Abstract
Defensins are antimicrobial peptides that are important in the innate immune defense of mammals. In contrast to mammalian α- and β-defensins, rhesus theta defensin-1 (RTD-1) comprises only 18 amino acids stabilized by three disulfide bonds and an unusual backbone cyclic topology. In this work we report for the first time the recombinant expression of the fully folded θ-defensin RTD-1 using a bacterial expression system. This was accomplished using an intramolecular native chemical ligation in combination with a modified protein-splicing unit. RTD-1 was produced either in vitro or in vivo. In-cell production of RTD-1 was estimated to reach an intracellular concentration of ≈ 4 μM. Recombinant RTD-1 was shown to be correctly folded as characterized by heteronucelar-NMR and by its ability to specifically inhibit Lethal Factor protease. The recombinant production of folded θ-defensins opens the possibility to produce peptide libraries based on this peptide scaffold that could be used to develop in-cell screening and directed evolution technologies.
Introduction
Defensins are cysteine-rich antimicrobial peptides that are important in the innate immune defense of mammals.1–3 They are classically known for their antimicrobial activities, but they are also involved in other defense mechanisms including wound healing, immune modulation, neutralization of endotoxin, and anti-cancer activities.3, 4 Mammalian defensins are cationic peptides with largely β-sheet structures and six conserved cysteines. They can be classified into three structurally distinct groups, α-, β- and θ-defensins. The overall fold of α- and β-defensins is quite similar despite differences in disulfide connectivities, and the presence of an N-terminalα-helix segment in β-defensins that is missing in α-defensins.5θ-Defensins, on the other hand, are backbone cyclized peptides formed by the head-to-tail covalent assembly of two nonapeptides derived from α-defensin related precursors,1 and to date, are the only known cyclic polypeptides expressed in animals.1
Rhesus θ-defensin-1 (RTD-1) was the first θ-defensin to be discovered from an extract of leukocytes from Rhesus macaques,1 and in contrast with α- and β-defensins was shown to have a β-hairpin-like structure with two anti-parallel β-strands stabilized by three disulfides in a ladder configuration (Fig. 1A).6 Since then, other less abundant RTD variants, named RTD-2 through RTD-6, were also found in Rhesus macaques.7–9 Circular θ-defensins have also been isolated from other primate species.10–12 Interestingly, humans possess genes encoding θ-defensins, but they have lost the ability to produce the peptides due to a stop codon mutation within the signal sequence that prevents subsequent translation.13
Figure 1.
A. Primary and tertiary structure of rhesus θ-defensin 1 (RTD-1) (PDB ID code: 1HVZ).6 The backbone cyclized peptide (connecting bond shown in blue) is stabilized by the three disulfide bonds in a ladder formation (disulfide bonds shown in yellow). B. Design of the two RTD-intein precursors used in this work, RTD-C3 and RTD-C7. The precursors consist of an RTD-1 based linear peptide attached to the engineered intein (represented by an asterisk) and the chitin binding domain (CBD). An N-terminal Met was added right in front of the required N-terminal Cys residue. The N-terminal Met residue is removed in vivo by endogeneous methionine aminopeptidases (AMPs) producing an N-terminal Cys available for cyclization through intramolecular native chemical ligation. The sequence RRGV is underlined for reference.
θ-Defensins have both Gram-positive and Gram-negative antibacterial activity,1 although this activity strongly depends on the buffer conditions used in the assays.14 For example, the antimicrobial activity of RTD-1 is negatively affected by the presence of 10% human serum 14. θ-Defensins also have anti-fungal1 and anti-HIV13, 15 activities. Chemically-synthesized θ-defensins (called retrocyclins), which are derived from the human pseudogene sequences, have been shown to protect human cells from infection by HIV-113 and have been evaluated as a topical anti-HIV agent for the prevention of HIV transmission,16–18 showing promise when compared to other topical anti-HIV drugs in pre-clinical development.15 It is likely that the ability of θ-defensins to bind gp120 and CD4 glycoproteins is integrally related to its ability to protect cells from HIV-1 infection.19θ-Defensins have also been shown to inactivate germinating anthrax spores and act as a competitive inhibitor of anthrax lethal factor protease.20
Although the precursor genes associated with θ-defensins have been identified,1 the biochemical mechanism responsible for their post-translational biosynthesis has not been elucidated yet. Our group has recently developed a method for the biosynthesis of backbone cyclized peptides using intramolecular Native Chemical Ligation (NCL) in combination with a modified protein-splicing unit or intein.21, 22 This process requires the presence, within the same polypeptide sequence, of an N-terminal Cys residue and a C-terminal α-thioester function.23, 24 We have successfully used this approach for the recombinant production of several naturally occurring backbone cyclized polypeptides using standard bacterial expression systems.25, 26 Encouraged by these results we decided to explore the potential of this approach for the biological production of folded θ-defensins using bacterial expression systems.
Using RTD-1 as a model system, we show here that folded backbone cyclized θ-defensins can be produced recombinantly using bacterial expression systems. Folded RTD-1 can be either produced in vitro or in vivo with similar yields. In-cell production of RTD-1 can reach intracellular concentrations ≈ 4 μM. Recombinant RTD-1 was shown to adopt a native folded structure as determined by heteronuclear NMR and was fully active as shown by inhibition of Anthrax lethal factor protease. Recombinant expression of θ-defensins makes it possible to introduce NMR active isotopes (15N and/or 13C) in a very inexpensive fashion, thus facilitating the use of NMR to study any molecular interaction between θ-defensins and their potential biomolecular targets. These results also open the intriguing possibility for in-cell production of genetically-encoded peptide libraries based on this peptide scaffold that could be used to develop in-cell screening and directed evolution technologies.
Results and Discussion
The recombinant expression of RTD-1 was carried out by using a modified protein splicing unit to assist the intramolecular native chemical ligation (NCL) required for backbone cyclization.23, 27 RTD-1 has six Cys residues that may be used for cyclization. In order to facilitate the cyclization reaction, we decided to use the Cys residues located in positions 3 and 7 (Fig. 1). These Cys residues are the only ones in the sequence of RTD-1 that do not have a charged or β-branched residue N-terminally adjacent, which should facilitate the kinetics of the cyclization reaction without affect the splicing activity of the intein. Accordingly, two different RTD-1 linear precursors (RTD-C3 and RTD-C7) were cloned in frame with a modified Mxe Gyrase intein (Fig. 1B). The N-terminal Met residue in both constructs is efficiently removed by endogenoeus methionine aminopeptidases when expressed in Escherichia coli21, 22 therefore yielding the required N-terminal Cys for intramolecular NCL. At the same time, the modified intein allows the generation of the required α-thioester at the junction between the C-terminal end of RTD-1 and the intein (Fig. 1)
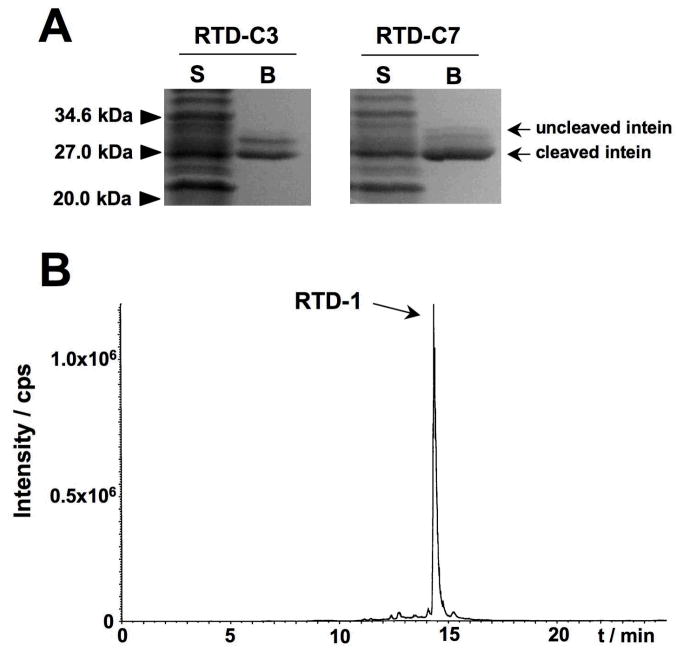
Both RTD-1 intein fusion protein precursors (RTD-C3 and RTD-C7) were expressed in E. coli BL21(DE3) cells at 30°C for 3 h, and purified by affinity chromatography using chitin-sepharose beads. The RTD-intein precursors have a chitin binding domain (CBD) fused at the C-terminus of the intein domain to facilitate purification. As shown in Fig. 2A, both intein precursors had comparable levels of expression in E. coli cells (≈ 5 mg/L as estimated by UV spectroscopy), and showed similar rates and propensities for in-vivo cleavage (≈ 65%). It is worth noting that under these conditions most of the intein fusion precursors in both cases were expressed as soluble proteins (Fig. 2A).
Figure 2.
In vitro production of RTD-1. A. SDS-PAGE analysis of cell lysates from BL21 E. coli cells expressing precursors RTD-C3 and RTD-C7. The identity of the bands corresponding to intein precursors and in vivo cleaved proteins are shown on the right. The positions and molecular weights of the molecular markers used are shown on the left of the gel (P: insoluble cell lysate, S: soluble cell lysate, B: purified intein, B/GSH: purified intein after cleavage with 100 mM glutathione at pH 7.2). B. Reverse-phase C18-HPLC traces for the GSH-induced cyclization folding of purified precursors RTD-C3 and RTD-C7. The peak corresponding to the folded RTD-1 is indicated with an arrow in each case. The asterisk denotes the cleaved intein-CBD fragment. C. Reverse-phase C18-HPLC of purified RTD-1. D. ES-MS spectrum of purified RTD-1. Calculated mass corresponds to the average isotopic mass.
We next tested the ability of the different precursors to be cleaved in vitro by using reduced glutathione (GSH) to produce folded RTD-1. GSH has been shown to promote cyclization and concomitant folding when used in the biosynthesis of Cys-rich cyclic polypeptides.21, 25, 26 The cyclization/folding reaction was performed on the chitin beads where the corresponding precursors had been purified. The best cleavage/cyclization conditions were accomplished using 100 mM of GSH in phosphate buffer at pH 7.2 for 48 h at room temperature. Under these conditions both RTD-intein precursors were completely cleaved (Fig. 2A). HPLC analysis of the crude cyclization mixture revealed that in both cases the main peptide product was the corresponding folded RTD-1 (Fig. 2B) as revealed by HPLC and ES-MS analysis (Figs. 2C and 2D). Other peptide minor peaks in the HPLC chromatograms were identified as not correctly folded GSH-adducts. Precursor RTD-C3 gave the best cyclization/folding yield producing around 10 μg/L of purified RTD-1. This yield corresponds to ≈10% of the theoretical value. The in vitro cyclization/folding of precursor RTD-C7 gave a slightly lower yield (≈7 μg/L of purified RTD-1). The lower yield observed for this precursor can be attributed to the slightly more complex cyclization/folding crude (Fig. 2B)
Recombinant RTD-1 was purified by HPLC and its biological activity tested using an Anthrax lethal factor (LF) protease inhibitory assay.28 In vitro produced RTD-1 was able to inhibit anthrax LF with a IC50 of 384 ± 33 nM (Fig. 3A), which corresponds to a Ki ≈ 0.4 μM under the conditions used in the inhibitory assay.36,37 This IC50 value closely parallels previously reported values,20, 29 thus confirming the biological activity of the recombinantly produced θ-defensin.
Figure 3.
Characterization of recombinantly produced θ-defensin RTD-1. A. Inhibition assay of RTD-1 against Anthrax lethal factor (LF). Different concentrations of RTD-1 were tested against LF. At each concentration, residual LF activity was measured and divided by the activity of LF in the absence of inhibitor. Activity was measured as the rate LF protease cleaves a fluorescence LF substrate28 and determined by the rate of fluorescence signal decaying (see experimental). B. 15N-HSQC spectra of 15N-labeled RTD-1 defensin in water at pH 6.0. The identity of the crosspeaks is indicated by the number of the residue according to Figure 1.
We also used NMR spectroscopy to confirm that recombinant RTD-1 adopted a native θ-defensin fold. The structure of native RTD-1 has been previously reported by Craik in 10% MeCN aqueous buffer at pH 4.5.6 We decided to carry out the NMR experiments in more physiological conditions using an aqueous buffer containing no organic solvents at pH 6.5. The rationale to use more physiological conditions was to allow the future study of biologically relevant interactions between RTD-1 and potential biomolecular targets by NMR. The chemical shifts of the assigned backbone amide and alpha protons (HNα and HCα) for recombinant RTD-1 at pH 6.5 were very similar to those reported earlier by Craik at pH 4.5 (Table S2). No significant (≥0.3 ppm) differences were found in the backbone amide protons. We also saw a uniform shift rather than variable changes for the backbone alpha protons (≈0.2 ppm) at pH 6.0 (Table S2). Since backbone alpha protons usually reflect the secondary structure of the peptide backbone, this uniform offset in the resonances of the backbone HCα protons could be attributed to the different buffer conditions used in the two samples rather than changes in secondary structure.
We also produced recombinant 15N-labeled RTD-1 by expressing RTD-C3 precursor in minimal M9 medium containing 15NH4Cl as the only source of nitrogen. Under these conditions the expression yield was around 7 μg/L of 15N-labeled RTD-1 after purification by HPLC. The HSQC spectra of recombinant RTD-1 was very well dispersed, indicating a well-folded structure (Fig. 3B). Having access to the recombinant expression of θ-defensins allows the introduction of NMR active isotopes (15N and/or 13C) in a very inexpensive fashion, thus facilitating the use of heteronuclear NMR to study intermolecular interactions between θ-defensins and their biomolecular targets.
Encouraged by the results obtained with the in vitro GSH-induced cyclization/folding of the RTD-intein precursors we also decided to explore the expression of folded RTD-1 inside E. coli cells. RTD-1 has been shown to be antimicrobial against both Gram-positive and Gram-negative bacteria. However, the antimicrobial activity of RTD-1 has been shown to strongly depend on the conditions used in the antimicrobial assays. The presence of 10% human serum has been shown to significantly decrease the antimicrobial properties of RTD-1 especially against Gram-negative bacteria such as E. coli 14. We anticipated that the high molecular complexity of the bacterial cytosol could decrease the antimicrobial activity of RTD-1 when produced intracellularly and therefore allow its production in the cellular cytosol.
In order to test this hypothesis we used the RTD-C7 precursor (Fig. 1B). In-cell expression of RTD-1 was accomplished in Origami2(DE3). These cells have mutations in the thioredoxin and glutathione reductase genes, which facilitates the formation of disulfide bonds in the bacterial cytosol.30 We have recently used these cells for the in vivo production of several disulfide-containing backbone cyclized polypeptides.22, 25, 26
Precursor RTD-C7 was expressed in Origami2(DE3) overnight at room temperature giving a total yield of precursor protein of ≈3 mg/L. Under these conditions the precursor was completely cleaved in vivo (Fig. 4A). In contrast when precursor RTD-C3 was expressed under these conditions only ≈74% of the precursor protein was cleaved in vivo (Fig. 4A). This difference could be attributed to the proximity of Cys5 to the RTD-intein junction in the precursor RTD-C7, which is only one residue away from the RTD-intein junction therefore facilitating the intramolecular cleavage. In the precursor RTD-C3, the closest Cys residue within the sequence (Cys16) is four residues away from the RTD-intein junction (Fig. 1). After lysing the cells expressing RTD-C7, both the insoluble and soluble cellular fractions were analyzed by HPLC-MS/MS in multiple reaction mode to identify and quantify the amount of folded RTD-1. Interestingly, almost all of the folded RTD-1 produced inside living E. coli cells was found in the insoluble fraction (≈22 μg/L, Fig. 4B). θ-Defensins are known to interact with model phospholipid membranes, and have been shown to have low nM affinity for glyco-proteins (gp120 and cd4) and glyco-lipids (galactosylceramide).19 This could explain the higher affinity of recombinant RTD-1 for the insoluble cellular lysate fraction, where it is likely to find insoluble membrane fragments containing glyco-lipids and/or membrane-bound peptidoglycan precursors. In fact, peptidoglycan precursor and membrane-anchored lipid II has been shown to bind to human α-and β-defensins.31 We are now also studying the potential interaction between several peptidoglycan precursors and θ-defensins.
Figure 4.
In vivo production of RTD-1 in E. coli cells. A. SDS-PAGE analysis of the soluble cell lysate fraction (S) of Origami E. coli cells expressing precursors RTD-C3 and RTD-C7. Expression was induced as described in the text at room temperature for 18 h (S: soluble cell lysate and B: purified intein). B. HPLC-MS/MS analysis in multiple reaction mode of in vivo produced RTD-1 in the insoluble fraction of the cell lysate. Specific MRM for RTD-1 identified ≈22 μg/L of folded RTD-1.
In summary, we report in this work the first recombinant expression of the θ-defensin RTD-1 in E. coli cells. RTD-1 can be produced either in vitro, by processing of the corresponding RTD-intein precursor, or in vivo by using longer expression times in combination with Origami or similar E. coli cells. The yield of purified RTD-1 was greater in vivo than in vitro (≈ 22 μg vs. ≈ 10 μg of purified peptide/L of bacterial culture). Although the yields are modest, our approach represents an improvement over the approach reported by Suga for the ribosomal synthesis of RTD-1 using a cell-free expression system in combination with genetic code reprogramming.29 The bacterial expression yields could be easily improved by using richer media and/or fermentors to allow expression at higher cellular densities. The bacterial expression of RTD-1 allows the incorporation of NMR-active isotopes in a very inexpensive manner to facilitate the structural study of the interactions between θ-defensin and potential biomolecular targets through heteronuclear NMR. For example, we are using 15N-labeled RTD-1 to study the potential interaction of this defensin to a soluble form of lipid II. More interestingly, in-cell expression of RTD-1 gave a yield of ≈22 μg/L, which corresponds to an intracellular concentration of ≈4 μM. This result opens the intriguing possibility for the generation of genetically-encoded libraries using the θ-defensin scaffold. These libraries could be used for the development of in-cell screening and directed evolution technologies to further study the biological activity of θ-defensins or to engineer new biological activities on the defensin scaffold.
Experimental
Analytical reverse phase (RP)-HPLC was performed on a HP1100 series instrument with 220 nm and 280 nm detection using a Vydac C18 column (5 mm, 4.6 × 150 mm) at a flow rate of 1 mL/min. Semipreparative RP-HPLC was performed on a Waters Delta Prep system fitted with a Waters 2487 Ultraviolet-Visible (UV-vis) detector using a Vydac C18 column (5 μm, 10 × 250 mm) at a flow rate of 5 mL/min. All runs used linear gradients of 0.1% aqueous trifluoroacetic acid (TFA, solvent A) vs. 0.1% TFA, 90% MeCN in H2O (solvent B). UV-vis spectroscopy was carried out on an Agilent 8453 diode array spectrophotometer. Electro-spray mass spectrometry (ES-MS) was performed on an Applied Biosystems API 3000 triple quadrupole mass spectrometer using Analyst 1.4.2. Calculated masses were obtained using Analyst 1.4.2. Protein samples were analyzed by SDS-PAGE 4–20% Tris-Glycine Gels (Lonza, Rockland, ME). The gels were then stained with Pierce (Rockford, IL) Gelcode Blue, photographed/digitized using a Kodak (Rochester, NY) EDAS 290, and quantified using NIH Image-J software (http://rsb.info.nih.gov/ij/). The integrity of all plasmids was confirmed by DNA sequencing. DNA sequencing was performed by the DNA Sequencing and Genetic Analysis Core Facility at the University of Southern California using an ABI 3730 DNA sequencer, and the sequence data was analyzed with DNAStar (Madison, WI) Lasergene v8.0.2. Amino acid analysis was performed at the Amino Acid Laboratory in the Department of Molecular Biosciences, School of Veterinary Medicine, University of California at Davis. All chemicals were obtained from Sigma-Aldrich (Milwaukee, WI) unless otherwise indicated.
Cloning of RTD-C3 and RTD-C7 and in vitro production of RTD-1
Synthetic DNA oligos (Integrated DNA technologies, Coralville, IA) encoding RTD-1 (Table S1) were annealed and ligated into the pTXB1 vector (New England Biolabs, Ipswich, MA) using the NdeI and SapI restriction sites as described previously.21, 22 The resulting plasmids were transformed into either BL21(DE3) or Origami2(DE3) cells (EMD Chemicals, Gibbstown, NJ) and grown in LB broth. Transformed BL21(DE3) cells were induced with 0.3 mM IPTG for 3 h at 30 °C and transformed Origami2(DE3) cells with 0.1 mM IPTG for 20 h at 22 °C. Cells were lysed in 0.1 mM EDTA, 1 mM PMSF, 50 mM sodium phosphate, 250 mM sodium chloride buffer at pH 7.2 containing 5% glycerol by sonication. The soluble fraction was incubated with chitin beads (New England Biolabs) for 1 h at 4°C and the beads were washed with column buffer (0.1 mM EDTA, 50 mM sodium phosphate, 250 mM sodium chloride buffer at pH 7.2) containing 0.1% Triton X-100 followed by washes with column buffer without Triton X-100. The peptide was cyclized and folded in vitro using column buffer at pH 7.2 containing 100 mM reduced glutathione (GSH) for 2–3 days at room temperature with gentle rocking. We found that under these conditions RTD-1 binds strongly to the chitin column, and therefore was eluted using 8 M GdmCl in water. The corresponding supernatant and washes were pooled, and RTD-1 was purified by semipreparative HPLC using a linear gradient of 17–39% solvent B over 30 min. Purified products were characterized by HPLC and ES-MS. RTD-1 was quantified by amino acid analysis. The expression of RTD-intein fusion precursors were quantified by first desorption of the proteins from an aliquot of chitin beads using 8 M GdmCl and then measurement by UV-VIS using a molar absorption coefficient value of 36,660 M−1 cm−1.
Expression of 15N-labeled RTD-1
Expression was carried out using BL21(DE3) cells as described above except grown in M9 minimal medium containing 0.1% 15NH4Cl as the nitrogen source.25, 32 Cyclization and folding was performed as described above. The 15N-labeled RTD-1 was purified by semipreparative HPLC as before. Purified products were characterized by HPLC and ES-MS (Fig. S1).
In-cell expression of RTD-1
Origami2(DE3) cells transformed with the plasmid encoding the RTD-C7 precursor were grown, induced, harvested and lysed as described above. The insoluble pellet was first washed three times with column buffer containing 0.1% Triton X-100 and then twice with just column buffer. The resulting pellet was dissolved in minimal amount of 8 M GdmCl in water. Both the soluble cell lysate and solubilized cell lysate pellet were extracted using C18 SepPak cartridges (Waters, Milford, MA) with elution in MeCN:H2O (3:2 vol.) containing 0.1% TFA and lyophilized. The samples were dissolved in H2O containing 0.1% formic acid and analyzed by HPLC-tandem mass spectrometry using a C18-HPLC column (5 mm, 2.1 × 100 mm), and H2O-MeCN buffers containing 0.1% formic acid as mobile phase. Typical analysis used a linear gradient of 0%-90% MeCN in H2O over 10 min. Detection was performed on an API 3000 ES-MS using a multiple reaction-monitoring (MRM) mode. Data were collected and processed using Analyst software (Applied Biosystems). The calibration curve using pure RTD-1 was found to be linear in the range of 10–40 ng. Loss of RTD-1 during extraction was quantified by spiking a control sample with a known amount of purified RTD-1 and analysis by HPLC-MS/MS. The recovery was found to be approximately 45%.
NMR spectroscopy
NMR samples were prepared by dissolving 15N-labeled backbone cyclized RTD-1 into 80 mM sodium phosphate buffer at pH 6.0 in 90% H2O/10% 2H2O (v/v) or 100% D2O to a concentration of approximately 0.2 mM. All 1H NMR data were recorded on a Bruker Avance II 500 MHz spectrometer equipped with a cryoprobe. Data were acquired at 27 °C, and 2,2-dimethyl-2-silapentane-5-sulfonate, DSS, was used as an internal reference. All All 2D 1H{1H}-TOCSY and 1H{1H}-NOESY and 3D experiments, 1H{15N}-TOCSY-HSQC and 1H{15N}-NOESY-HSQC, were performed according to standard procedures33 with spectral widths of 14 ppm in proton dimensions and 35 ppm in nitrogen dimension. The carrier frequency was centered on the water signal, and the solvent was suppressed by using WATERGATE pulse sequence. TOCSY (spin lock time 80 ms) and 2D-NOESY (mixing time 250 ms) spectra were collected using 4096 t2 points and 512 t1 blocks of 64 transients. 3D-TOCSY-HSQC (spin lock time 80 ms) and 3D-NOESY-HSQC (mixing time 250 ms) spectra were collected using 1024 t3 points, 256 t2 and 128 t1 blocks of 16 transients. Spectra were processed using Topspin 1.3 (Bruker). Each 3D data set was apodized by 90°-shifted sinebell-squared in all dimensions, and zero filled to 1024 × 512 × 256 points prior to Fourier transformation. Assignments for the backbone nitrogens, Hα and H′ protons (Tables S2) were obtained using standard procedures.33,34
Anthrax Lethal Factor protease inhibition assay
Lethal factor (LF) protease and FRET-based substrate containing fluorescent proteins CyPet and YPet linked by consensus sequence (RRKKVYPYPMEGTIA) were expressed and purified as previously described.28, 35 RTD-1 concentrations were quantified by amino acid analysis. LF inhibition assay was performed in LF reaction buffer (10 μM CaCl2, 10 μM MgCl2, 20 μM ZnCl2, 20 mM sodium phosphate, and 100 mM NaCl at pH 7.2). Samples of 50 nM LF in LF reaction buffer (100 μL) were preincubated with different concentrations of RTD-1 ranging frm 1 nM to 10 μM. After incubation at room temperature for 30 min, residual LF activity was measured by adding the FRET-based substrate to a final concentration of 10 nM and the decrease in FRET signal was observed.28 Measurements of FRET signal were taken every 2 min for 3 h. FRET was measured using an Envision 2103 plate reader (PerkinElmer) using an excitation wavelength of 405 nm. The relative FRET change was calculated using: FRC = It535/I0535, where I0 and It are the fluorescence intensities at time zero and at a particular time (t), at 535 nm. The initial velocities for the hydrolysis of substrate Lethal Factor in the presence of different concentrations of RTD-1 were fitted to a one-site competitive binding equation using the software package Prism (GraphPad Software). Ki was calculated using the equation of Cheung and Prusoff36 and a Km value of 40 μM.37
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Research Grant R01-GM090323 (JAC) and NIH Grant 5R01GM085006-02 (AS); and by the Department of Defense Congressionally Directed Medical Research Program Grant PC09305 (JAC).
Notes and References
- 1.Tang YQ, Yuan J, Osapay G, Osapay K, Tran D, Miller CJ, Ouellette AJ, Selsted ME. Science. 1999;286:498–502. doi: 10.1126/science.286.5439.498. [DOI] [PubMed] [Google Scholar]
- 2.Tanabe H, Yuan J, Zaragoza MM, Dandekar S, Henschen-Edman A, Selsted ME, Ouellette AJ. Infect Immun. 2004;72:1470–1478. doi: 10.1128/IAI.72.3.1470-1478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selsted ME, Ouellette AJ. Nat Immunol. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 4.Lehrer RI. Nature reviews. 2004;2:727–738. doi: 10.1038/nrmicro976. [DOI] [PubMed] [Google Scholar]
- 5.Rosengren KJ, Daly NL, Fornander LM, Jonsson LM, Shirafuji Y, Qu X, Vogel HJ, Ouellette AJ, Craik DJ. J Biol Chem. 2006;281:28068–28078. doi: 10.1074/jbc.M604992200. [DOI] [PubMed] [Google Scholar]
- 6.Trabi M, Schirra HJ, Craik DJ. Biochemistry. 2001;40:4211–4221. doi: 10.1021/bi002028t. [DOI] [PubMed] [Google Scholar]
- 7.Leonova L, Kokryakov VN, Aleshina G, Hong T, Nguyen T, Zhao C, Waring AJ, Lehrer RI. J Leukoc Biol. 2001;70:461–464. [PubMed] [Google Scholar]
- 8.Tran D, Tran PA, Tang YQ, Yuan J, Cole T, Selsted ME. J Biol Chem. 2002;277:3079–3084. doi: 10.1074/jbc.M109117200. [DOI] [PubMed] [Google Scholar]
- 9.Tongaonkar P, Tran P, Roberts K, Schaal J, Osapay G, Tran D, Ouellette AJ, Selsted ME. J Leukoc Biol. 2011;89:283–290. doi: 10.1189/jlb.0910535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen TX, Cole AM, Lehrer RI. Peptides. 2003;24:1647–1654. doi: 10.1016/j.peptides.2003.07.023. [DOI] [PubMed] [Google Scholar]
- 11.Garcia AE, Osapay G, Tran PA, Yuan J, Selsted ME. Infect Immun. 2008;76:5883–5891. doi: 10.1128/IAI.01100-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stegemann C, Tsvetkova EV, Aleshina GM, Lehrer RI, Kokryakov VN, Hoffmann R. Rapid Commun Mass Spectrom. 2010;24:599–604. doi: 10.1002/rcm.4424. [DOI] [PubMed] [Google Scholar]
- 13.Cole AM, Hong T, Boo LM, Nguyen T, Zhao C, Bristol G, Zack JA, Waring AJ, Yang OO, Lehrer RI. Proc Natl Acad Sci U S A. 2002;99:1813–1818. doi: 10.1073/pnas.052706399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran D, Tran P, Roberts K, Osapay G, Schaal J, Ouellette A, Selsted ME. Antimicrob Agents Chemother. 2008;52:944–953. doi: 10.1128/AAC.01090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penberthy WT, Chari S, Cole AL, Cole AM. Cell Mol Life Sci. 2011;68:2231–2242. doi: 10.1007/s00018-011-0715-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munk C, Wei G, Yang OO, Waring AJ, Wang W, Hong T, Lehrer RI, Landau NR, Cole AM. AIDS Res Hum Retroviruses. 2003;19:875–881. doi: 10.1089/088922203322493049. [DOI] [PubMed] [Google Scholar]
- 17.Owen SM, Rudolph D, Wang W, Cole AM, Sherman MA, Waring AJ, Lehrer RI, Lal RB. J Pept Res. 2004;63:469–476. doi: 10.1111/j.1399-3011.2004.00155.x. [DOI] [PubMed] [Google Scholar]
- 18.Gallo SA, Wang W, Rawat SS, Jung G, Waring AJ, Cole AM, Lu H, Yan X, Daly NL, Craik DJ, Jiang S, Lehrer RI, Blumenthal R. J Biol Chem. 2006;281:18787–18792. doi: 10.1074/jbc.M602422200. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, Cole AM, Hong T, Waring AJ, Lehrer RI. J Immunol. 2003;170:4708–4716. doi: 10.4049/jimmunol.170.9.4708. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, Mulakala C, Ward SC, Jung G, Luong H, Pham D, Waring AJ, Kaznessis Y, Lu W, Bradley KA, Lehrer RI. J Biol Chem. 2006;281:32755–32764. doi: 10.1074/jbc.M603614200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura RH, Tran AT, Camarero JA. Angew Chem Int Ed. 2006;45:973–976. doi: 10.1002/anie.200503882. [DOI] [PubMed] [Google Scholar]
- 22.Camarero JA, Kimura RH, Woo YH, Shekhtman A, Cantor J. Chembiochem. 2007;8:1363–1366. doi: 10.1002/cbic.200700183. [DOI] [PubMed] [Google Scholar]
- 23.Camarero JA, Muir TW. J Am Chem Soc. 1999;121:5597–5598. [Google Scholar]
- 24.Camarero JA, Fushman D, Cowburn D, Muir TW. Bioorg Med Chem. 2001;9:2479–2484. doi: 10.1016/s0968-0896(01)00217-6. [DOI] [PubMed] [Google Scholar]
- 25.Austin J, Wang W, Puttamadappa S, Shekhtman A, Camarero JA. Chembiochem. 2009;10:2663–2670. doi: 10.1002/cbic.200900534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Austin J, Kimura RH, Woo YH, Camarero JA. Amino Acids. 2010;38:1313–1322. doi: 10.1007/s00726-009-0338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camarero JA, Pavel J, Muir TW. Angew Chem Int Ed. 1998;37:347–349. doi: 10.1002/(SICI)1521-3773(19980216)37:3<347::AID-ANIE347>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 28.Kimura RH, Steenblock ER, Camarero JA. Anal Biochem. 2007;369:60–70. doi: 10.1016/j.ab.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 29.Kawakami T, Ohta A, Ohuchi M, Ashigai H, Murakami H, Suga H. Nat Chem Biol. 2009;5:888–890. doi: 10.1038/nchembio.259. [DOI] [PubMed] [Google Scholar]
- 30.Bessette PH, Aslund F, Beckwith J, Georgiou G. Proc Natl Acad Sci U S A. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilmes M, Cammue BP, Sahl HG, Thevissen K. Nat Prod Rep. 2011 doi: 10.1039/c1np00022e. [DOI] [PubMed] [Google Scholar]
- 32.Puttamadappa SS, Jagadish K, Shekhtman A, Camarero JA. Angew Chem Int Ed Engl. 2010;49:7030–7034. doi: 10.1002/anie.201002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavanagh J, Rance M. J Magn Res. 1992;96:670–678. [Google Scholar]
- 34.Wuthrich K. NMR of Proteins and Nucleic Acids. 1986. [Google Scholar]
- 35.Kwon Y, Welsh K, Mitchell AR, Camarero JA. Org Lett. 2004;6:3801–3804. doi: 10.1021/ol048417n. [DOI] [PubMed] [Google Scholar]
- 36.Cheng Y, Prusoff WH. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 37.Tonello F, Ascenzi P, Montecucco C. J Biol Chem. 2003;278:40075–40078. doi: 10.1074/jbc.M306466200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.