Genome sequencing has changed. Once a slow, expensive process—the exclusive realm of large groups with enviable funding—it is now an achievable goal for smaller, individual labs. As the price decreases and the technological capacity increases, it is still important for the community to consider how it is directing its efforts to maximize the scientific knowledge gained. Here, we argue for the importance of sequencing both phylogenetically and phenotypically diverse species in order to capture the points at which ecologically important traits arose. First, we should recognize the current taxonomic bias in genome sequencing and attempt to capture more genomic information on underrepresented groups. Second, sets of genomes should be chosen for sequencing based on hypotheses that can be tested across taxa. With concerted effort into targeted sequencing, we can address questions such as the evolution of immune systems, a topic that we highlight here. Each genome will of course still provide useful tools to study single species, but within a broader comparative framework.
Where are we now?
Genome sequencing first focused on small microbial genomes, a strategy based on fiscal and technical feasibility. The addition of eukaryotic genomes, slow at first, has increased dramatically over the last 2 years with the rapid advancement of new sequencing and annotation technologies. The associated reduction in sequencing cost is dramatic, dropping from over 5000 dollars per megabase in 2001 to 23 cents at the start of 2011 (http://www.genome.gov/sequencingcosts). At first, researchers used genome sequencing mostly to provide tools for geneticists and developmental biologists to study model organisms. Researchers then sequenced some genomes for comparative needs (for example, chimpanzee). Now, we can ask how genomics can be effectively utilized to explore questions about the evolutionary and ecological adaptations that shape all life. From the point of view of studying ecology and evolution, we suggest two strategies to better develop an approximation of the ideal genomics toolbox.
Capturing genomic diversity
Sir Robert May famously asked “how many species?”, highlighting the importance of studying biodiversity (May, 1990). May concluded that we should investigate the biodiversity of our earth for three major reasons: first, to gain from this biodiversity in a concrete sense (such as, finding new foods, drugs and sources of energy); second, to learn about biological systems so that we can assess how they respond to our perturbations; and third, as a central pursuit of human knowledge. What are the benefits of capturing genomic diversity, and what is the strategy for continuing forward?
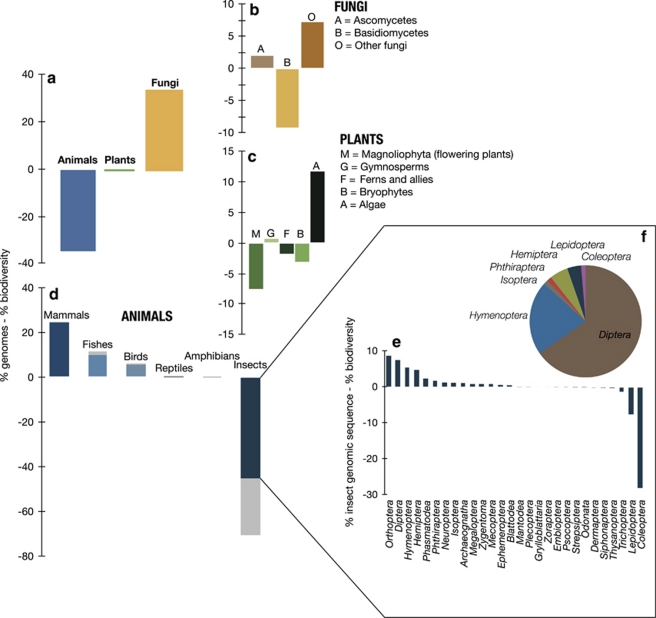
There is a strong taxonomic bias to the available genomes. Although researchers have sequenced more animal genomes (45% of all non-protist eukaryote genome projects are animals) than plant (17%) or fungal (38%) genomes, animals are grossly underrepresented when we consider the number of described species (Figure 1a) (genome data from http://www.ncbi.nlm.nih.gov/genomes/leuks.cgi and described species data from the 2011 IUCN red list http://www.iucnredlist.org/documents/summarystatistics/2011_1_RL_Stats_Table_1.pdf). When we look at these groups at a slightly finer level we see further bias in sequencing effort. Basidiomycetes are underrepresented relative to Ascomycetes and other fungi (Figure 1b). Similarly, flowering plants have been more fully sequenced than any other group of plants but are so diverse that sequencing relative to their biodiversity is still below any of the other groups of plants examined here (Figure 1c). However, the strongest case of inequity between sequencing effort and biodiversity comes from animals, and is entirely driven by insects (Figures 1d–f). (Data regarding the number of genomic sequences are from http://www.ncbi.nlm.nih.gov/Taxonomy/, and accepted species numbers within insect orders are from Chapman (2009), using the middle of the range when a range was given.) Including estimated species numbers within animals only exacerbates this pattern in animals (Figure 1d), but has negligible effects in plants and fungi (results not shown). Strikingly, the most common group of insects, beetles (Coleoptera), which consist of approximately 380 000 described species (38% of all described insects, and notably more than all described plant and fungi species combined) has only a single fully sequenced genome (Tribolium castaneum) and minimal other sequence data. Based on these biases, we suggest prioritizing genome sequencing in groups whose genomic resources are underrepresented relative to their biodiversity. We suggest coupling this approach with targeted sequencing in a few groups that, while taxonomically not as diverse (for example, amphibian and reptiles), have great ecological importance and evolutionary novelty relative to the paucity of their available genomic resources.
Figure 1.
Disparity between genomic interest and contribution to biodiversity based on full genome sequencing projects and the number of described species for each group across (a) three eukaryote kingdoms; (b) fungi; (c) plants; (d) animals; (e) insects based on the number of genomic sequence resources and described species number (dark portion of bars) and predicted species number (light portion of bars); (f) percent of full insect genome projects by order. (a–e) Positive numbers means that these classes of organisms are overrepresented in the genomic research.
May's benefits of understanding the diversity of life on earth also apply to understanding the genomes underlying this diversity. First, we can gain from genome sequence availability in a concrete sense. For example, the search for antibiotics is becoming increasingly desperate (Palumbi, 2001), and researchers have turned to insect immune genes to discover novel antimicrobial compounds (Hancock and Lehrer, 1998). Widely surveying broad taxonomic groups will reveal far greater novelty of such compounds than staying within the bounds of known model taxa. Second, having access to genome diversity will provide insight into the genetic capacity that organisms may have to respond to environmental perturbations, which is of particular importance as we enter another great extinction event (Wake and Vredenburg, 2008). Third, genome sequence availability across diverse taxa will facilitate scientific exploration and discovery into novel questions about diverse taxa, furthering the pursuit of human knowledge and awareness of biodiversity.
Hypothesis-targeted comparative sequencing: a case study in immunity
Capturing more phylogenetic diversity through genome sequencing will provide more evolutionary and ecological insight, but it will not necessarily provide the data necessary to address all intriguing ecological and evolutionary questions. Thus, we suggest coupling the phylogenetic diversity strategy with a hypothesis-driven strategy. As an example, we can turn to our current understanding of the evolution of innate immunity. Although invertebrates do not have the adaptive immune system of vertebrates, the innate components of immunity are strongly conserved across most animals (Flajnik and Du Pasquier, 2004), and many of the functions and features of this branch of immunity have been discovered and best characterized in insects. Most of our mechanistic understanding of invertebrate immunity has been gleaned from a few well-studied species, and is built on work in a single species of fruit fly (Park and Lee, 2011). As new, annotated genomes have become available to the scientific community, a number of discrepancies have begun to emerge between these newly sequenced genomes and those of the existing model organisms. Specifically, immune gene repertoires seem to vary considerably across insects sequenced thus far. The honeybee genome revealed a considerably reduced immune system compared with fruit flies, mosquitoes, and the single beetle with an available genome (Evans et al., 2006). The authors of the honeybee genome project quite reasonably proposed that honeybees do not need the fortified immunity of these better-characterized insects because bees have the benefits of sociality and behavioral adaptations to combat infection. This makes considerable sense, and behavioral genes have indeed been linked to immunity in bees (Wilson-Rich et al., 2009). Since the honeybee genome was sequenced, the genome sequences of several other Hymenoptera (the group including wasps, bees and ants), have revealed a similar pattern of reduced immune gene numbers (for example, Werren et al., 2010; Suen et al., 2011). Unfortunately, as genomes from only one non-social hymenopteran genus (Nasonia wasps) are available, it is currently impossible to assess how sociality has shaped immunity in this group.
The recent sequencing of the pea aphid genome revealed an even more reduced immune gene repertoire than honeybees (Gerardo et al., 2010). Pea aphids lack a central immune pathway and all known antibacterial peptides (although they do have a relatively uncommon class of potentially antifungal peptides). Aphids can harbor several species of intracellular mutualistic bacteria. These mutualists can produce needed amino acids (Douglas, 1998) and can protect their hosts against parasites and other environmental stressors (reviewed in Gerardo et al., 2010). Researchers have hypothesized that aphids may lack these immune components because they are intimately dependent on bacterial symbionts for survival (Gerardo et al., 2010). Alternatively, the loss of a strong immune response towards bacteria may have facilitated the establishment of these symbiotic associations. With few genomes available for symbiont-associated insects, and no genomes available for other aphids or their close relatives, it is currently impossible to determine the link between immune gene repertoire and symbiosis.
Here, we see two evolutionary hypotheses in which targeted-genome sequencing could answer fundamental questions about the evolution of immunity. First, do social defenses relax selection maintaining expensive immune responses? Second, does a reduced immune response facilitate the establishment or maintenance of symbiotic associations? For the first question, we have very little genomic data about the link between sociality and immunity, as almost all of the sequenced hymenoptera are highly social. Sequencing solitary bees such as sweat bees, some of which have lost sociality (Wcislo and Danforth, 1997), and others of which are plastic in their sociality (Soucy and Danforth, 2002), and more wasps (social and non-social) would help clarify this relationship. Similarly, sequencing other social organisms, such as termites, social aphids, social beetles and naked mole rats, could shed light on the commonality of immune reduction with sociality, and would also allow exploration of other traits linked to sociality. To study the connection between immunity and symbiosis, within aphids and their close relatives, we need to determine when the reduced immune system arose (before or after association with obligate symbionts), and we need to work across phylogenetically diverse groups in order to compare the genomes of symbiont-associated hosts relative to hosts with less intimate microbial associations. Overall, picking phylogenetically diverse targets will maximize the potential to reveal important differences within groups, but this should also be paired with sequencing of closely-related species that differ in specific ecological traits, such as sociality or symbiosis, in order to have appropriate comparisons.
An ecological and evolutionary strategy for picking targets
If genomics is to inform studies in ecology and evolution, a strategy of targeting species that fulfill ecological traits as counterpoints or comparisons to existing, genomically known, species will provide valuable, specific, insight into the evolution of these traits, but may or may not directly expand our taxonomic base of genomic tools. Thus, we should also sequence widely across taxa, prioritizing currently genomically underrepresented groups, which will ultimately allow us to identify interesting patterns of genome evolution and sources of evolutionary novelty. We, of course, still must take into account the amenability of follow-up field and laboratory experiments, as a genome is only as powerful as the context in which it can be placed.
Conclusions
As sequencing gets ever cheaper, the number of fully sequenced organisms will grow dramatically. We will achieve greater understanding of evolution with every genome, but we can do so more effectively by prioritizing our efforts. Recent initiatives to sequence 5000 arthropod genomes (Robinson et al., 2011) (I5K: http://arthropodgenomes.org/wiki/i5K), 10 000 more vertebrate genomes (http://genome10k.soe.ucsc.edu/), and the Beijing Genomics Institute initiative to sequence 1000 additional plant and animal genomes are a very encouraging start, especially as there has been a community approach to species selection. Groups already sequencing or preparing to sequence their study organisms should interact with these initiatives to avoid unintentional duplication of research effort. Individual labs, research groups and funding agencies should carefully allocate their available funds in order to maximize the number of questions that can be addressed by including evolutionary and ecological criteria in their decision-making. Extending genomic sequencing efforts into question-based comparisons and broader-scale phylogenetic sampling of genomes will not only answer specific questions but will provide tools for applied technology development, will identify sources of biological novelty, and will frame our understanding of genome evolution as a whole.
Acknowledgments
We thank Ben Sadd, members of the Gerardo lab and two anonymous reviewers for constructive suggestions on this manuscript. SMB is supported by the Swiss NSF (grant number 31003A-116057 to Paul Schmid-Hempel) and NMG by NSF award IOS-1025853.
The authors declare no conflict of interest.
References
- Chapman A.2009Numbers of living species in Australia and the world2nd edn.Report for the Australian Biological Resources Study
- Douglas AE. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol. 1998;43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- Evans JD, Aronstein K, Chen YP, Hetru C, Imler JL, Jiang H, et al. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol Biol. 2006;15:645–656. doi: 10.1111/j.1365-2583.2006.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik MF, Du Pasquier L. Evolution of innate and adaptive immunity: can we draw a line. Trends Immunol. 2004;25:640–644. doi: 10.1016/j.it.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Gerardo NM, Altincicek B, Anselme C, Atamian H, Barribeau SM, De Vos M, et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010;11:R21. doi: 10.1186/gb-2010-11-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock REW, Lehrer R. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 1998;16:82–88. doi: 10.1016/s0167-7799(97)01156-6. [DOI] [PubMed] [Google Scholar]
- May RM. How many species. Philos T Roy Soc B. 1990;330:293–304. [Google Scholar]
- Palumbi SR. Evolution—humans as the world's greatest evolutionary force. Science. 2001;293:1786–1790. doi: 10.1126/science.293.5536.1786. [DOI] [PubMed] [Google Scholar]
- Park JW, Lee BL.2011Insect immunologyIn: Gilbert LI (ed).Insect Molecular Biology and Biochemistry Elsevier; 480–512. [Google Scholar]
- Robinson GE, Hackett KJ, Purcell-Miramontes M, Brown SJ, Evans JD, Goldsmith MR, et al. Creating a buzz about insect genomes. Science. 2011;331:1386. doi: 10.1126/science.331.6023.1386. [DOI] [PubMed] [Google Scholar]
- Soucy SL, Danforth BN. Phylogeography of the socially polymorphic sweat bee Halictus rubicundus (Hymenoptera: Halictidae) Evolution. 2002;56:330–341. doi: 10.1111/j.0014-3820.2002.tb01343.x. [DOI] [PubMed] [Google Scholar]
- Suen G, Teiling C, Li L, Holt C, Abouheif E, Bornberg-Bauer E, et al. The genome sequence of the leaf-cutter ant Atta cephalotes reveals insights into its obligate symbiotic lifestyle. PLoS Genet. 2011;7:e1002007. doi: 10.1371/journal.pgen.1002007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake DB, Vredenburg VT. Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc Natl Acad Sci USA. 2008;105 (suppl 1:11466–11473. doi: 10.1073/pnas.0801921105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wcislo WT, Danforth BN. Secondarily solitary: the evolutionary loss of social behavior. Trends Ecol Evol. 1997;12:468–474. doi: 10.1016/s0169-5347(97)01198-1. [DOI] [PubMed] [Google Scholar]
- Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, Colbourne JK, et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327:343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Rich N, Spivak M, Fefferman NH, Starks PT. Genetic, individual, and group facilitation of disease resistance in insect societies. Annu Rev Entomol. 2009;54:405–423. doi: 10.1146/annurev.ento.53.103106.093301. [DOI] [PubMed] [Google Scholar]