Abstract
Cyclotides are a unique and growing family of backbone cyclized peptides that also contain a cystine knot motif built from six conserved cysteine residues. This unique circular backbone topology and knotted arrangement of three disulfide bonds makes them exceptionally stable to thermal, chemical, and enzymatic degradation compared to other peptides of similar size. Aside from the conserved residues forming the cystine knot, cyclotides have been shown to have high variability in their sequence. Consisting of over 160 known members, cyclotides have many biological activities, ranging from anti-HIV, antimicrobial, hemolytic, and uterotonic capabilities; additionally, some cyclotides have been shown to have cell penetrating properties. Originally discovered and isolated from plants, cyclotides can be also produced synthetically and recombinantly. The high sequence variability, stability, and cell penetrating properties of cyclotides make them potential scaffolds to be used to graft known active peptides or engineer peptide-based drug design. The present review reports recent findings in the biological diversity and therapeutic potential of natural and engineered cyclotides.
Keywords: Cyclotides, cyclic peptides, peptide therapeutics, drug discovery, drug design
Introduction
Cyclotides are fascinating backbone-cyclized or circular proteins ranging from 28 to 37 amino acid residues that are naturally expressed in plants. They all share a unique head-to-tail circular knotted topology of three disulfide bridges, with one disulfide bond penetrating through a macrocycle formed by the other two disulfides bonds and interconnecting peptide backbones, forming what is called a cystine knot topology (Fig. 1) [1]. This cyclic cystine knot (CCK) framework gives cyclotides a compact, highly rigid structure [2], which confers exceptional resistance to thermal/chemical denaturation, and enzymatic degradation [3, 4]. In fact, the use of cyclotide-containing plants in indigenous medicine first highlighted the fact that these peptides are resistant to boiling and are apparently orally bioavailable [5].
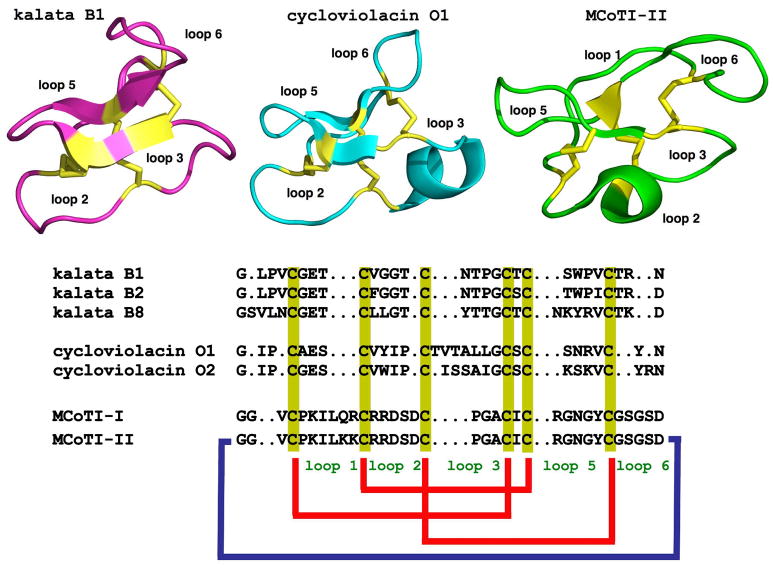
Figure 1.
Primary and tertiary structures of cyclotides from the Möbius (kalata B1, pdb code: 1NB1), bracelet (cycloviolacin O1, pdb code: 1NBJ) and trypsin inhibitor (MCoTI-II, pdb code: 1IB9) subfamilies. The sequence of kalata B8, a novel hybrid cyclotide isolated from the plant O. affinis is also shown. Conserved cysteine residues are marked in yellow and disulfide connectivities in red. The circular backbone topology is shown with a blue line. Figure adapted from Ref. [4].
More than 160 cyclotides have been isolated from plants in the Violacea (violet), Rubiaceae (coffee) and Cucurbitaceae (squash) families [6, 7]; and more recently in the Fabaceae (legume) family [8–10]. It has been estimated, however, that around 50,000 cyclotides might exist [6]. The discovery of cyclotides in the plant Clitoria ternatea from the Fabaceae family represents an important discovery [9, 10]. The Fabaceae family is one the largest families of plants on Earth, representing ≈18,000 species, some of which are widely used as crops in human nutrition and food supply [11]. Due to the high diversity of is also expected that more cyclotides will be discovered in the near future.
Most of the cyclotides reported so far have been found in the Violaceae and Rubiaceae families. All the plants studied so far from the Violaceae family have shown to contain cyclotides. In contrast, only 5% of the Rubiaceae plants analyzed thus far have been shown to have cyclotides [6]. The only two cyclotides found to date from the Cucurbitaceae plant family are Momordica cochinchinensis trypsin inhibitor I and II (MCoTI-I/II; Fig. 1). These cyclotides are found in the seeds of M. cochinchinensis (a tropical squash plant) and are potent trypsin inhibitors. MCoTI cyclotides, however, do not share significant sequence homology with the other cyclotides beyond the presence of the three-cystine bridges that adopt a similar backbone-cyclic cystine-knot topology (Figs. 1 and 2). They are more related to linear cystine-knot squash trypsin inhibitors and sometimes are considered as circular knottins [7].
Figure 2.
General features of the cyclic cystine knot (CCK) topology found in cyclotides. A. Detailed structure the cystine knot core and the connecting loops. The six Cys residues are labeled I through VI whereas loops connecting the different Cys residues are designated as loop 1 through 6, in numerical order from the N- to the C-terminus. B. Möbius (right) and bracelet (left) cyclotides are defined by the presence (Möbius) or absence (bracelet) of a Pro residue in loop 5 that introduces a twist in the circular backbone topology.
Despite the sequence diversity, all cyclotides share the same CCK motif (Figs.1 and 2). Hence, these micro-proteins can be considered natural combinatorial peptide libraries structurally constrained by the cystine-knot scaffold [12] and head-to-tail cyclization but in which hypermutation of most of the residues may be tolerated with the exception of the strictly conserved cysteines that comprise the knot.
All of the cyclotides reported so far from the Violaceae and Rubiaceae families are biosynthesized via processing from dedicated genes that in some cases encode multiple copies of the same cyclotide, and in others, mixtures of different cyclotide sequences [13]. Cyclotides from the Fabaceae family, however, are biosynthesized from evolved albumin-1 genes [9, 10].
Cyclotides can also be produced chemically using solid-phase peptide synthesis in combination with native chemical ligation [14–17] or recombinantly in bacteria by using modified protein splicing units or inteins [18–20]. The latter method can generate folded cyclotides either in vivo or in vitro using standard bacterial expression systems [18–20] and opens the possibility of producing large libraries of genetically encoded cyclotides which can be analyzed by high throughput cell-based screening for selection of specific sequences able to bind particular biomolecular targets [19, 20].
Naturally occurring cyclotides show various biological activities including insecticidal [15, 21, 22], uterotonic [23, 24], anti-viral [25, 26], antimicrobial [15, 24], antitumor [27], antihelminthic [28, 29] and protease inhibitory activity [30]. Their insecticidal and antihelminthic properties suggest that they may function as defense molecules in plants.
All these features make cyclotides ideal drug development tools [17, 31–33]. They are remarkably stable due to the cyclic cystine knot [2]. They are relatively small, making them readily accessible to chemical synthesis [14], and they can be encoded within standard cloning vectors and expressed in cells [18–20]. They are amenable to substantial sequence variation [34], which make them ideal substrates for molecular grafting of peptide epitopes [4] or for molecular evolution strategies to enable generation and selection of compounds with optimal binding and inhibitory characteristics [20, 34]. Even more importantly, some cyclotides have been shown recently to be able to cross cell membranes [35, 36].
In the present review, we discuss the structural and more pharmacologically-relevant biological properties of cyclotides as well as the latest developments in the use of the cyclotide scaffold to design novel peptide-based therapeutics.
The discovery
The discovery of the first cyclotide occurred indirectly when Norwegian researchers isolated and partially characterized a peptide, kalata B1, from the African plant Oldenlandia affinis [23, 37]. This plant was used by the natives to make a tea extract that was used to accelerate childbirth during labor, and thereafter kalata B1 was identified as the main uterotonic component of the plant [23, 37, 38]. Kalata B1 was initially characterized as a likely cyclic peptide of ≈30 amino acids in size.
It was not until 1995, however, that the structure of kalata B1 was confirmed to be a 29-amino acid head-to-tail macrocyclic peptide containing a cystine knot motif by using a combination of mass spectrometry and NMR techniques [5]. The structural determination of kalata B1 coincided in time with the discovery of several other macrocyclic peptides of similar size and sequence from other Rubiaceae [39, 40] and Violaceae [41] plants. By 1999, another 20 different macrocyclic peptides featuring similar characteristics were described at the National Cancer Institute at USA [42] and by a Swedish group that was looking for novel cyclic peptides from plant biomass [43, 44]. In order to group this growing family of peptides, Craik and co-workers coined the term “cyclotides” (cyclo peptides) to refer to this interesting family of cyclic peptides. Since then, around 160 different cyclotides have been discovered in plants of the Rubiaceae, Violaceae, Cucurbitaceae and Fabaceae families and it has been estimated that the cyclotide family might contain over 50,000 different members [6, 45].
Cyclotides can be also classified in the larger family of knottins, a group of microproteins that are characterized for containing a cystine knot [46]. Several web-based databases have been created to curate the sequences of knottins (knottin.cbs.cnrs.fr) [47, 48] and cyclotides as well as other circular proteins (www.cybase.org.au) [49], both at the amino acid and nucleic acid levels.
Three-dimensional structure
Nearly all three dimensional structures of cyclotides elucidated to date have been performed by solution NMR spectroscopy [50]. The only exception is the cyclotide varv F, for which both an NMR and an X-ray crystal structure exists [51]. There are also several reports where the structures of cyclotides have been modeled [52, 53].
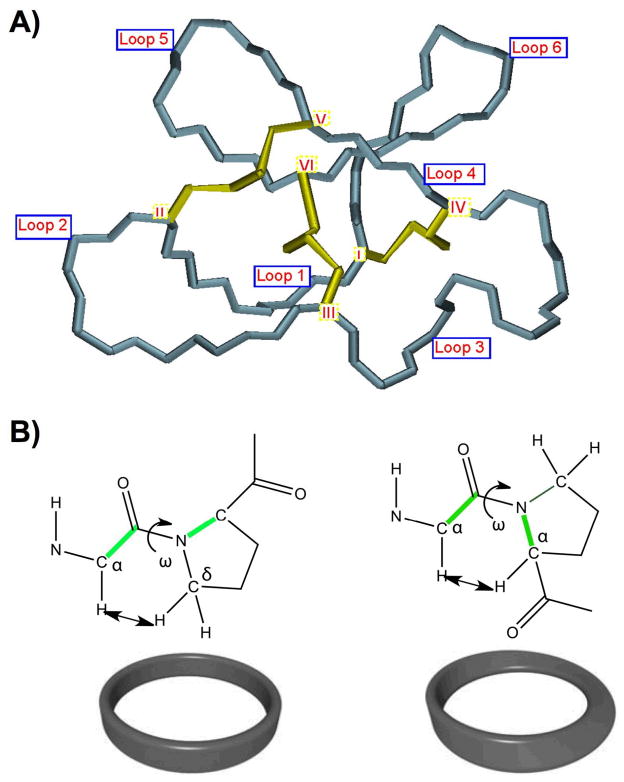
All of the natural cyclotides isolated thus far contain between 28 and 37 amino acids, including six cysteine residues (labeled CysI through CysVI), and are backbone cyclized (Figs. 1 and 2). The Cys residues are oxidized to form a cystine knot core in which the embedded ring formed by two disulfide bonds (CysI-CysIV and CysII-CysV) is penetrated by a third disulfide bond (CysIII-CysVI). This unique topology of circular backbone and interlocked cystine core is usually referred to as a cyclic cystine knot (CCK) motif (Fig. 2) [54, 55]. The CCK motif is decorated by six interconnecting segments, or loops, which are successively numbered loop 1 through 6, starting at CysI (see Fig. 2). In most of the cyclotides, loops 1 and 4 are highly conserved in length and composition. In contrast, the other loops show more variation in size and sequence.
The first CCK structure was reported for the prototypic Möbius cyclotide kalata B1 in 1995 [5]. Since then NMR and X-ray studies have confirmed that all cyclotides retain a similar CCK scaffold but with high sequence variability in the six loops. The validity of the embedded knotted cystine core of cyclotides has been further confirmed by chemical means [56]. The CCK framework gives cyclotides a compact, highly rigid structure [2], which confers exceptional resistance to thermal/chemical denaturation and enzymatic degradation [3] thereby making cyclotides a promising molecular scaffold for drug discovery [1, 4, 57]. For example, cyclotides are more resistant to high temperatures approaching boiling when compared to their linear counterparts [3]. The stabilizing abilities of the CCK motif do not end there; the cystine knot also confers chemical stability to the protein allowing it to retain its native structure under typical denaturing conditions involving 8 M urea or 6 M guanidine hydrochloride [3]. Furthermore, the CCK motif also aids in protecting the proteins from enzymatic degradation by trypsin, pepsin, and thermolysin [3].
Cyclotides have been classified into three main subfamilies. The Möbius and bracelet cyclotide subfamilies differ in the presence or absence of a cis-Pro residue in loop 5, which introduces a twist in the circular backbone topology (Fig. 2B) [58]. Möbius and bracelet subfamilies of cyclotides may also be distinguished by the amount of hydrophobic residues and their location on the surface. For example, around 60% of the surface residues in the bracelet cyclotide cycloviolacin O2 are hydrophobic and are located primarily in loops 2 and 3. In contrast, only 40% of the surface residues in the Möbius cyclotide kalata B1 are hydrophobic and mainly localized in loops 2, 5 and 6 [59]. Nevertheless both major subfamilies of cyclotides contain some highly conserved residues such as Glu3 in loop 1 and the natural site of cyclization, Asn/Asp in loop 6. It is also interesting to remark that cyclotides that share properties of both Möbius and bracelet such as kalata B8 and psyle C are also starting to emerge [60, 61]. These cyclotides are usually referred to as hybrid cyclotides.
A third subfamily of cyclotides comprises the cyclic trypsin inhibitors MCoTI-I/II (Fig. 1), which have been isolated from the dormant seeds of Momordica cochinchinensis, a plant member of the Cucurbitaceae family, and are powerful trypsin inhibitors (Ki ≈ 20 –30 pM) [62]. These cyclotides do not share significant sequence homology with other cyclotides beyond the presence of the three-cystine bridges, but structural analysis by NMR has shown that they adopt a similar backbone-cyclic cystine-knot topology [63, 64].
Cyclotide plant biosynthesis
All the cyclotides reported so far from the Violaceae and Rubiaceae families are biosynthesized via processing from dedicated genes that, in some cases, encode multiple copies of the same cyclotide, and in others, mixtures of different cyclotide sequences [13, 21]. The first genes encoding cyclotides were discovered in the plant Oldenlandia affinis (Rubiaceae family) for the kalata cyclotides (Fig. 3A) [21]. For example, the gene encoding the precursor protein of kalata B1 (Oak 1) consists of an endoplasmic reticulum (ER)-targeting sequence, a pro-region, a highly conserved N-terminal repeat (NTR) region, a mature cyclotide domain, and a hydrophobic C-terminal tail (Fig. 3A). In contrast, the gene precursor of kalata B2 encodes up to three copies of the NTR-cyclotide region, which allows the formation of up to three mature kalata B2 cyclotides per molecule of precursor. Similar genes have been also found in plants from the Violaceae family [13, 45].
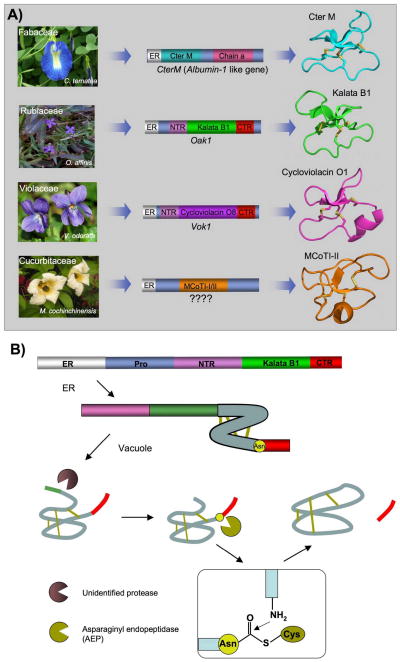
Figure 3.
Genetic origin and biosynthesis of cyclotides in plants. A. Rubiacea and Violaceae plants have dedicated genes for the production of cyclotides [13]. These cyclotide precursors comprise an ER signal peptide, an N-terminal Pro region, the N-terminal repeat (NTR), the mature cyclotide domain and a C-terminal flanking region (CTR). Cyclotides from the Fabaceae family of plants isolated recently from C. ternatea [9, 10], show an ER signal peptide immediately followed by the cyclotide domain, which is flanked at the C-terminus by a peptide linker and the albumin a-chain. In this case, the cyclotide domain replaces albumin-1 b-chain. The genetic origin of the trypsin inhibitor cyclotides MCoTI-I/II (found in the seeds of M. cochinchinensis) remains yet to be determined. B. Scheme representing the proposed mechanism of protease-catalyzed cyclotide cyclization. It has been proposed that the cyclization step is mediated by an asparaginyl endopeptidase (AEP), a common Cys protease found in plants. The cyclization takes place at the same time as the cleavage of the C-terminal pro-peptide from the cyclotide precursor protein through a transpeptidation reaction [66]. The transpeptidation reaction involves an acyl-transfer step from the acyl-AEP intermediate to the N-terminal residue of the cyclotide domain [67]. Figure adapted from Refs. [4, 11].
More recently, Craik and co-workers have revealed that the gene encoding the protein precursor of a novel cyclotide (Cter M) isolated from the leaf of butterfly pea (Clitoria ternatea), a representative member of the Fabaceae plant family, is embedded within the albumin-1 gene (Fig. 3A) [9, 10]. Similar findings have been also recently reported by Tam and co-workers [10]. Generic albumin-1 genes are comprised of an ER signal sequence followed by an albumin chain-b, a linker, and an albumin chain-a. In the precursor of cyclotide Cter M, the cyclotide domain replaces the albumin chain-b domain. The discovery that albumin genes can evolve into protein precursors that can be subsequently processed to become cyclic has also been described in a recent report on the biosynthesis of the peptide sunflower trypsin inhibitor 1 (SFTI-1) [65]. In this case the SFTI-1 precursor gene was identified as seed napin-like albumin [65]. SFTI-1 is a 14-residue peptide isolated from sunflower seeds with a head-to-tail cyclic backbone structure having only a single disulfide bond. In this context, it is worth noting that the protein precursors of the only two cyclotides isolated so far from the Cucurbitaceae plant family (MCoTI-I/II) have not been identified yet.
The complete mechanism of how cyclotide precursors are processed and cyclized has not been completely elucidated yet. However, recent studies suggest that an asparaginyl endopeptidase (AEP) is a key element in the cyclization of cyclotides [66, 67]. It has been proposed that the cyclization step mediated by AEP takes place at the same time as the cleavage of the C-terminal pro-peptide from the cyclotide precursor protein through a transpeptidation reaction (Fig. 3B) [66]. The transpeptidation reaction involves an acyl-transfer step from the acyl-AEP intermediate to the N-terminal residue of the cyclotide domain [67]. AEPs are Cys proteases that are very common in plants and specifically cleave the peptide bond at the C-terminus of Asn and, less efficiently, Asp residues. All of the cyclotide precursors identified so far contain a well-conserved Asn/Asp residue at the C-terminus of the cyclotide domain in loop 6, which is consistent with the idea that cyclotides are cyclized by a transpeptidation reaction mediated by AEP (Fig. 3B) [66]. It is also worth noting that the only naturally linear ‘cyclotide’ isolated to date is violacin A from Viola odorata [68]. Interestingly, the gene encoding violacin A lacks the key Asn/Asp residue in loop 6, which has been replaced by a stop codon, and is required for the backbone cyclization reaction [68]. As a result, the peptide remains linear after translation and folding. Studies using transgenic plants expressing cyclotide precursors also support the involvement of AEP and requirement for a C-terminal Asn residue in the cyclotide sequence [66, 67]. For example, it has been shown that the introduction of a mutation at the C-terminal Asn residue of the cyclotide domain in transgenic plants resulted in no circular peptide production [67]. AEP has also been shown to be involved in the biosynthesis of the cyclic peptide SFTI-1 [65].
The expression of cyclotides in transgenic plants like Arabidopsis and tobacco is, however, highly inefficient giving rise to mostly acyclic or truncated proteins [66, 67], thus indicating that the processing and cyclization of cyclotides may involve other enzymes or may be species dependent. Craik and co-workers have recently isolated a protein disulfide isomerase (PDI) from the plant O. affinis showing that at least in vitro it is able to increase the folding yield of cyclotide-related molecules, including linear and cyclic cyclotides [69]. The relevance of this result in vivo still remains to be established.
Chemical Synthesis of cyclotides
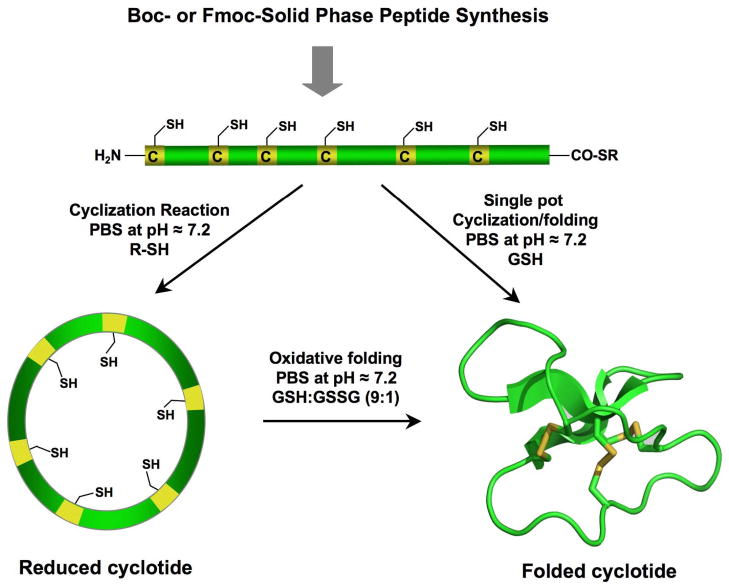
Cyclotides are relatively small polypeptides, ≈30–40 amino acids long, and therefore the linear precursors can be readily synthesized by chemical methods using solid-phase peptide synthesis (SPPS) [70]. Backbone cyclization of the corresponding linear precursor can readily be accomplished in aqueous buffers under physiological conditions by using an intramolecular version of native chemical ligation [71, 72] (Fig. 4). This approach has been successfully used to chemically generate native and engineered cyclotides [14, 16, 17, 73–76]. The only requirements for NCL are an N-terminal cysteine and a α-thioester group at the C-terminus of the linear precursor [77–80]. Both tert-butyloxyxarbonyl (Boc)- and 9-fluorenyloxycarbonyl (Fmoc)-based chemistries have been used to incorporate C-terminal thioesters during chain assembly (Boc) [81–83] or using safety-catch based linkers (Fmoc) [80, 84–87]. Once the peptide is cleaved from the resin, both cyclization and folding can be carried out sequentially [14, 15, 74] or in a single pot reaction [16–18, 36].
Figure 4.
Chemical synthesis of cyclotides by means of an intramolecular native chemical ligation (NCL). This approach requires the chemical synthesis of a linear precursor bearing an N-terminal Cys residue and an α-thioester moiety at the C-terminus. The linear precursor can be cyclized first under reductive conditions and then folded using a redox buffer containing reduced and oxidized gluthathione (GSH). Alternatively, the cyclization and folding can be efficiently accomplished in a single pot reaction when the cyclization is carried out in the presence of reduced GSH as the thiol cofactor [18, 20].
Typically Möbius cyclotides such as kalata B1 are able to readily fold into its native structure quite efficiently [14] although during the oxidative folding a stable two-disulfide native-like intermediate accumulates. This intermediate is not the immediate precursor of the three-disulfide native peptide and is not observed during the reductive unfolding of kalata B1 [88, 89]. MCoTI-cyclotides also fold very efficiently under oxidative conditions [16, 17, 36]. MCoTI-II also accumulates a native-like partially folded intermediate during its oxidative folding, but in contrast to kalata B1, this intermediate is a direct precursor to the fully folded native form [90].
The oxidative folding of bracelet cyclotides, on the other hand, has proven to be more challenging. For example, Tam and co-workers used orthogonal thiol protecting groups for the Cys residues in the synthesis of the bracelet cyclotides circulins A/B and cyclopsychotride [15, 74]. This approach allowed a more controlled step-wise folding strategy thereby improving the yield for the natively folded products. More recently, Göransson and co-workers have also shown that the bracelet cyclotide cycloviolacin O2 can be folded with yields over 50% using the mildly oxidizing agent dimethyl sulfoxide as co-solvent and a non-ionic detergent (Brij35) as a hydrophobic surface stabilizing agent [76].
Craik and co-workers have also recently reported that several analogs of the bracelet cyclotide cycloviolacin O1 resulting by the introduction of mutations on loops 2 and 6 can be efficiently folded [75]. The best analog in folding terms was accomplished by replacement of Ile in loop 2 with Gly together with the insertion of a Thr residue preceding Tyr in loop 6 [75]. These studies have allowed a better understanding of the structural elements influencing the efficiency of oxidative folding in Möbius and bracelet cyclotides thereby demonstrating that they are mostly localized within loops 2 and 6.
The oxidative folding of cyclotides is greatly influenced by the reaction conditions including the redox buffer used and the concentration of organic co-solvents. The Cys-containing tripeptide glutathione (GSH) is by far the most commonly employed reagent to accomplish the oxidative folding of cyclotides although other oxidizing reagents have also been used. Redox buffers containing GSH make possible the formation of disulfide bonds under thermodynamic control allowing disulfide reshuffling to recycle non-productive or misfolded intermediates [18, 76, 88, 89, 91]. GSH has been also used as a thiol additive as well as redox buffer for the cyclization and concomitant folding of cyclotides in a single pot reaction [18]. This has been successfully accomplished for Möbius [18, 20] and MCoTI [16, 17, 36] cyclotides (Fig. 3).
Leatherbarrow and coworkers have also reported the chemoenzymatic synthesis of several MCoTI cyclotides by using a protease-mediated ligation [17]. In this approach, the linear precursor was synthesized using Fmoc-SPPS, folded in oxidative conditions and cyclized using polymer-supported trypsin. This approach of enzyme-mediated in vitro cyclization has been also used for the cyclization of a linearized version of the cyclic peptide sunflower trypsin inhibitor, SFTI-1 [92]. These two studies suggest that protease-mediated cyclization can be a general and efficient process for producing cyclic peptides.
Recombinant expression of cyclotides
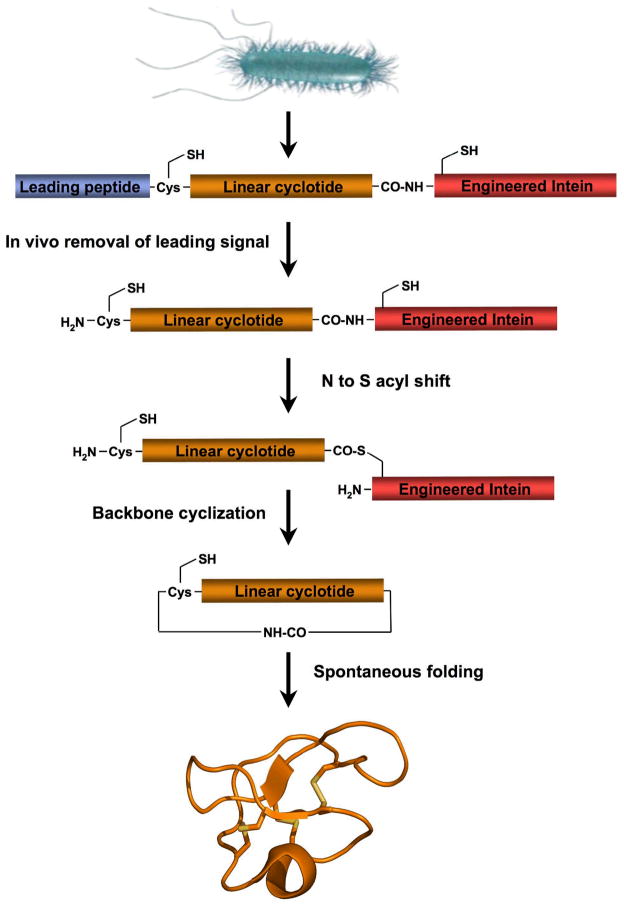
Our group has pioneered the recombinant production of cyclotides in bacteria through intramolecular NCL (see above) by using a modified protein-splicing unit or intein (Fig. 5) (see reference [34] for a recent review). Inteins are self-processing domains that undergo post-translational processing to splice together flanking external polypeptide domains (exteins) [93]. This approach uses a modified intein fused to the C-terminus of the cyclotide sequence to facilitate the formation of the required α-thioester at the C-terminus of the recombinant linear precursor (Fig. 5) [34]. The introduction of the required N-terminal Cys for cyclization can be easily accomplished by expressing the precursor protein with an N-terminal leading peptide, which can be cleaved by proteolysis or autoproteolysis (Fig. 5). The simplest way to achieve this is to introduce a Cys residue downstream to the initiating Met residue. Once the translation step is completed, the endogeneous methionyl aminopeptidases (MAP) removes the Met residue, thereby generating in vivo an N-terminal Cys residue [94–98]. The N-terminal Cys can then capture the reactive thioester in an intramolecular fashion to form a backbone cyclized polypeptide (Fig. 5).
Figure 5.
Biosynthetic approach for the recombinant expression of cyclotides using E. coli expression systems. Cleavage of the leading signal either in vitro [18] or in vivo [19, 20] by appropriate proteases provides the N-terminal Cys residue required for the cyclization. The backbone cyclization of the linear precursor is then mediated by a modified protein splicing unit or intein. Once the linear precursor is cyclized, folding is spontaneous for kalata B1 and MCoTI-I/II cyclotides [18–20].
Additional methods to generate N-terminal Cys proteases involve the use of exogeneous proteases to cleave the leading signal after purification or in vivo by co-expressing the protease. Proteases that have been used so far include Factor Xa [79, 99], ubiquitin C-terminal hydrolase [100, 101], tobacco etch virus (TEV) protease [102], and thrombin [103]. The N-terminal pelB leader sequence has been also used recently to direct newly synthesized fusion proteins to the E. coli periplasmic space where the corresponding endogenous leader peptidases [104, 105] can generate the desired N-terminal cysteine-containing protein fragment [106]. Protein splicing can be also used to generate recombinant N-terminal Cys-containing polypeptides. For example, some inteins have been modified to allow cleavage at the C-terminal splice junction in a pH- and temperature-dependent fashion [107–109].
Intein-mediated backbone cyclization of polypeptides has also been recently used for the biosynthesis of the trypsin inhibitor SFTI-1 [110]. This method can also be applied to other Cys-rich peptides. The biosynthesis of backbone-cyclized α-defensins and naturally occurring cyclic θ-defensins is currently underway in our laboratory. It is worth noting that the recombinant expression of cyclotides facilitates the incorporation of NMR active isotopes such as 15N and 13C in a very inexpensive fashion, thus facilitating the use of the SAR by NMR (structure–activity relationship by nuclear magnetic resonance) [111] technique to characterize interactions between cyclotides and their biomolecular targets [19, 20] as well as carry out studies on the dynamics of the cyclotide scaffold [2]. The incorporation of 15N into the cyclotide kalata B1 has been also recently reported by Craik and co-workers by whole plant labeling, i.e. by extracting the cyclotide from plants grown in the laboratory in media enriched in 15N [112].
Biological activities of cyclotides
The natural function of cyclotides in plants seems to be as host-defense agents as deduced from their activity against insects [21, 22], nematodes [28, 29, 113, 114], and mollusks [60]. For example, it has been shown that cyclotides can efficiently inhibit the growth and development of insect and nematode larvae [115]. Although their mechanism of action is not totally well understood much of these activities seem to involve interaction of the cyclotide with membranes [9, 115]. Aside from their insecticidal and nematocidal activities, cyclotides have also been shown to have potential pharmacologically relevant activities, which include antimicrobial, anti-HIV, anti-tumor and neurotensin activities.
Antimicrobial activity
Most cyclotides have hydrophobic and hydrophilic patches located in different regions of their surface resembling to some extent the amphipathic character of classical antimicrobial peptides. The antimicrobial activities of cyclotides have been reported by two groups with conflicting results on the potency of kalata B1 against Escherichia coli and Staphylococcus aureus. In one study performed by Tam and co-workers, kalata B1 was active against S. aureus, but not E. coli [15], and in the second study, led by Gran and co-workers the peptide had the reverse effect [24]. This controversy has not been fully resolved yet, but it is most likely that different techniques were employed by both research teams. More recently, Tam and co-workers have also isolated several cyclotides from the plant Clitoria ternatea of the Fabaceae family with antimicrobial properties. In this work cyclotides CT1 and CT4 showed antimicrobial activity against strains of E. coli, Klebsiella pneumonia and Pseudomonas aeruginosa with minimal inhibitory concentrations ranging from 1 to 4 μM [10]. Craik and co-workers have also identified similar cyclotides in the same plant [8, 9]. In this work, cyclotides CT1 and CT4 are referred as Cter P and Cter O, respectively. These bracelet cyclotides have high sequence homology with the cyclotide cycloviolacin O2, which has been shown to have antimicrobial activity against P. aeruginosa and a multiple drug resistant strain of K. pneumonia [116].
Antimicrobial peptides that target bacterial membranes typically have an amphipathic structure with a large number of positively charged residues, which explains their affinities for negatively charged membranes over more neutral mammalian cell membranes. Most cyclotides, however, have an overall charge close to zero at physiological pH, thereby making it unlikely that they interact with bacterial membranes through electrostatic interactions like classical cationic antimicrobial peptides [117]. Hence, further studies are required to investigate the antimicrobial properties and mechanism of action of cyclotides as well as its clinical relevance. This is particular important given the growing occurrence of antibiotic resistance by microorganisms to current antibiotics.
Anti-HIV activity
The anti-HIV activity of cyclotides has been one of the most extensively studied so far due to its potential pharmacological applications [25, 39, 58, 118, 119]. The first account of a cyclotide with anti-HIV activity was reported by Gustafson and co-workers as part of a screening program at the U.S. National Cancer Center to search for novel natural products with anti-viral activity [39, 42]. In this work, several cyclotides isolated from the bark of the African tree Chassalia parvifolia, called circulins A-F, were shown to inhibit HIV infection in different host cell lines. Since then, several other cyclotides from the bracelet and Möbius subfamilies were also shown to have anti-HIV activity [25, 58, 118, 119].
The exact mode of action of these compounds remains still unclear although the inability of cyclotides to affect HIV reverse transcriptase activity combined with their cytoprotective effect suggests that the antiviral activity occurs before the entry of the virus into the host cell [39]. Recent studies have also suggested a strong correlation between the hydrophobic character of cyclotides and their anti-HIV activities [119, 120]. Moreover, surface plasma resonance and NMR studies have also shown that cyclotides can bind to model lipid membranes and that this interaction occurs primarily through their surface-exposed hydrophobic patches [121–123]. All these data suggest that the probable mode of anti-HIV action of cyclotides occurs through a mechanism that affects the binding and/or fusion of the virus to the target membrane. It is still premature, however, to conclude if cyclotide anti-HIV activity is the result of binding to the viral envelope, host cell membrane or both.
None of the cyclotides with anti-HIV activity, however, is being considered as candidates for anti-HIV therapy so far. This is due to their low therapeutic index (i.e. the ratio between the dose required for therapeutic effects versus toxic effects), which is typically too small (≤ 10) to be clinically useful. For example the therapeutic index of cyclotides kalata B1 and Varv E is 9 and 11, respectively. On the other hand, recent studies carried out on the cyclotides isolated from the plant Viola yedoensis (cycloviolacins Y4 and Y5) have shown therapeutic indexes of 45 and 14, respectively, which show some promise on the potential clinical use of these peptides to treat HIV infection [119].
Neurotensin antagonism
Cyclopsychotride (Cpt) A is a natural cyclotide isolated in 1994 from the South American tree Psychotropia longipes that has been reported to have neurotensin inhibition properties [40]. Neurotensin is a 13 amino acid neuropeptide that exerts its function by interacting with specific extracellular receptors increasing inositol triphosphate (IP3) production and inducing Ca2+ mobilization from intracellular stores. Witherup and co-workers reported that Cpt A was able to inhibit neurotensin binding to its receptor in HT-29 cell membranes with an IC50 ≈ 3 μM [40]. The direct neurotensin antagonism of Cpt A, however, was contradicted by the facts that it also increased intracellular Ca2+ levels, which could not be blocked by neurotensin antagonists [40]. Furthermore, Cpt A showed similar activity in two unrelated cell lines that did not express neurotensin receptors indicating that the mechanism of action is unlikely to be mediated through an interaction with the neurotensin receptor [40]. Recent studies using the cyclotide kalata B1 have shown that cyclotides are able to modulate membrane permeability by the formation of membrane pores with channel-like activity and no or little selectivity for specific cations [124]. The formation of similar channels by Cpt A could explain the increase on intracellular Ca2+ levels, although this has not been yet tested. Unfortunately there has been no follow-up on the neurotensin antagonism of Cpt A beyond the original report.
Antitumor activity
Several studies have reported the selective cytotoxicity of some cyclotides against cancer cells compared to normal cells [27, 125, 126]. In addition, the cytotoxic activity has been demonstrated using primary cancer cell lines [27]. Although the mechanism for cytotoxic activity of cyclotides is not totally well understood, it has been suggested that disturbance of the membrane integrity may be the cause for the cytotoxic activity. This is supported by the ability of cycloviolacin O2, one of the most active anticancer cyclotides, to disrupt tumor cell membranes [127]. Cancer cells differ from normal cells in the lipid and glycoprotein composition, which alters the overall net charge. For example, it has been shown that cancer cells express larger amounts of anionic phosphatidylserine phospholipids and O-glycosylated mucins, which typically confers a net negative charge to their membranes [128]. These differences are believed to play a major role in the cytotoxic selectivity of peptides with anticancer activity [128]. It is worth noting that cyclotide cytotoxicity is not only related to the three-dimensional structure but also to specific amino acid residues within the sequence [126, 129, 130]. For example, small modifications in the sequence of the bracelet cyclotide cycloviolacin O2 have been shown to have a great impact on cytotoxicity [129, 130]. Modification of the three positively charged residues in loops 5 and 6 as well as the Glu residue in loop 1 decreased the cytotoxic activity sevenfold and 48-fold, respectively [129]. More recent studies have also revealed that modification of the Trp residue in loop 2 has a detrimental effect on the cytotoxic activity of cycloviolacin O2 [130]. Similar results were also obtained for the cyclotide varv A, a Möbius cyclotide with high anticancer activity isolated from the plant Viola arvensis [130].
All of these studies suggest that differences in membrane composition and cyclotide primary sequence modulate membrane binding and the cytotoxic effects of cyclotides. A more comprehensive structural study on the membrane-binding properties of cyclotides will be required to have a better understanding on their antitumor mechanism of action.
Toxicity
Some cyclotides have been reported to have several toxic effects. For example, some cyclotides have been found to cause extensive hemolysis of human and rat erythrocytes, with HD50 (hemolytic dose) values ranging from 5 μM to 300 μM [15, 119, 131]. The large variation in HD50 value reflects different experimental conditions such as temperature and incubation time. The cyclotide kalata B1 has strong hemolytic activity and its lethal dose (LD50) in rats and rabbits has been reported to be 1.0 mg/kg and 1.2 mg/kg, respectively, when it was injected intravenously. This cyclotide has been also reported to produce cardiotoxic effects associated with increased arterial blood pressure and tachycardia [38]. Interestingly, the strong hemolytic activity of kalata B1 can be eliminated by mutation to Ala of any one of eight residues located in the bioactive face of the molecule [132]. This suggests that it may be possible to eliminate other toxic effects by single mutation although this has yet to be tested.
It is worth noting that some other naturally occurring cyclotides have markedly reduced or no toxic effects at all. For example, the trypsin inhibitor cyclotides MCoTI-I and -II are non-hemolytic and non-toxic to human cells up to a concentration of 100 μM [35], and therefore provide an excellent scaffold for the design of novel cyclotides with new biological activities. MCoTI-cyclotides are also particularly interesting from a pharmaceutical perspective because of their ability to penetrate cells, and therefore interact with intracellular targets [35, 36, 133].
Engineering cyclotides with novel activities
The unique properties associated with the cyclotide scaffold make them extremely valuable tools in drug discovery [1, 4, 117]. For example, the cyclic cystine knot (CCK) framework gives cyclotides a compact, highly rigid structure [2], which confers exceptional resistance to thermal/chemical denaturation, and enzymatic degradation [3]. Cyclotide can be also readly produced by chemical synthesis [14] and expressed in cells using standard cloning vectors [18–20]. Moreover, their high tolerance to mutations makes them ideal scaffolds for molecular grafting and evolution in order to generate novel cyclotides with new biological activities [20, 34]. Even more importantly, MCoTI-cyclotides have been shown recently to be able to enter human macrophages, breast and ovarian cancer cell lines [35, 36].
The high plasticity and tolerance to substitution of the cyclotide scaffold was first demonstrated by replacing some of the hydrophobic residues in loop 5 of cyclotide kalata B1 with polar and charged residues [31]. The mutated cyclotides retained the native fold of kalata B1, but were no longer hemolytic [31]. More recently our group has also preformed a similar study on the cyclotide MCoTI-I, where all residues located in loops 1 through 5 were replaced by different amino acids. In this study only 2 from a total of 26 mutants were not able to fold efficiently [20]. These studies demonstrate the high plasticity of the cyclotide scaffold to mutations thus opening the possibility to introduce or graft foreign sequences onto them without affecting the native fold. Figure 6 highlights several studies that have used the cyclotide molecular scaffold to graft peptide sequences and to generate libraries for the purpose of engineering cyclotides with novel biological functions. It should be noted however, that the great stability and robustness of cyclotide framework makes necessary to be careful when grafting a peptide sequence into the peptide scaffold. It is important to be sure that the structure of the peptide displayed on the cyclotide will not be distorted by the conformation requirements of the cyclotide leading to a non-biologically active conformation of the grafted sequence.
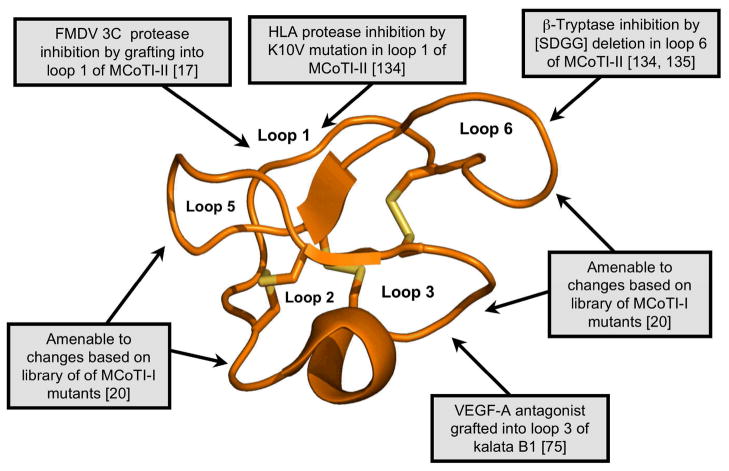
Figure 6.
Summary of some of the modifications made to kalata B1 and MCoTI-II to elicit novel biological activities. Modifications include the grafting of peptides onto various loops of kalata B1 [75] and MCoTI-II [17, 134, 135]. Cyclotide based libraries have been also generated using loops 1, 2, 3, 4 and 5 of MCoTI-I to study the effect of individual mutations on the biological activity and folding ability of MCoTI-I mutants [20]. The locations of the changes introduced into the cyclotide framework are illustrated using the MCoTI-II structure (pdb ID: 1IB9).
The potential pharmaceutical applications of grafted cyclotides was first demonstrated in two recent studies aimed to develop novel anti-cancer [75] and anti-viral peptide-based therapeutics [17] (Fig. 6). The development of anti-cancer cyclotides involved grafting a peptide antagonist of angiogenesis onto the kalata B1 scaffold [75]. Tumor growth is usually associated with unregulated angiogenesis and therefore molecules with anti-angiogenic activity have potential applications in cancer treatment. In this study an Arg-rich peptide antagonist for the interaction of vascular endothelial growth factor A (VEGF-A) and its receptor was individually grafted into loops 2, 3 and 5 of kalata B1 [75]. The cyclotide grafted into loop 3 showed the highest activity in blocking VEGF-A receptor binding (IC50 ≈ 12 μM) when compared with that of the isolated grafted peptide epitope as well as the other grafted cyclotides. Although this is the first example of a successful functional redesign of a cyclotide, it should be noted that the biological activity would still need to be improved by several orders of magnitude for a potential application in vivo.
Additionally, the hemolytic activity of the grafted katala B1 was completely removed and the proteolytic susceptibility of the Arg-rich grafted peptide was greatly diminished when compared to that of the isolated peptide sequence. This study clearly demonstrates that cyclotides can be used as molecular scaffolds for displaying and stabilizing pharmacologically relevant active peptide sequences.
The utility of the cyclotide scaffold in drug design has also been recently shown by engineering non-native activities into the cyclotide MCoTI-II [17]. MCoTI-II is a member of the trypsin inhibitor subfamily. Trypsin inhibitor cyclotides are strong inhibitors of trypsin (Ki ≈ 20 pM) and other trypsin-like proteases. In these cyclotides loop 1 is responsible for binding to the selectivity pocket of trypsin [62]. In this work, the loop 1 of MCoTI-II was engineered to be able to inhibit other proteases [17]. Interestingly, several analogs showed activity against the foot-and-mouth-disease virus (FMDV) 3C protease in the low μM range [17]. The FMDV 3C protease is a Cys protease key for viral replication and therefore a potential target for the development of novel anti-viral therapeutics. This is the first reported peptide-based inhibitor for this protease. Although the potency was relatively low, this study demonstrates the potential of using MCoTI-based cyclotides for designing novel protease inhibitors [17].
In a more recent study, the same authors were also able to generate inhibitors of the serine proteases β-tryptase and human leukocyte elastase (HLE) using the cyclotide MCoTI-II as a molecular scaffold [134]. These two proteases are pharmacologically relevant drug targets that have been associated with respiratory and pulmonary disorders (HLE) and implicated in allergic and inflammatory disorders (β-tryptase). In this work Leatherbarrow and co-workers replaced the P1 residue in loop 1 to produce several MCoTI-II mutants (K6A and K6V) that showed activity against HLE in the low nM range (Ki ≈ 25 nM) and relatively low activity against trypsin (Ki ≥ 1 μM) [134]. Interestingly, the same authors also showed that removal of the SDGG peptide segment in loop 6 yielded a relative potent β-tryptase inhibitor (Ki ≈ 10 nM) without significantly altering the three-dimensional structure of the resulting cyclotide as determined by NMR [134]. The authors hypothesized that the removal of the Asp residue from loop 6 removes repulsive electrostatic and steric interactions with β-tryptase thus improving the inhibitory constant 160-fold when compared to the wild-type MCoTI-II.
Kolmar and co-workers have also recently reported the design of a series of inhibitors of human mast cell tryptase beta using the cyclotide MCoTI-II as scaffold [135]. In this interesting work, the authors introduced additional positive charge in the loop 6 of MCoTI-II. The resulting engineered cyclotides were able to inhibit all the monomers of the tryptase beta tetramer with Ki values around 1 nM.
Proteases are well-recognized drug targets and many diseases including inflammatory and pulmonary diseases, cancer, cardiovascular and neurodegenerative conditions have been associated with abnormal expression levels of proteases [136]. These two examples demonstrate that trypsin inhibitor cyclotides can be re-engineered to tailor their specificity for proteases other than trypsin, which has potential applications in drug development for protease targeting. The availability
Screening of cyclotide-based libraries
The ability to produce natively folded cyclotides in vivo discussed earlier opens up the intriguing possibility of generating large libraries of genetically-encoded cyclotides potentially containing billions of members. This tremendous molecular diversity should allow the selection strategies mimicking the evolutionary processes found in nature to select novel cyclotide sequences able to target specific molecular targets.
The potential for generating cyclotide libraries was first explored by our group using the kalata B1 scaffold [18]. In this work wild-type and several mutants of kalata B1 were biosynthesized using an intramolecular native chemical ligation facilitated by a modified protein splicing unit (Fig. 5). The study generated six different linear versions of kalata B1, which were expressed in E. coli as fusions to a modified version of the yeast vacuolar membrane ATPase (VMA) intein. The in vitro folding and cyclization of the different kalata B1 linear precursors did not occur equally, suggesting the predisposition to adopt a native fold of the corresponding linear precursor may determine the efficiency of the cleavage/cyclization step [18]. This information was used to express a small library based on the kalata B1 scaffold. Cyclization and concomitant folding of the library was successfully performed in vitro using a redox buffer containing reduced GSH as a thiol co-factor therefore mimicking the intracellular conditions [18].
We have also demonstrated that this approach can be used for the production of cyclotides inside live E. coli cells [19]. In this study, the cyclotide MCoTI-II was efficiently produced in living E. coli cells by in vivo processing of the corresponding intein fusion precursor. In order to improve the expression yield of the precursor protein and boost the expression of folded cyclotide the bacterial gyrase A intein from Mycobacterium xenopus was used in this study instead [19]. This intein has been shown to express at higher yields than the yeast VMA intein in E. coli expression systems [19]. Using this approach, folded MCoTI-I can be expressed in E. coli cells to an intracellular concentration of low μM [19].
More recently, our group has extended this technology to the biosynthesis of a genetically encoded library of MCoTI-I based cyclotides [20]. In this case, the cyclization/folding of the library was performed either in vitro, by incubation with a redox buffer containing GSH, or by in vivo self-processing of the corresponding cyclotide-intein precursors. The cyclotide library was purified and screened for activity using trypsin-immobilized sepharose beads. The library was designed to mutate every single amino acid in loops 1, 2, 3, 4 and 5 to explore the effects on folding and trypsin binding activity of the resulting mutants. Interestingly, only two mutations (G27P and I22G) out of the 26 substitutions studied were able to negatively affect the folding of the resulting cyclotides. The I22G mutation affects loop 4, which is only formed by one residue. This loop forms part of the cyclotide scaffold, which may explain the deficient folding of this mutant. The G27P mutation is located at the end of loop 5. Intriguingly, this position is occupied by a Pro residue in Möbius cyclotides and is required for efficient folding. It is also interesting to remark that although these two mutants were not able to fold efficiently, the natively folded form was still able to bind trypsin [20]. The rest of the mutants were able to cyclize and fold with similar yields. As expected, the mutant K6A-MCoTI-I was not able to bind trypsin under the conditions used in the experiment although adopted a native cyclotide fold as determined by NMR [20]. As mentioned before, this residue is key for binding to the specificity pocket of trypsin, ant it can be modified to change the inhibitory specificity of the resulting MCoTI-cyclotide to target other proteases [17, 134]. The affinity of each member of the MCoTI-library for trypsin was assayed using a competitive trypsin-binding assay [20]. The mutant cyclotides with less affinity were mostly found in loop 1 and the C-terminal region of loop 6, both well conserved among other squash trypsin inhibitors. These results combined with similar studies performed in kalata B1 [132] confirm the high plasticity and tolerance to mutations of the cyclotide framework, thus providing an ideal scaffold for the biosynthesis of large combinatorial libraries inside living bacterial cells. These genetically-encoded libraries can be screened in-cell for biological activity using high-throughput flow cytometry techniques for the rapid selection of novel biologically active cyclotides [18, 137, 138].
Concluding remarks
Cyclotides are a new emerging family of highly stable plant-derived backbone-cyclized polypeptides that share a disulfide-stabilized core characterized by an unusual knotted structure. Their unique circular backbone topology and knotted arrangement of three disulfide bonds provides a compact, highly rigid structure [2] which confers exceptional resistance to thermal/chemical denaturation, and enzymatic degradation [3]. They can be also chemically synthesized allowing the introduction of chemical modifications such as non-natural amino acids and PEGylation to improve their pharmacological properties [14, 16, 17, 139]. Cyclotides can also be expressed in bacterial cells, and are amenable to substantial sequence variation, thus making them ideal substrates for molecular evolution strategies to enable generation and selection of compounds with optimal binding and inhibitory characteristics [18–20]. Finally, cyclotides have been shown to be able to cross human cell membranes [35, 36, 133]. Folded cyclotides are extremely resistant to chemical, physical and proteolytic degradation [3, 4]. Cyclotides have been also shown to fold inside bacterial cells [19, 20], which have a more reductive cytosolic environment than eukaryotic cells and therefore is highly unlikely that they were reduced in the cytosol of mammalian cells. Altogether, these characteristics make them promising leads or frameworks for peptide drug design [4, 31, 32, 117].
Acknowledgments
This work was supported by National Institutes of Health Research Grant R01-GM090323 and Department of Defense Congressionally Directed Medical Research Program Grant PC09305.
References
- 1.Daly NL, Rosengren KJ, Craik DJ. Discovery, structure and biological activities of cyclotides. Adv Drug Deliv Rev. 2009;61:918–930. doi: 10.1016/j.addr.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Puttamadappa SS, Jagadish K, Shekhtman A, Camarero JA. Backbone Dynamics of Cyclotide MCoTI-I Free and Complexed with Trypsin. Angew Chem Int Ed Engl. 2010;49:7030–7034. doi: 10.1002/anie.201002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colgrave ML, Craik DJ. Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: the importance of the cyclic cystine knot. Biochemistry. 2004;43:5965–5975. doi: 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- 4.Garcia AE, Camarero JA. Biological activities of natural and engineered cyclotides, a novel molecular scaffold for peptide-based therapeutics. Curr Mol Pharmacol. 2010;3:153–163. doi: 10.2174/1874467211003030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saether O, Craik DJ, Campbell ID, Sletten K, Juul J, Norman DG. Elucidation of the primary and three-dimensional structure of the uterotonic polypeptide kalata B1. Biochemistry. 1995;34:4147–4158. doi: 10.1021/bi00013a002. [DOI] [PubMed] [Google Scholar]
- 6.Gruber CW, Elliott AG, Ireland DC, Delprete PG, Dessein S, Goransson U, et al. Distribution and evolution of circular miniproteins in flowering plants. Plant Cell. 2008;20:2471–2483. doi: 10.1105/tpc.108.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiche L, Heitz A, Gelly JC, Gracy J, Chau PT, Ha PT, et al. Squash inhibitors: from structural motifs to macrocyclic knottins. Curr Protein Pept Sci. 2004;5:341–349. doi: 10.2174/1389203043379477. [DOI] [PubMed] [Google Scholar]
- 8.Poth AG, Colgrave ML, Philip R, Kerenga B, Daly NL, Anderson MA, et al. Discovery of cyclotides in the fabaceae plant family provides new insights into the cyclization, evolution, and distribution of circular proteins. ACS chemical biology. 2010;6:345–355. doi: 10.1021/cb100388j. [DOI] [PubMed] [Google Scholar]
- 9.Poth AG, Colgrave ML, Lyons RE, Daly NL, Craik DJ. Discovery of an unusual biosynthetic origin for circular proteins in legumes. Proc Natl Acad Sci U S A. 2011;108:1027–1032. doi: 10.1073/pnas.1103660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen GK, Zhang S, Nguyen NT, Nguyen PQ, Chiu MS, Hardjojo A, et al. Discovery and characterization of novel cyclotides originated from chimeric precursors consisting of albumin-1 chain a and cyclotide domains in the fabaceae family. J Biol Chem. 2011 doi: 10.1074/jbc.M111.229922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camarero JA. Legume cyclotides shed light on the genetic origin of knotted circular proteins. Proc Natl Acad Sci U S A. 2011;108:10025–10026. doi: 10.1073/pnas.1107849108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craik DJ, Cemazar M, Wang CK, Daly NL. The cyclotide family of circular miniproteins: nature’s combinatorial peptide template. Biopolymers. 2006;84:250–266. doi: 10.1002/bip.20451. [DOI] [PubMed] [Google Scholar]
- 13.Dutton JL, Renda RF, Waine C, Clark RJ, Daly NL, Jennings CV, et al. Conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins. J Biol Chem. 2004;279:46858–46867. doi: 10.1074/jbc.M407421200. [DOI] [PubMed] [Google Scholar]
- 14.Daly NL, Love S, Alewood PF, Craik DJ. Chemical synthesis and folding pathways of large cyclic polypeptides: studies of the cystine knot polypeptide kalata B1. Biochemistry. 1999;38:10606–10614. doi: 10.1021/bi990605b. [DOI] [PubMed] [Google Scholar]
- 15.Tam JP, Lu YA, Yang JL, Chiu KW. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc Natl Acad Sci U S A. 1999;96:8913–8918. doi: 10.1073/pnas.96.16.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thongyoo P, Tate EW, Leatherbarrow RJ. Total synthesis of the macrocyclic cysteine knot microprotein MCoTI-II. Chem Commun (Camb) 2006:2848–2850. doi: 10.1039/b607324g. [DOI] [PubMed] [Google Scholar]
- 17.Thongyoo P, Roque-Rosell N, Leatherbarrow RJ, Tate EW. Chemical and biomimetic total syntheses of natural and engineered MCoTI cyclotides. Org Biomol Chem. 2008;6:1462–1470. doi: 10.1039/b801667d. [DOI] [PubMed] [Google Scholar]
- 18.Kimura RH, Tran AT, Camarero JA. Biosynthesis of the cyclotide kalata B1 by using protein splicing. Angew Chem Int Ed. 2006;45:973–976. doi: 10.1002/anie.200503882. [DOI] [PubMed] [Google Scholar]
- 19.Camarero JA, Kimura RH, Woo YH, Shekhtman A, Cantor J. Biosynthesis of a fully functional cyclotide inside living bacterial cells. Chembiochem. 2007;8:1363–1366. doi: 10.1002/cbic.200700183. [DOI] [PubMed] [Google Scholar]
- 20.Austin J, Wang W, Puttamadappa S, Shekhtman A, Camarero JA. Biosynthesis and biological screening of a genetically encoded library based on the cyclotide MCoTI-I. Chembiochem. 2009;10:2663–2670. doi: 10.1002/cbic.200900534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jennings C, West J, Waine C, Craik D, Anderson M. Biosynthesis and insecticidal properties of plant cyclotides: the cyclic knotted proteins from Oldenlandia affinis. Proc Natl Acad Sci U S A. 2001;98:10614–10619. doi: 10.1073/pnas.191366898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jennings CV, Rosengren KJ, Daly NL, Plan M, Stevens J, Scanlon MJ, et al. Isolation, solution structure, and insecticidal activity of kalata B2, a circular protein with a twist: do Mobius strips exist in nature? Biochemistry. 2005;44:851–860. doi: 10.1021/bi047837h. [DOI] [PubMed] [Google Scholar]
- 23.Gran L. Isolation of oxytocic peptides from Oldenlandia affinis by solvent extraction of tetraphenylborate complexes and chromatography on Sephadex LH-20. Lloydia. 1973;36:207–208. [PubMed] [Google Scholar]
- 24.Gran L, Sletten K, Skjeldal L. Cyclic Peptides from Oldenlandia affinis DC. Molecular and Biological Properties. Chem Biodivers. 2008;5:2014–2022. doi: 10.1002/cbdv.200890184. [DOI] [PubMed] [Google Scholar]
- 25.Gustafson KR, McKee TC, Bokesch HR. Anti-HIV cyclotides. Curr Protein Pept Sci. 2004;5:331–340. doi: 10.2174/1389203043379468. [DOI] [PubMed] [Google Scholar]
- 26.Hallock YF, Sowder RC, Pannell LK, Hughes CB, Johnson DG, Gulakowski R, et al. Cycloviolins A-D, anti-HIV macrocyclic peptides from Leonia cymosa. J Org Chem. 2000;65:124–128. doi: 10.1021/jo990952r. [DOI] [PubMed] [Google Scholar]
- 27.Lindholm P, Göransson U, Johansson S, Claeson P, Gullbo J, Larsson R, et al. Cyclotides: A novel type of cytotoxic agents. Mol Cancer Ther. 2002;1:365–369. [PubMed] [Google Scholar]
- 28.Colgrave ML, Kotze AC, Ireland DC, Wang CK, Craik DJ. The anthelmintic activity of the cyclotides: natural variants with enhanced activity. Chembiochem. 2008;9:1939–1945. doi: 10.1002/cbic.200800174. [DOI] [PubMed] [Google Scholar]
- 29.Colgrave ML, Kotze AC, Kopp S, McCarthy JS, Coleman GT, Craik DJ. Anthelmintic activity of cyclotides: In vitro studies with canine and human hookworms. Acta Trop. 2009;109:163–166. doi: 10.1016/j.actatropica.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Avrutina O, Schmoldt HU, Gabrijelcic-Geiger D, Le Nguyen D, Sommerhoff CP, Diederichsen U, et al. Trypsin inhibition by macrocyclic and open-chain variants of the squash inhibitor MCoTI-II. Biol Chem. 2005;386:1301–1306. doi: 10.1515/BC.2005.148. [DOI] [PubMed] [Google Scholar]
- 31.Clark RJ, Daly NL, Craik DJ. Structural plasticity of the cyclic-cystine-knot framework: implications for biological activity and drug design. The Biochemical journal. 2006;394:85–93. doi: 10.1042/BJ20051691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Craik DJ, Cemazar M, Daly NL. The cyclotides and related macrocyclic peptides as scaffolds in drug design. Current opinion in drug discovery & development. 2006;9:251–260. [PubMed] [Google Scholar]
- 33.Craik DJ, Daly NL, Mulvenna J, Plan MR, Trabi M. Discovery, structure and biological activities of the cyclotides. Curr Protein Pept Sci. 2004;5:297–315. doi: 10.2174/1389203043379512. [DOI] [PubMed] [Google Scholar]
- 34.Sancheti H, Camarero JA. “Splicing up” drug discovery. Cell-based expression and screening of genetically-encoded libraries of backbone-cyclized polypeptides. Adv Drug Deliv Rev. 2009;61:908–917. doi: 10.1016/j.addr.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenwood KP, Daly NL, Brown DL, Stow JL, Craik DJ. The cyclic cystine knot miniprotein MCoTI-II is internalized into cells by macropinocytosis. Int J Biochem Cell Biol. 2007;39:2252–2264. doi: 10.1016/j.biocel.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 36.Contreras J, Elnagar AYO, Hamm-Alvarez S, Camarero JA. Cellular Uptake of cyclotide MCoTI-I follows multiple endocytic pathways. J Control Release. 2011 doi: 10.1016/j.jconrel.2011.1008.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gran L. Oxytocic principles of Oldenlandia affinis. Lloydia. 1973;36:174–178. [PubMed] [Google Scholar]
- 38.Gran L. On the effect of a polypeptide isolated from “Kalata-Kalata” (Oldenlandia affinis DC) on the oestrogen dominated uterus. Acta Pharmacol Toxicol (Copenh) 1973;33:400–408. doi: 10.1111/j.1600-0773.1973.tb01541.x. [DOI] [PubMed] [Google Scholar]
- 39.Gustafson KR, Sowder RC, Louis E, Henderson LE, Parsons IC, Kashman Y, Cardellina JH, et al. Circulins A and B. Novel human immunodeficiency virus (HIV)-inhibitory macrocyclic peptides from the tropical tree Chassalia parvifolia. J Am Chem Soc. 1994;116:9337–9338. [Google Scholar]
- 40.Witherup KM, Bogusky MJ, Anderson PS, Ramjit H, Ransom RW, Wood T, et al. Cyclopsychotride A, a biologically active, 31-residue cyclic peptide isolated from Psychotria longipes. J Nat Prod. 1994;57:1619–1625. doi: 10.1021/np50114a002. [DOI] [PubMed] [Google Scholar]
- 41.Schöpke T, Hasan Agha MI, Kraft R, Otto A, Hiller K. Hämolytisch aktive komponenten aus Viola tricolor L. und Viola arvensis Murray. Sci Pharm. 1993;61:145–153. [Google Scholar]
- 42.Gustafson KR, Walton LK, Sowder RC, Jr, Johnson DG, Pannell LK, Cardellina JH, Jr, et al. New circulin macrocyclic polypeptides from Chassalia parvifolia. J Nat Prod. 2000;63:176–178. doi: 10.1021/np990432r. [DOI] [PubMed] [Google Scholar]
- 43.Claeson P, Goransson U, Johansson S, Luijendijk T, Bohlin L. Fractionation Protocol for the Isolation of Polypeptides from Plant Biomass. J Nat Prod. 1998;61:77–81. doi: 10.1021/np970342r. [DOI] [PubMed] [Google Scholar]
- 44.Goransson U, Luijendijk T, Johansson S, Bohlin L, Claeson P. Seven novel macrocyclic polypeptides from Viola arvensis. J Nat Prod. 1999;62:283–286. doi: 10.1021/np9803878. [DOI] [PubMed] [Google Scholar]
- 45.Simonsen SM, Sando L, Ireland DC, Colgrave ML, Bharathi R, Goransson U, et al. A continent of plant defense peptide diversity: cyclotides in Australian Hybanthus (Violaceae) Plant Cell. 2005;17:3176–3189. doi: 10.1105/tpc.105.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolmar H, Skerra A. Alternative binding proteins get mature: rivalling antibodies. FEBS J. 2008;275:2667. doi: 10.1111/j.1742-4658.2008.06437.x. [DOI] [PubMed] [Google Scholar]
- 47.Gelly JC, Gracy J, Kaas Q, Le-Nguyen D, Heitz A, Chiche L. The KNOTTIN website and database: a new information system dedicated to the knottin scaffold. Nucleic Acids Res. 2004;32:D156–159. doi: 10.1093/nar/gkh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gracy J, Le-Nguyen D, Gelly JC, Kaas Q, Heitz A, Chiche L. KNOTTIN: the knottin or inhibitor cystine knot scaffold in 2007. Nucleic Acids Res. 2008;36:D314–319. doi: 10.1093/nar/gkm939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang CK, Kaas Q, Chiche L, Craik DJ. CyBase: a database of cyclic protein sequences and structures, with applications in protein discovery and engineering. Nucleic Acids Res. 2008;36:D206–210. doi: 10.1093/nar/gkm953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daly NL, Rosengren KJ, Henriques ST, Craik DJ. NMR and protein structure in drug design: application to cyclotides and conotoxins. Eur Biophys J. 2011;40:359–370. doi: 10.1007/s00249-011-0672-9. [DOI] [PubMed] [Google Scholar]
- 51.Wang CK, Hu SH, Martin JL, Sjogren T, Hajdu J, Bohlin L, et al. Combined X-ray and NMR analysis of the stability of the cyclotide cystine knot fold that underpins its insecticidal activity and potential use as a drug scaffold. J Biol Chem. 2009;284:10672–10683. doi: 10.1074/jbc.M900021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Svangard E, Goransson U, Smith D, Verma C, Backlund A, Bohlin L, et al. Primary and 3-D modelled structures of two cyclotides from Viola odorata. Phytochemistry. 2003;64:135–142. doi: 10.1016/s0031-9422(03)00218-8. [DOI] [PubMed] [Google Scholar]
- 53.Sze SK, Wang W, Meng W, Yuan R, Guo T, Zhu Y, et al. Elucidating the structure of cyclotides by partial acid hydrolysis and LC-MS/MS analysis. Anal Chem. 2009;81:1079–1088. doi: 10.1021/ac802175r. [DOI] [PubMed] [Google Scholar]
- 54.Craik DJ, Daly NL, Bond T, Waine C. Plant cyclotides: A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J Mol Biol. 1999;294:1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- 55.Rosengren KJ, Daly NL, Plan MR, Waine C, Craik DJ. Twists, knots, and rings in proteins. Structural definition of the cyclotide framework. J Biol Chem. 2003;278:8606–8616. doi: 10.1074/jbc.M211147200. [DOI] [PubMed] [Google Scholar]
- 56.Goransson U, Craik DJ. Disulfide mapping of the cyclotide kalata B1. Chemical proof of the cystic cystine knot motif. J Biol Chem. 2003;278:48188–48196. doi: 10.1074/jbc.M308771200. [DOI] [PubMed] [Google Scholar]
- 57.Jagadish K, Camarero JA. Cyclotides, a promising molecular scaffold for peptide-based therapeutics. Biopolymers. 2010;94:611–616. doi: 10.1002/bip.21433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daly NL, Gustafson KR, Craik DJ. The role of the cyclic peptide backbone in the anti-HIV activity of the cyclotide kalata B1. FEBS Lett. 2004;574:69–72. doi: 10.1016/j.febslet.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 59.Wang CK, Colgrave ML, Ireland DC, Kaas Q, Craik DJ. Despite a conserved cystine knot motif, different cyclotides have different membrane binding modes. Biophys J. 2009;97:1471–1481. doi: 10.1016/j.bpj.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Plan MR, Saska I, Cagauan AG, Craik DJ. Backbone cyclised peptides from plants show molluscicidal activity against the rice pest Pomacea canaliculata (golden apple snail) J Agric Food Chem. 2008;56:5237–5241. doi: 10.1021/jf800302f. [DOI] [PubMed] [Google Scholar]
- 61.Daly NL, Clark RJ, Plan MR, Craik DJ. Kalata B8, a novel antiviral circular protein, exhibits conformational flexibility in the cystine knot motif. Biochem J. 2006;393:619–626. doi: 10.1042/BJ20051371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hernandez JF, Gagnon J, Chiche L, Nguyen TM, Andrieu JP, Heitz A, et al. Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry. 2000;39:5722–5730. doi: 10.1021/bi9929756. [DOI] [PubMed] [Google Scholar]
- 63.Heitz A, Hernandez JF, Gagnon J, Hong TT, Pham TT, Nguyen TM, et al. Solution structure of the squash trypsin inhibitor MCoTI-II. A new family for cyclic knottins. Biochemistry. 2001;40:7973–7983. doi: 10.1021/bi0106639. [DOI] [PubMed] [Google Scholar]
- 64.Felizmenio-Quimio ME, Daly NL, Craik DJ. Circular proteins in plants: solution structure of a novel macrocyclic trypsin inhibitor from Momordica cochinchinensis. J Biol Chem. 2001;276:22875–22882. doi: 10.1074/jbc.M101666200. [DOI] [PubMed] [Google Scholar]
- 65.Mylne JS, Colgrave ML, Daly NL, Chanson AH, Elliott AG, McCallum EJ, et al. Albumins and their processing machinery are hijacked for cyclic peptides in sunflower. Nat Chem Biol. 2011;7:257–259. doi: 10.1038/nchembio.542. [DOI] [PubMed] [Google Scholar]
- 66.Saska I, Gillon AD, Hatsugai N, Dietzgen RG, Hara-Nishimura I, Anderson MA, et al. An asparaginyl endopeptidase mediates in vivo protein backbone cyclization. J Biol Chem. 2007;282:29721–29728. doi: 10.1074/jbc.M705185200. [DOI] [PubMed] [Google Scholar]
- 67.Gillon AD, Saska I, Jennings CV, Guarino RF, Craik DJ, Anderson MA. Biosynthesis of circular proteins in plants. Plant J. 2008;53:505–515. doi: 10.1111/j.1365-313X.2007.03357.x. [DOI] [PubMed] [Google Scholar]
- 68.Ireland DC, Colgrave ML, Nguyencong P, Daly NL, Craik DJ. Discovery and characterization of a linear cyclotide from Viola odorata: implications for the processing of circular proteins. J Mol Biol. 2006;357:1522–1535. doi: 10.1016/j.jmb.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 69.Gruber CW, Cemazar M, Heras B, Martin JL, Craik DJ. Protein disulfide isomerase: the structure of oxidative folding. Trends Biochem Sci. 2006;31:455–464. doi: 10.1016/j.tibs.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 70.Marglin A, Merrifield RB. Chemical synthesis of peptides and proteins. Annu Rev Biochem. 1970;39:841–866. doi: 10.1146/annurev.bi.39.070170.004205. [DOI] [PubMed] [Google Scholar]
- 71.Dawson PE, Muir TW, Clark-Lewis I, Kent SBH. Synthesis of Proteins by Native Chemical Ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 72.Camarero JA, Pavel J, Muir TW. Chemical Synthesis of a Circular Protein Domain: Evidence for Folding-Assisted Cyclization. Angew Chem Int Ed. 1998;37:347–349. doi: 10.1002/(SICI)1521-3773(19980216)37:3<347::AID-ANIE347>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 73.Tam JP, Lu Y-A. Synthesis of Large Cyclic Cystine-Knot Peptide by Orthogonal Coupling Strategy Using Unprotected Peptide Precursor. Tetrathedron Lett. 1997;38:5599–5602. [Google Scholar]
- 74.Tam JP, Lu YA. A biomimetic strategy in the synthesis and fragmentation of a cyclic protein. Prot Sci. 1998;7:1583–1592. doi: 10.1002/pro.5560070712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gunasekera S, Daly NL, Clark RJ, Craik DJ. Dissecting the Oxidative Folding of Circular Cystine Knot Miniproteins. Antioxid Redox Sign. 2008;11:971–980. doi: 10.1089/ars.2008.2295. [DOI] [PubMed] [Google Scholar]
- 76.Aboye TL, Clark RJ, Craik DJ, Göransson U. Ultra-stable peptide scaffolds for protein engineering-synthesis and folding of the circular cystine knotted cyclotide cycloviolacin O2. Chembiochem. 2008;9:103–113. doi: 10.1002/cbic.200700357. [DOI] [PubMed] [Google Scholar]
- 77.Camarero JA, Muir TW. Chemoselective backbone cyclization of unprotected peptides. Chem Comm. 1997;1997:1369–1370. [Google Scholar]
- 78.Shao Y, Lu WY, Kent SBH. A novel method to synthesize cyclic peptides. Tetrahedron Lett. 1998;39:3911–3914. [Google Scholar]
- 79.Camarero JA, Muir TW. Biosynthesis of a Head-to-Tail Cyclized Protein with Improved Biological Activity. J Am Chem Soc. 1999;121:5597–5598. [Google Scholar]
- 80.Camarero JA, Mitchell AR. Synthesis of proteins by native chemical ligation using Fmoc-based chemistry. Protein Pept Lett. 2005;12:723–728. doi: 10.2174/0929866054864166. [DOI] [PubMed] [Google Scholar]
- 81.Camarero JA, Cotton GJ, Adeva A, Muir TW. Chemical ligation of unprotected peptides directly from a solid support. J Pept Res. 1998;51:303–316. doi: 10.1111/j.1399-3011.1998.tb00428.x. [DOI] [PubMed] [Google Scholar]
- 82.Beligere GS, Dawson PE. Conformationally assisted ligation using C-terminal thioester peptides. J Am Chem Soc. 1999;121:6332–6333. [Google Scholar]
- 83.Camarero JA, Adeva A, Muir TW. 3-Thiopropionic acid as a highly versatile multidetachable thioester resin linker. Lett Pept Sci. 2000;7:17–21. [Google Scholar]
- 84.Ingenito R, Bianchi E, Fattori D, Pessi A. Solid phase synthesis of peptide C-terminal thioesters by Fmoc/t-Bu chemistry Source. J Am Chem Soc. 1999;121:11369–11374. [Google Scholar]
- 85.Shin Y, Winans KA, Backes BJ, Kent SBH, Ellman JA, Bertozzi CR. Fmoc-Based Synthesis of Peptide-aThioesters: Application to the Total Chemical Synthesis of a Glycoprotein by Native Chemical Ligation. J Am Chem Soc. 1999;121:11684–11689. [Google Scholar]
- 86.Camarero JA, Hackel BJ, de Yoreo JJ, Mitchell AR. Fmoc-based synthesis of peptide alpha-thioesters using an aryl hydrazine support. J Org Chem. 2004;69:4145–4151. doi: 10.1021/jo040140h. [DOI] [PubMed] [Google Scholar]
- 87.Blanco-Canosa JB, Dawson PE. An efficient Fmoc-SPPS approach for the generation of thioester peptide precursors for use in native chemical ligation. Angew Chem Int Ed Engl. 2008;47:6851–6855. doi: 10.1002/anie.200705471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Daly NL, Clark RJ, Craik DJ. Disulfide folding pathways of cystine knot proteins. Tying the knot within the circular backbone of the cyclotides. J Biol Chem. 2003;278:6314–6322. doi: 10.1074/jbc.M210492200. [DOI] [PubMed] [Google Scholar]
- 89.Daly NL, Clark RJ, Göransson U, Craik DJ. Diversity in the disulfide folding pathways of cystine knot peptides. Lett Pept Sci. 2003;10:523–531. [Google Scholar]
- 90.Cemazar M, Daly NL, Haggblad S, Lo KP, Yulyaningsih E, Craik DJ. Knots in rings. The circular knotted protein Momordica cochinchinensis trypsin inhibitor-II folds via a stable two-disulfide intermediate. J Biol Chem. 2006;281:8224–8232. doi: 10.1074/jbc.M513399200. [DOI] [PubMed] [Google Scholar]
- 91.Aboye TL, Clark RJ, Burman R, Roig MB, Craik DJ, Göransson U. Interlocking Disulfides in Circular Proteins: Toward Efficient Oxidative Folding of Cyclotides. Antioxid Redox Sign. 2011;14:77–86. doi: 10.1089/ars.2010.3112. [DOI] [PubMed] [Google Scholar]
- 92.Marx UC, Korsinczky ML, Schirra HJ, Jones A, Condie B, Otvos L, Jr, et al. Enzymatic cyclization of a potent bowman-birk protease inhibitor, sunflower trypsin inhibitor-1, and solution structure of an acyclic precursor peptide. J Biol Chem. 2003;278:21782–21789. doi: 10.1074/jbc.M212996200. [DOI] [PubMed] [Google Scholar]
- 93.Perler FB, Adam E. Protein splicing and its applications. Curr Opin Biotechnol. 2000:377–383. doi: 10.1016/s0958-1669(00)00113-0. [DOI] [PubMed] [Google Scholar]
- 94.Hirel PH, Schmitter MJ, Dessen P, Fayat G, Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc Natl Acad Sci U S A. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dwyer MA, Lu W, Dwyer JJ, Kossiakoff AA. Biosynthetic phage display: a novel protein engineering tool combining chemical and genetic diversity. Chem Biol. 2000;7:263–274. doi: 10.1016/s1074-5521(00)00102-2. [DOI] [PubMed] [Google Scholar]
- 96.Iwai H, Pluckthum A. Circular b-lactamase: stability enhancement by cyclizing the backbone. FEBS Lett. 1999:166–172. doi: 10.1016/s0014-5793(99)01220-x. [DOI] [PubMed] [Google Scholar]
- 97.Cotton GJ, Ayers B, Xu R, Muir TW. Insertion of a Synthetic Peptide into a Recombinant Protein Framework; A Protein Biosensor. J Am Chem Soc. 1999;121:1100–1101. [Google Scholar]
- 98.Camarero JA, Fushman D, Cowburn D, Muir TW. Peptide chemical ligation inside living cells: in vivo generation of a circular protein domain. Bioorg Med Chem. 2001;9:2479–2484. doi: 10.1016/s0968-0896(01)00217-6. [DOI] [PubMed] [Google Scholar]
- 99.Erlandson DA, Chytil M, Verdine GL. The leucine zipper domain controls the orientation of AP-1 in the NFAT AP-1 DNA complex. Chem Biol. 1996;3:981–991. doi: 10.1016/s1074-5521(96)90165-9. [DOI] [PubMed] [Google Scholar]
- 100.Baker RT, Smith SA, Marano R, McKee J, Board PG. Protein expression using cotranslational fusion and cleavage of ubiquitin. Mutagenesis of the glutathione-binding site of human Pi class glutathione S-transferase. J Biol Chem. 1994;269:25381–25386. [PubMed] [Google Scholar]
- 101.Varshavsky A. Ubiquitin fusion technique and related methods. Methods Enzymol. 2005;399:777–799. doi: 10.1016/S0076-6879(05)99051-4. [DOI] [PubMed] [Google Scholar]
- 102.Tolbert TJ, Wong C-H. New methods for proteomic research: preparation of proteins with N-terminal cysteines for labeling and conjugation. Angew Chem Int Ed. 2002;41:2171–2174. doi: 10.1002/1521-3773(20020617)41:12<2171::aid-anie2171>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 103.Liu D, Xu R, Dutta K, Cowburn D. N-terminal cysteinyl proteins can be prepared using thrombin cleavage. FEBS Lett. 2008;582:1163–1167. doi: 10.1016/j.febslet.2008.02.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dalbey RE, Lively MO, Bron S, van Dijl JM. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 1997;6:1129–1138. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Paetzel M, Dalbey RE, Strynadka NC. Crystal structure of a bacterial signal peptidase apoenzyme: implications for signal peptide binding and the Ser-Lys dyad mechanism. J Biol Chem. 2002;277:9512–9519. doi: 10.1074/jbc.M110983200. [DOI] [PubMed] [Google Scholar]
- 106.Hauser PS, Ryan RO. Expressed protein ligation using an N-terminal cysteine containing fragment generated in vivo from a pelB fusion protein. Protein Expr Purif. 2007;54:227–233. doi: 10.1016/j.pep.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Evans TC, Benner J, Xu M-Q. The in Vitro Ligation of Bacterially Expressed Proteins Using an Intein from Metanobacterium thermoautotrophicum. J Biol Chem. 1999;274:3923–3926. doi: 10.1074/jbc.274.7.3923. [DOI] [PubMed] [Google Scholar]
- 108.Southworth MW, Amaya K, Evans TC, Xu MQ, Perler FB. Purification of proteins fused to either the amino or carboxy terminus of the Mycobacterium xenopi gyrase A intein. Biotechniques. 1999;27:110–114. 116, 118–120. doi: 10.2144/99271st04. [DOI] [PubMed] [Google Scholar]
- 109.Mathys S, Evans TC, Chute IC, Wu H, Chong S, Benner J, et al. Characterization of a self-splicing mini-intein and its conversion into autocatalytic N- and C-terminal cleavage elements: facile production of protein building blocks for protein ligation. Gene. 1999;231:1–13. doi: 10.1016/s0378-1119(99)00103-1. [DOI] [PubMed] [Google Scholar]
- 110.Austin J, Kimura RH, Woo YH, Camarero JA. In vivo biosynthesis of an Ala-scan library based on the cyclic peptide SFTI-1. Amino Acids. 2010;38:1313–1322. doi: 10.1007/s00726-009-0338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 112.Mylne JS, Craik DJ. 15N cyclotides by whole plant labeling. Biopolymers. 2008;90:575–580. doi: 10.1002/bip.21012. [DOI] [PubMed] [Google Scholar]
- 113.Colgrave ML, Kotze AC, Huang YH, O’Grady J, Simonsen SM, Craik DJ. Cyclotides: natural, circular plant peptides that possess significant activity against gastrointestinal nematode parasites of sheep. Biochemistry. 2008;47:5581–5589. doi: 10.1021/bi800223y. [DOI] [PubMed] [Google Scholar]
- 114.Colgrave ML, Huang YH, Craik DJ, Kotze AC. Cyclotide interactions with the nematode external surface. Antimicrob Agents Chemother. 2010;54:2160–2166. doi: 10.1128/AAC.01306-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Barbeta BL, Marshall AT, Gillon AD, Craik DJ, Anderson MA. Plant cyclotides disrupt epithelial cells in the midgut of lepidopteran larvae. Proc Natl Acad Sci U S A. 2008;105:1221–1225. doi: 10.1073/pnas.0710338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pranting M, Loov C, Burman R, Goransson U, Andersson DI. The cyclotide cycloviolacin O2 from Viola odorata has potent bactericidal activity against Gram-negative bacteria. J Antimicrob Chemother. 2010;65:1964–1971. doi: 10.1093/jac/dkq220. [DOI] [PubMed] [Google Scholar]
- 117.Henriques ST, Craik DJ. Cyclotides as templates in drug design. Drug Discov Today. 2010;15:57–64. doi: 10.1016/j.drudis.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 118.Chen B, Colgrave ML, Daly NL, Rosengren KJ, Gustafson KR, Craik DJ. Isolation and characterization of novel cyclotides from Viola hederaceae: solution structure and anti-HIV activity of vhl-1, a leaf-specific expressed cyclotide. J Biol Chem. 2005;280:22395–22405. doi: 10.1074/jbc.M501737200. [DOI] [PubMed] [Google Scholar]
- 119.Wang CK, Colgrave ML, Gustafson KR, Ireland DC, Goransson U, Craik DJ. Anti-HIV cyclotides from the Chinese medicinal herb Viola yedoensis. J Nat Prod. 2008;71:47–52. doi: 10.1021/np070393g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ireland DC, Wang CK, Wilson JA, Gustafson KR, Craik DJ. Cyclotides as natural anti-HIV agents. Biopolymers. 2008;90:51–60. doi: 10.1002/bip.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kamimori H, Hall K, Craik DJ, Aguilar MI. Studies on the membrane interactions of the cyclotides kalata B1 and kalata B6 on model membrane systems by surface plasmon resonance. Anal Biochem. 2005;337:149–153. doi: 10.1016/j.ab.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 122.Shenkarev ZO, Nadezhdin KD, Lyukmanova EN, Sobol VA, Skjeldal L, Arseniev AS. Divalent cation coordination and mode of membrane interaction in cyclotides: NMR spatial structure of ternary complex Kalata B7/Mn2+/DPC micelle. J Inorg Biochem. 2008;102:1246–1256. doi: 10.1016/j.jinorgbio.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 123.Shenkarev ZO, Nadezhdin KD, Sobol VA, Sobol AG, Skjeldal L, Arseniev AS. Conformation and mode of membrane interaction in cyclotides. Spatial structure of kalata B1 bound to a dodecylphosphocholine micelle. FEBS J. 2006;273:2658–2672. doi: 10.1111/j.1742-4658.2006.05282.x. [DOI] [PubMed] [Google Scholar]
- 124.Huang YH, Colgrave ML, Daly NL, Keleshian A, Martinac B, Craik DJ. The biological activity of the prototypic cyclotide kalata b1 is modulated by the formation of multimeric pores. J Biol Chem. 2009;284:20699–20707. doi: 10.1074/jbc.M109.003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Svangard E, Goransson U, Hocaoglu Z, Gullbo J, Larsson R, Claeson P, et al. Cytotoxic cyclotides from Viola tricolor. J Nat Prod. 2004;67:144–147. doi: 10.1021/np030101l. [DOI] [PubMed] [Google Scholar]
- 126.Herrmann A, Burman R, Mylne JS, Karlsson G, Gullbo J, Craik DJ, et al. The alpine violet, Viola biflora, is a rich source of cyclotides with potent cytotoxicity. Phytochemistry. 2008;69:939–952. doi: 10.1016/j.phytochem.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 127.Svangard E, Burman R, Gunasekera S, Lovborg H, Gullbo J, Goransson U. Mechanism of action of cytotoxic cyclotides: cycloviolacin O2 disrupts lipid membranes. J Nat Prod. 2007;70:643–647. doi: 10.1021/np070007v. [DOI] [PubMed] [Google Scholar]
- 128.Hoskin DW, Ramamoorthy A. Studies on anticancer activities of antimicrobial peptides. Biochim Biophys Acta. 2008;1778:357–375. doi: 10.1016/j.bbamem.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Herrmann A, Svangard E, Claeson P, Gullbo J, Bohlin L, Goransson U. Key role of glutamic acid for the cytotoxic activity of the cyclotide cycloviolacin O2. Cell Mol Life Sci. 2006;63:235–245. doi: 10.1007/s00018-005-5486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Burman R, Herrmann A, Tran R, Kivela JE, Lomize A, Gullbo J, et al. Cytotoxic potency of small macrocyclic knot proteins: Structure-activity and mechanistic studies of native and chemically modified cyclotides. Org Biomol Chem. 2011;9:4306–4314. doi: 10.1039/c0ob00966k. [DOI] [PubMed] [Google Scholar]
- 131.Chen B, Colgrave ML, Wang C, Craik DJ. Cycloviolacin H4, a hydrophobic cyclotide from Viola hederaceae. J Nat Prod. 2006;69:23–28. doi: 10.1021/np050317i. [DOI] [PubMed] [Google Scholar]
- 132.Simonsen SM, Sando L, Rosengren KJ, Wang CK, Colgrave ML, Daly NL, et al. Alanine scanning mutagenesis of the prototypic cyclotide reveals a cluster of residues essential for bioactivity. J Biol Chem. 2008;283:9805–9813. doi: 10.1074/jbc.M709303200. [DOI] [PubMed] [Google Scholar]
- 133.Cascales L, Henriques ST, Kerr MC, Huang YH, Sweet MJ, Daly NL, et al. Identification and characterization of a new family of cell penetrating peptides: Cyclic cell penetrating peptides. J Biol Chem. 2011 doi: 10.1074/jbc.M111.264424. [DOI] [PMC free article] [PubMed]
- 134.Thongyoo P, Bonomelli C, Leatherbarrow RJ, Tate EW. Potent inhibitors of beta-tryptase and human leukocyte elastase based on the MCoTI-II scaffold. Journal of medicinal chemistry. 2009;52:6197–6200. doi: 10.1021/jm901233u. [DOI] [PubMed] [Google Scholar]
- 135.Sommerhoff CP, Avrutina O, Schmoldt HU, Gabrijelcic-Geiger D, Diederichsen U, Kolmar H. Engineered cystine knot miniproteins as potent inhibitors of human mast cell tryptase beta. J Mol Biol. 2010;395:167–175. doi: 10.1016/j.jmb.2009.10.028. [DOI] [PubMed] [Google Scholar]
- 136.Gialeli C, Theocharis AD, Karamanos NK. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011;278:16–27. doi: 10.1111/j.1742-4658.2010.07919.x. [DOI] [PubMed] [Google Scholar]
- 137.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 138.You X, Nguyen AW, Jabaiah A, Sheff MA, Thorn KS, Daugherty PS. Intracellular protein interaction mapping with FRET hybrids. Proc Natl Acad Sci U S A. 2006;103:18458–18463. doi: 10.1073/pnas.0605422103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tam JP, Yu Q, Miao Z. Orthogonal ligation strategies for peptide and protein. Biopolymers. 1999;51:311–332. doi: 10.1002/(SICI)1097-0282(1999)51:5<311::AID-BIP2>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]