouses, denoted by Wright's coefficient of inbreeding F. Similarly, if there is random mating with respect to the gene responsible for a trait, then the genetic covariance between spouse trait values is zero; however, if there is population stratification, then the genetic covariance between spouses for the same trait is 4a2Var(pi)+ad(2Var(p2i)−2Var(q2i))+d2(−4Var(pi)+2Var(p2i)+2Var(q2i)) using the parameterization in Model 1, or 4a2Fpq+ad(Fpq(16p−8)+8ϕ3)+d2(Fpq(4−16pq−4Fpq)+(16p−8)ϕ3+4ϕ4) using the parameterization in Model 2. If we assume there is no dominance variance (d=0), then the genetic covariance between spouses for the same trait is 4a2Var(pi) using the parameterization in Model 1 and 4a2Fpq using the parameterization in Model 2. It is well appreciated that there is apparent homogamy for several physical characteristics including weight and height; however, the magnitude of the positive correlation seen between spouses could be partially due to population stratification.
There is increased genetic covariance between relatives in the presence of population stratification. If we assume no dominance variance, then the genetic covariance between MZ twins is 2pqa2(1+F) and is increased by a factor of F relative to that calculated assuming HWE. The genetic covariance between sib-pairs is pqa2(1+3F) and is increased by a factor of 3F compared with HWE. The genetic covariance between second-degree relatives is 1/2pqa2(1+7F), increased by a factor of 7F relative to that calculated assuming HWE. Finally, the genetic covariance between third-degree relatives 1/4pqa2(1+15F) is increased by a factor of 15F relative to that calculated assuming HWE. Because F is generally much smaller than 1 and on the order of 0.001 to 0.01 for most stratified populations, the ratio of the genetic covariance between MZ twins to the genetic covariance between first-degree relatives to that between second-degree relatives to that between third-degree relatives remains almost 1:1/2:1/4:1/8, which is exactly that predicted assuming a population in HWE.
Our results show that the heritability of a trait in a stratified population is higher than the heritability of the trait in a population in HWE if there is no dominance variance. This finding could explain some of the variation in heritability estimates from different studies.
The results are similar when extended to the two-locus model. If two genes contribute to the phenotypic value of a trait and the population is in HWE (random mating), then contributions to the variance of the trait from each locus can be summed. If we assume no dominance variance and HWE, then the genetic correlation between MZ twins is 1, the genetic correlation between sib-pairs as well as parent-offspring pairs is ½, and the correlation between unrelated individuals is 0.
The genetic correlation between MZ twins is always 1, and is not affected by population stratification, or dominance variance. When there is population stratification and no dominance variance, then the genetic correlation between first-degree relative pairs is greater than ½. The magnitude of the increase in genetic correlation over ½ is on the order of F1, and increases slightly with an increase in linkage disequilibrium between loci. The genetic correlation between sib-pairs is also greater than ½, and the magnitude of the increase in genetic correlation over ½ is on the order of F1, and increases slightly with an increase in linkage disequilibrium between loci. An increase in either F1 or D increases the genetic correlation between unrelated individuals.
If there is dominance variance at either or both loci, then the genetic correlation between sib-pairs and parent-offspring varies significantly, and can be less than ½ or less than ½ depending on the ratio of the dominance variance to the additive variance. Although it is currently thought that the dominance variance is generally minimal or almost 0, this finding may prove to be more significant as we learn more about the Human Genome.
Discussion
We show theoretically that if there is no dominance variance, then the heritability of a trait in the presence of population stratification is greater than that in a population in HWE. Although population stratification does not affect the genetic correlation between MZ twins, small amounts of population stratification can significantly affect the genetic correlation noted between sib-pairs and parent-offspring pairs. When a two-locus model with no dominance variance is considered, the findings are similar—a higher degree of population stratification measured by F1 is associated with a higher genetic correlation between parent-offspring and sib-pairs. Similarly, in the absence of dominance variance, an increase in the linkage disequilibrium induced by population stratification measured by D results in an increase in the genetic correlation between parent-offspring and sib-pairs.
Our findings are in concert with that expected based on the literature, as it is well known that positive assortative mating increases the heritability of a trait. Our work is novel, because we build on the recently discovered fact that there is genetic positive assortative mating at all loci that differ in allele frequency between sub-populations, whereas prior works consider phenotypic positive assortative mating. We do not assume the assortative mating is based on the phenotypic resemblance between spouse pairs. In true assortative mating, spouse selection is done at the phenotypic level. We assume there are intrinsic differences in allele frequencies across sub-populations due to religion, socioeconomic status, geographical restrictions or linguistic barriers, probably due to genetic drift. For example, in a European-derived sample from the Framingham Heart Study, there is a strong positive genetic correlation between spouse pairs around the lactase gene (LCT) (Sebro et al., 2010). Although possible, it is unlikely that spouse selection is based on adult lactase-persistence, which is what would be expected with phenotypic assortative mating.
The major clinical implication of our finding is that small amounts of population stratification can have significant impact on the correlation between different relative pairs. This finding is important because the correlation of a trait between relative pairs is often used to estimate heritability and twin data are often used in the study of the genetic determination of traits. If researchers are interested in the ratio of (VA.HWE) to the total variance of the trait (VP) when a population is in HWE, then principal components (Price et al., 2006) or STRUCTURE analysis (Pritchard et al., 2000) could be performed to cluster individuals into sub-populations, and the narrow-sense heritability could be estimated in each sub-population. One limitation of this principal component/STRUCTURE analysis is that spouses may belong to different sub-populations and these discordant pairs should then be excluded from the analysis.
Our study has a few limitations. The model used for population stratification does not allow for admixture or for spouses to belong to different sub-populations. This assumption is likely violated in practice. Immediate admixture (where the spouses belong to different sub-populations) attenuates ancestrally related positive assortative mating, and therefore should slightly decrease the effect of population stratification. Another limitation is that the two-locus model did not allow for epistasis. There are several dozen different models of epistasis for the two-locus case and further work needs to be done to understand the impact of population stratification and the genetic correlation between relatives when there is underlying epistasis. Finally, we assume that there is no genetic heterogeneity and the effect of the gene on the phenotype is the same for all sub-populations. This final assumption is not always correct in practice. Risk heterogeneity exists between ApoE and Alzheimer's disease, where the association exists pan-ethnically but is strongest in Caucasians and Asians, and weaker in Hispanics and African-Americans (Risch, 2000).
Identifying the genes involved in complex quantitative traits remains a challenge. Genomewide-association studies have yielded some success in identifying genes associated with complex traits. However, in some cases the identified variants only explain a small proportion of the variation of the trait. For example, the GIANT consortium have pooled genomewide-association studies involving over 180 000 research participants, and have identified ∼200 variants associated with stature/height; however, these 200 variants only explain ∼10% of the variance of height and most of these variants have very small effect sizes (Perola, 2011). The majority of the genetic variance of several traits remains unexplained despite using these large genomewide-association studies. This phenomenon has been termed ‘missing heritability' (Perola, 2011).
Population stratification may be in part responsible for some of the ‘missing heritability' however, other factors such as gene–gene interactions and gene–environment interactions may also be involved. In summary, further research needs to be done to better understand the effect of population structure on quantitative trait analysis.
Data Archiving
There was no data to deposit.
Acknowledgments
Ronnie Sebro was supported by a Stanford Medical School Medical Scholars Project and in part by a Howard Hughes Predoctoral Fellowship.
The authors declare no conflict of interest.
References
- Crow TJ. The missing genes: what happened to the heritability of psychiatric disorders. Mol Psychiatry. 2011;16:362–364. doi: 10.1038/mp.2010.92. [DOI] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. Longman: Harlow, UK; 1996. [Google Scholar]
- Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Trans R Soc Edinburgh. 1918;52:399–433. [Google Scholar]
- Hill WG, Goddard ME, Visscher PM. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 2008;4:e000008. doi: 10.1371/journal.pgen.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perola M. Genome-wide association approaches for identifying loci for human height genes. Best Pract Res Clin Endocrinol Metab. 2011;25:19–23. doi: 10.1016/j.beem.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- Risch N, Choudhry S, Via M, Basu A, Sebro R, Eng C, et al. Ancestry-related assortative mating in Latino populations. Genome Biol. 2009;10:R132. doi: 10.1186/gb-2009-10-11-r132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebro R, Hoffman TJ, Lange C, Rogus JJ, Risch NJ. Testing for non-random mating: evidence for ancestry-related assortative mating in the Framingham heart study. Genet Epidemiol. 2010;34:674–679. doi: 10.1002/gepi.20528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebro R, Rogus JJ. The power of the transmission disequilibrium test in the presence of population stratification. Eur J Hum Genet. 2010;18:1032–1038. doi: 10.1038/ejhg.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]