Abstract
Context
TSPY1 is a tandemly-repeated gene on the human Y chromosome forming an array of approximately 21–35 copies. The testicular expression pattern and the inferred function of the TSPY1 protein suggest possible involvement in spermatogenesis. However, data are scarce on TSPY1 copy number variation in different Y lineages and its role in spermatogenesis.
Objectives
We sought to define: 1) the extent of TSPY1 copy number variation within and among Y chromosome haplogroups; and 2) the role of TSPY1 dosage in spermatogenic efficiency.
Materials and Methods
A total of 154 idiopathic infertile men and 130 normozoospermic controls from Central Italy were analyzed. We used a quantitative PCR assay to measure TSPY1 copy number and also defined Y haplogroups in all subjects.
Results
We provide evidence that TSPY1 copy number shows substantial variation among Y haplogroups and thus that population stratification does represent a potential bias in case-control association studies. We also found: 1) a significant positive correlation between TSPY1 copy number and sperm count (P < 0.001); 2) a significant difference in mean TSPY1 copy number between patients and controls (28.4 ± 8.3 vs. 33.9 ± 10.7; P < 0.001); and 3) a 1.5-fold increased risk of abnormal sperm parameters in men with less than 33 copies (P < 0.001).
Conclusions
TSPY copy number variation significantly influences spermatogenic efficiency. Low TSPY1 copy number is a new risk factor for male infertility with potential clinical consequences.
The human testis-specific protein Y-encoded 1 (TSPY1) gene was originally identified as a gene on the short arm of the human Y chromosome with a testis-specific expression pattern (1, 2). The majority of TSPY1 copies are arranged in 20.4 kb tandemly repeated units, each containing one copy of TSPY1 and one of the CYorf16 pseudogene transcription unit, forming an approximately 400- to 800-kb cluster (DYZ5) corresponding to approximately 20–40 gene copies on proximal Yp (3, 4). Such a structure is highly unusual in the human genome: only 12 protein-coding genes are present in tandem clusters with more than three copies, and none of the others shows more than 16 copies in the current assembly (5). The TSPY1 cluster lies within the critical region for the gonadoblastoma locus on the Y chromosome (GBY) that predisposes dysgenetic gonads of intersex patients to gonadoblastoma development (6, 7), but the biological role of TSPY1 is otherwise poorly understood.
TSPY1 is a member of a protein superfamily that includes SET and NAP, which are activating factors of the replication process, as binding-partners of cyclin B (8, 9). Various TSPY1 isoforms are generated by alternative splicing events of transcripts originating from the same transcriptional units (10, 11). Full-length TSPY1 is expressed in the normal testis predominantly in spermatogonia and at a low level in primary spermatocytes, indicating a major role in the mitotic division of early germ cells (9). The role of the minor TSPY1 transcripts, which seem to be preferentially expressed in round spermatids, remains to be elucidated (12). Thus, the expression pattern of full-length TSPY1 suggests a potential role in male germ cell development as a proliferation factor during spermatogenesis (2, 9).
The number of TSPY1 gene copies observed in the two main studies ranged from 18 to 47 and from 23 to 64 copies, respectively (13, 14); this variation is likely to be generated by unequal sister chromatid exchange (8, 15). Maintenance of a minimum copy number through selection is suggested by the evolutionary conservation of multiple copies of the gene on the Y chromosomes of other mammals (16, 17) and by limited variation in copy number in humans, with pronounced modes and few excursions to low or high numbers of copies (18). The nature of the putative selective force remains entirely unknown but seems most likely to be related to spermatogenesis.
Thus far, only one study has addressed the question of whether or not TSPY1 copy number variation may influence spermatogenesis and reported an association between increased TSPY1 copy number and infertility (19). The interpretation of these results, however, is difficult for two reasons: 1) the quantification method employed was unable to provide an exact TSPY1 copy number because it was not validated against a gold standard method (e.g. pulsed-field gel electrophoresis); and 2) the size of the study population was small, and Y haplogroup (hg) matching was performed only in a minority of cases, so differences might reflect hg stratification.
We have now reinvestigated the role of TSPY1 dosage in spermatogenic efficiency by setting up a validated method able to provide an absolute number of TSPY1 copies and applying it to a large study population. For this purpose we compared TSPY1 copy number between men with normozoospermia (controls) and a heterogeneous group of idiopathic infertile men (cases) with different grades of spermatogenic failure. We controlled for Y lineage effects by determining the hg of participants and thus also provide the largest analysis to date of TSPY1 copy number variation in individuals belonging to six different Y lineages.
Subjects and Methods
Subjects
The study population consisted of a total of 284 subjects: 154 idiopathic infertile patients, and 130 normozoospermic men. The study was approved by the local ethics committee. Infertile patients included in the study were seeking complete andrological diagnostic work-up for couple infertility at the Andrology Unit and the Unit of Physiopathology of Reproduction of the University Hospital Careggi (Florence, Italy). Infertile patients were selected on the basis of a comprehensive andrological examination including medical history, semen analysis, scrotal ultrasound, hormone analysis, karyotype and Y chromosome microdeletion screening. Patients with mono- or bilateral cryptorchidism, varicocele grades 2 and 3, obstructive azoospermia, recurrent infections, iatrogenic infertility, hypogonadotrophic hypogonadism, karyotype anomalies, Y chromosome microdeletions including partial deletions of the AZFc region, and partial AZFc duplications and patients with non-Italian origin were excluded.
According to the three major sperm parameters, the infertile group could be divided as follows: azoospermia in 36 patients; cryptozoospermia (<1 million spermatozoa/ml) in 13 patients; severe oligozoospermia (1–5 million spermatozoa/ml) in 55 patients; moderate oligozoospermia (5–20 million spermatozoa/ ml) in 36 patients; astheno and/or teratozoospermia in 14 patients. The mean sperm concentrations in patients and controls were 4.0 ± 4.7 × 106 and 83.9 ± 53.6 × 106 sperm/ml, respectively. The mean total sperm numbers in patients and controls were 14.5 ± 19.8 × 106 and 271.5 ± 163.0 × 106 spermatozoa, respectively. Controls were selected on the basis of normal sperm parameters (sperm count, motility, and morphology) defined according to the World Health Organization criteria (20). Samples were collected using approved protocols, and the informed consent of all individuals was obtained.
Estimation of TSPY1 copy number
Relative TSPY1 copy number was determined by quantitative PCR, using a region of the single copy PMP22 gene as a control locus as described (21). Absolute numbers of TSPY1 genes were estimated by reference to DNA samples in which copy number was known from size measurement of the hybridizing XbaI fragment in pulsed-field gel analysis (13, 22).
Detection of the AMELY gene deletion
All patients and controls were screened for AMELY gene deletion by PCR amplification of specific Sequence Tagged Sites: sY70 (GenBank accession no. G66517) and sY276 (GenBank accession no. G38362). The amplified products were run on an agarose gel, and all showed the presence of both fragments, indicating that AMELY was present.
Y hg definition
To exclude recruitment bias, care was taken in the ethnic and geographic matching of the patients and controls. All patients and controls were asked for their paternal and maternal origin and were included only if they had Central Italian ancestry on both sides. The subjects were genotyped for six binary markers defining eight hgs (including paragroups): hgs A, DE, J, K*(xN, P), N, P*(xR1a), R1a, and the remaining Y*(xA, D, E, J, K) chromosomes. Y chromosome haplotyping was performed as previously described for the YAP, M9, SRY-1532, 92R7, LLY22 g, and 12f2 polymorphisms (23). Polymorphisms were visualized by size or presence/absence of fragments (YAP, 12f2) or restriction enzyme digestion pattern for: M9 (HinfI), SRY-1532 (DraIII), 92R7 and LLY22 g (HindIII). Further analysis showed that most P*(xR1a) chromosomes fell into R1b1b (M269-derived), but the P*(xR1a) classification is used here because M269 data are not available for all samples.
Statistical analysis
Statistical analysis was performed using the statistical package SPSS for Windows (version 15.0.1; SPSS Inc., Chicago, IL). Median and mean values between groups were compared using a nonparametric Mann-Whitney U test and Student’s t test (in case of normal distributions), respectively. Correlation between TSPY1 copy number and sperm count was ascertained by Spearman’s correlation test. The cutoff value of TSPY1 copy number for the comparison of different subgroups was defined on the basis of receiver operating characteristic curve analysis. Differences in the incidence of subjects with TSPY1 copy number above and below the selected cutoff between patients and controls were evaluated using the Fisher’s exact test. A P value of 0.05 was considered statistically significant for each test.
Results
Y hg distribution in the study population and sperm count in men belonging to distinct hgs
We performed Y hg analysis in the entire study population. Patients and controls showed a similar Y hg distribution with the exception of the low-frequency hg K*(xN, P) (Table 1). Hgs A and N were not found in our study population at all, whereas hg R1a was found only in one subject, who was excluded from subsequent analyses.
TABLE 1.
Sperm characteristics (expressed as mean ± SD of sperm concentration and total sperm number) of patients and controls stratified by Y-chromosome hg
| Patients (n = 154) |
Controls (n = 130) |
|||||
|---|---|---|---|---|---|---|
| Y hg | n (%) | Sperm concentration (n × 106/ml) |
Total sperm no. (n × 106) |
n (%) | Sperm concentration (n × 106/ml) |
Total sperm no. (n × 106) |
| Y*(xA, D, E, J, K) | 37 (24) | 3.2 ± 3.9 | 11.0 ± 15.0 | 31 (23.9) | 87.1 ± 64.8 | 257.7 ± 182.5 |
| P*(xR1a) | 76 (49.4) | 4.3 ± 5.3 | 16.1 ± 22.7 | 64 (49.2) | 83.5 ± 53.7 | 270.0 ± 160.9 |
| J | 20 (13) | 3.7 ± 4.7 | 13.7 ± 15.8 | 12 (9.2) | 97.8 ± 56.3 | 312.9 ± 196.5 |
| DE | 19 (12.3) | 4.3 ± 3.7 | 16.3 ± 20.7 | 16 (12.3) | 84.7 ± 30.1 | 298.6 ± 135.9 |
| K*(xN,P) | 1 (0.65) | 2.9 | 17.7 | 7 (5,4) | 47.0 ± 16.6 | 213.9 ± 72.5 |
| R1a | 1 (0.65) | 0.5 | 1.7 | 0 (0) | ||
The mean sperm concentration of normozoospermic men of hg K*(xN, P) was significantly lower than of those belonging to hgs Y* and DE (P = 0.04 and P = 0.001, respectively). However, when total sperm counts were compared, no significant differences were found between subjects belonging to these Y hgs.
TSPY1 copy number variation and Y hgs
The number of TSPY1 copies varied substantially within each hg (Table 2). Although the variance within different hgs was similar, the mean TSPY1 copy number differed significantly between the following Y lineages: 1) hg P*(xR1a) vs. hg J (29.7 ± 8.5 vs. 33.7 ± 11.4 copies; P = 0.025); and 2) hg P*(xR1a) vs. hg DE (29.7 ± 8.5 vs. 34.6 ± 11.1 copies; P = 0.005).
TABLE 2.
TSPY1 copy number distribution and comparison of mean ± SD TSPY1 copy number in different Y hgs with P values referred to hg P*(xR1a)
| Study population (n = 284) |
Patients (n = 154) |
Controls (n = 130) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Y hg | n |
TSPY1 copy no. |
Range | P a | n |
TSPY1 copy no. |
n |
TSPY1 copy no. |
P b |
| Y*(xA, D, E, J, K) | 68 | 30.7 ± 10.3 | 11–62 | ns | 37 | 27.9 ± 8.8 | 31 | 34.1 ± 11.1 | <0.05 |
| P*(xR1a) | 140 | 29.7 ± 8.5 | 13–58 | 76 | 27.2 ± 7.3 | 64 | 32.6 ± 9.0 | <0.001 | |
| J | 32 | 33.7 ± 11.4 | 18–72 | <0.05 | 20 | 30.7 ± 8.5 | 12 | 38.7 ± 14.1 | ns |
| DE | 35 | 34.6 ± 11.1 | 12–66 | <0.01 | 19 | 32.5 ± 10.0 | 16 | 37.1 ± 12.2 | ns |
| K*(xN,P) | 8 | 28.4 ± 12.1 | 19–56 | ns | 1 | 22 | 7 | 29.3 ± 12.8 | ns |
Comparison of mean TSPY1 copy numbers between patients and controls is shown separately for each Y hg with the respective P values. ns, Not significant.
P value relative to mean TSPY1 copy number in the different Y-hg vs. hg P*(xR1a).
P value relative to mean TSPY1 copy number in patients vs. controls, within each Y-hg.
TSPY1 copy number in patients vs. controls
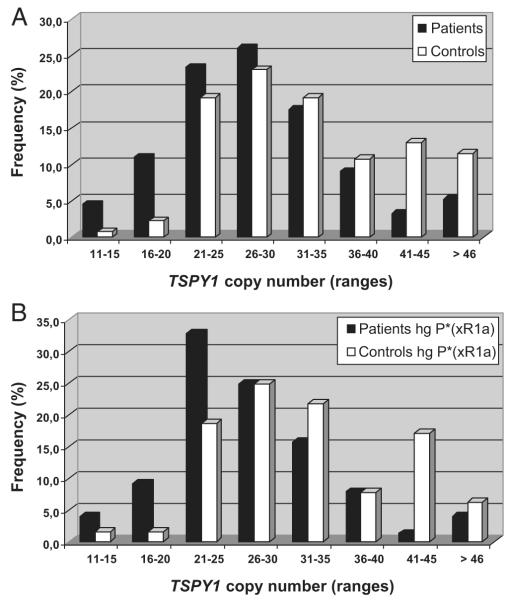
In our study population, the number of TSPY1 gene copies ranged from 11 to 72 (from 11 to 54 in patients and from 14 to 72 in controls), but the majority of subjects had a TSPY1 copy number within the range 21–35 (67% of patients and 62% of controls). We observed a shift toward higher copy numbers in controls and toward lower copy numbers in patients (Fig. 1A). The mean TSPY1 copy number was significantly lower in the patient group than in the controls (28.4 ± 8.3 vs. 33.9 ± 10.7; P < 0.001). The risk of an individual with less than 33 copies being a patient with impaired sperm parameters rather than a control was significantly increased [odds ratio (OR) = 1.5; 95% confidence interval (CI), 1.2–1.8; P < 0.001). We also calculated the risk for being a man with impaired sperm production after removing the 14 pure astheno/ teratozoospermic men (i.e. those men who had a normal total sperm count but abnormal motility and/or morphology): OR = 1.5; 95% CI, 1.3–1.8.
FIG. 1.
TSPY1 copy number ranges in patients and controls in the whole study population (A) and subjects belonging to hg P*(xR1a) (B). The majority of subjects have a TSPY1 copy number within the range 21–35.
To investigate whether or not the significant difference observed in the whole study population was limited to a specific hg, we performed—within each hg—a comparison of the mean TSPY1 copy number in patients vs. controls (Table 2). The mean TSPY1 copy number in patients was lower in each hg compared with controls, although a significant difference was observed only in hg P*(×R1a) (27.2 ± 7.3 vs. 32.6 ± 9.0 copies; P < 0.001) and in hg Y*(xA, D, E, J, K) (27.9 ± 8.8 vs. 34.1 ± 11.1 copies; P = 0.012). The lack of significance in the remaining Y hgs may be due to the low number of subjects.
Because hg P*(xR1a) showed the most significant difference between controls and patients we calculated the OR for this specific subgroup with the same threshold value of TSPY1 = 33 copies (OR = 1.5; 95% CI, 1.2–2.0; P = 0.001). The distribution of TSPY1 copy number in hg P*(xR1a) is reported in Fig. 1B.
Effect of TSPY1 gene copy number variation on spermatogenesis
Correlation between the TSPY1 copy number and sperm count
We found a positive correlation between the TSPY1 copy number and sperm count, both in the whole study population (Rho coefficient = 0.331, P < 0.001; and Rho coefficient = 0.315, P < 0.001 for sperm concentration and total sperm count, respectively) and separately in controls (Rho coefficient = 0.282, P = 0.001; and Rho coefficient = 0.178, P = 0.043 for sperm concentration and total sperm count, respectively) and in patients (Rho coefficient = 0.190, P = 0.018; and Rho coefficient = 0.214, P = 0.008 for sperm concentration and total sperm count, respectively) (Fig. 2).
FIG. 2.
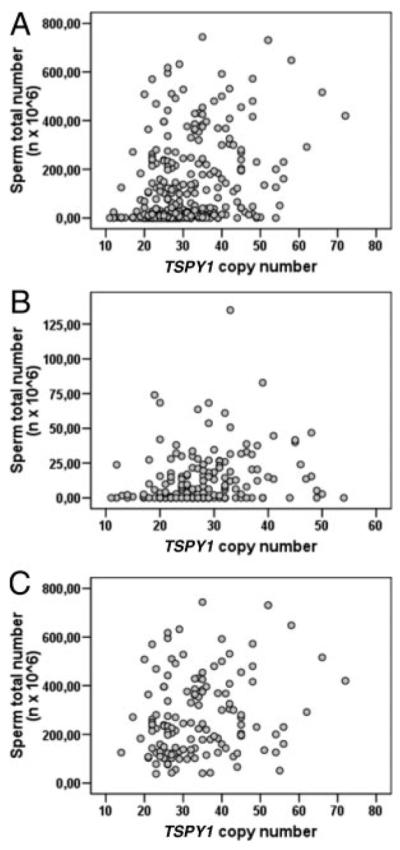
Scatter plots between TSPY1 copy number and total sperm count in the whole study population (A), patients (B), and controls (C). Spearman’s correlation coefficients are reported in the text.
Comparison of sperm concentration and total sperm count within subgroups defined on the basis of TSPY1 copy number
To investigate further the effect of TSPY1 copy number variation on spermatogenesis, patients and controls were grouped into quartiles on the basis of their TSPY1 copy number. The comparison of the median sperm concentration and total sperm count within quartiles is reported in Table 3. Sperm concentration and total sperm count were higher in the fourth quartile (with the highest TSPY1 copy numbers) than in the first quartile, and reached statistical significance for sperm concentration in both groups.
TABLE 3.
Comparison of the median values (with the respective ranges) of sperm concentration and total sperm count among quartiles I and IV in the two study populations (patients and controls)
| I Quartile |
IV Quartile |
|||||||
|---|---|---|---|---|---|---|---|---|
|
TSPY1 range (n) |
Sperm concentration (n × 106/ml) |
Total sperm no. (n × 106) |
TSPY1 range (n) |
Sperm concentration (n × 106/ml) |
Total sperm no. (n × 106) |
P a | P b | |
| Patients | 11–22 (n = 37) | 0.9 (0 –14.8) | 1.7 (0 –74) | 33–54 (n = 38) | 4.0 (0 –27) | 14.3 (0 –135) | 0.012 | 0.007 |
| Controls | 14 –25 (n = 29) | 53 (28 –188) | 224.0 (37.8 –570) | 40–72 (n = 36) | 80.0 (22–329) | 285.7 (51–730.8) | 0.001 | 0.041 |
P value obtained comparing the median sperm concentration, within patients and controls, between quartiles I and IV.
P value obtained comparing the median total sperm count, within patients and controls, between quartiles I and IV.
We compared sperm concentration and total sperm count within patients and controls, above and below the threshold value used for the calculation of OR, i.e. TSPY1 = 33. The analysis further confirmed that subjects with less than 33 TSPY1 copy number have a significantly lower sperm production (Fig. 3).
FIG. 3.
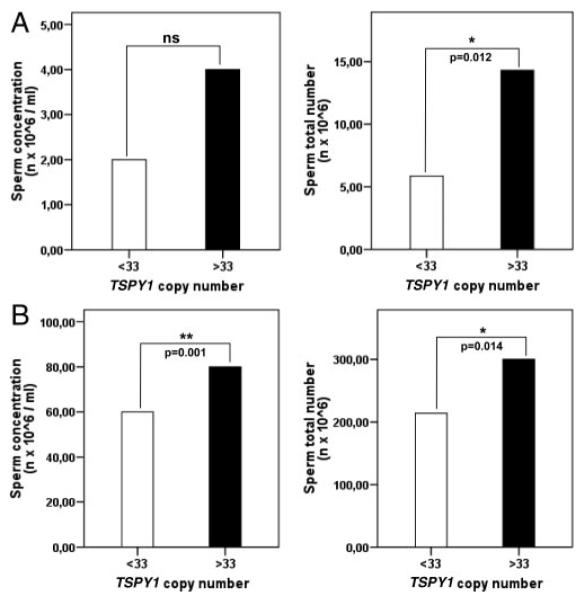
Comparison of the median values of sperm concentration and total sperm number above and below the selected threshold value (33 TSPY1 copies) used for the calculation of OR, within patients (A) and controls (B). *, P < 0.05; **, P = 0.001.
Discussion
The Y chromosome is enriched for genes involved in spermatogenesis, and its structure predisposes to it deletions and duplications, and thus to variation in the copy number of these genes. During the past 15 yr, much emphasis has been given to the AZF (azoospermia factor) regions on the long arm of the Y chromosome. The complete loss of AZF region(s) by deletion causes impairment of spermatogenesis and represents the most frequent known molecular genetic cause of male infertility (3, 24). In addition, partial loss of the AZFc region (gr/gr deletion) is a significant genetic risk factor for oligozoospermia (25, 26). We now provide evidence for a significant effect of the copy number of TSPY1, a multicopy gene situated on Yp, on spermatogenesis.
Because the Y chromosome shows marked population stratification and data in the literature concerning TSPY1 copy number variation in different Y lineages are scarce, we first investigated whether the different Y hgs that are common in the Italian population showed different TSPY1 copy numbers. Based on the analysis of 47 Y hgs, Repping et al. (14) had previously observed little variation in TSPY1 copy number within and between Y hgs, whereas an earlier analysis limited to the few hgs that could be identified in 1994 observed more variation within hgs (13). We now report substantial variation in the number of TSPY1 copies within the six common Y hgs and paragroups present in our study population. The variation was especially high in hg J (from 18 to 72 copies), although this was not significantly greater than in the other Y lineages. In all hgs we observed a peak of frequencies for TSPY1 copy number in the range 21–35, which may indicate an optimal range of copy number. Interestingly, hg P (the most frequent hg in Central Italy) contained significantly lower TSPY1 copy numbers than hgs J and DE; a P vs. DE difference is in line with the observations of Mathias et al. (13) on equivalent hgs then named 1 and 4, which were made using a different method (pulsed-field gel electrophoresis) on a smaller study population (20 subjects belonging to hg DE vs. 31 hg P).
Our finding indicates that the analysis of TSPY1 copy number variation in the context of case-control association studies is susceptible to stratification bias. This observation implies that data from unmatched study populations may not provide reliable information. The distribution of Y hgs in controls and patients in our study was similar, leading us to two major conclusions: 1) no Y hg predisposed to impaired spermatogenesis to a detectable extent; and 2) TSPY1 copy number differences between controls and cases were independent of the Y background. Concerning the former, the relationship between Y background and reproductive fitness has been controversial, which may partly be related to study design biases (small sample size, inappropriate inclusion criteria for patients and controls) (27). This is the first study of a large European sample in which Y hg distribution was compared between normozoospermic controls and idiopathic infertile men. The lack of a Yhg predisposing to impaired sperm production in our study population, together with the lack of significant differences in total sperm counts of men belonging to different Y hgs, provides strong evidence on the lack of a significant Y hg effect in the Italian population.
Concerning our second conclusion, we found a significantly lower number of TSPY1 copies in infertile men with abnormal sperm parameters compared with men with normozoospermia. As emphasized above, this difference was not related to a difference in the prevalence of hgs DE or J—the hgs that have a higher mean TSPY1 copy number than P—in the control group. We observed a trend toward lower TSPY1 copy number in the infertile group within each hg, suggesting that the contraction or expansion responsible for the observed differences between cases and controls occurs in all Y lineages. Accordingly to our analysis, the risk of having abnormal sperm parameters was increased 1.5-fold for a man with fewer than 33 TSPY1 copies. Other evidence for a significant contribution of TSPY1 dosage to spermatogenesis included: 1) positive correlation between sperm count and TSPY1 copy number in the whole study population and separately in controls and cases; and 2) significant differences in sperm production in men with more and less than 33 copies. Given the link between TSPY1 copy number and sperm count on the one hand and the difference in mean TSPY1 copy number between hgs on the other, we would expect to see a TSPY1-dependent relationship between hgs and sperm count. However, such an effect is small and is obscured by the large variation in TSPY1 within each hg, and would only be detected in substantially larger samples.
The maintenance of the evolutionarily unstable TSPY1 array across mammalian species suggested that low copy number must be disadvantageous, and an effect on spermatogenesis was likely on the basis of the observed expression pattern and inferred function of the TSPY1 protein. Spermatogenesis requires the concerted action of thousands of genes, all contributing to the efficiency of spermatogenesis to a different extent. Mutations in genes essential for spermatogenesis inevitably lead to impaired spermatogenesis, whereas mutations in genes acting as tuners/modulators of the efficiency of spermatogenesis may not necessarily lead to clinically overt conditions (28, 29). Infertility is a complex disorder. Hence, instead of highly penetrant mutations in single genes (until now very few causative mutations have been identified), the interaction of multiple factors, both genetic and environmental, with individually small effects is expected to play a major etiopathogenic role. We have now provided evidence for a link between low TSPY1 copy number and impaired spermatogenesis. Low TSPY1 copy number therefore represents a new genetic risk factor for male infertility with potential clinical consequences and should be taken into consideration in the context of a multigenic approach to idiopathic infertility.
Acknowledgments
We thank Grazia Matera, Francesca Gensini, and Valeri Tchepelev for technical assistance, and Vieri Boldi for expert help in statistical analysis. We also thank the Italian Ministry of Education and Research and Telethon-Italy for financial support.
This work was supported by the Italian Ministry of Education and Research (Grant PRIN 2008-2010, to C.K.) and by Telethon-Italy (Grant GGP08204, to C.K.). D.J.T., Y.X., and C.T.-S. were supported by The Wellcome Trust.
Abbreviations
- AZF
Azoospermia factor
- CI
confidence interval
- hg
haplogroup
- OR
odds ratio
- TSPY1
testis-specific protein Y-encoded 1
Footnotes
Disclosure Summary: The authors have nothing to disclose.
References
- 1.Arnemann J, Epplen JT, Cooke HJ, Sauermann U, Engel W, Schmidtke J. A human Y-chromosomal DNA sequence expressed in testicular tissue. Nucleic Acids Res. 1987;15:8713–8724. doi: 10.1093/nar/15.21.8713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang JS, Yang-Feng TL, Muller U, Mohandas TK, de Jong PJ, Lau YF. Molecular isolation and characterization of an expressed gene from the human Y chromosome. Hum Mol Genet. 1992;1:717–726. doi: 10.1093/hmg/1.9.717. [DOI] [PubMed] [Google Scholar]
- 3.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou SF, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S, Rohlfing T, Scott K, Schultz B, Strong C, Tin-Wollam A, Yang SP, Waterston RH, Wilson RK, Rozen S, Page DC. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 4.Tyler-Smith C, Taylor L, Müller U. Structure of a hypervariable tandemly repeated DNA sequence on the short arm of the human Y chromosome. J Mol Biol. 1988;203:837–848. doi: 10.1016/0022-2836(88)90110-6. [DOI] [PubMed] [Google Scholar]
- 5.Warburton PE, Hasson D, Guillem F, Lescale C, Jin X, Abrusan G. Analysis of the largest tandemly repeated DNA families in the human genome. BMC Genomics. 2008;9:533. doi: 10.1186/1471-2164-9-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau YF. Gonadoblastoma, testicular and prostate cancers, and the TSPY gene. Am J Hum Genet. 1999;64:921–927. doi: 10.1086/302353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsuchiya K, Reijo R, Page DC, Disteche CM. Gonadoblastoma: molecular definition of the susceptibility region on the Y chromosome. Am J Hum Genet. 1995;57:1400–1407. [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Lau YF. TSPY and its X-encoded homologue interact with cyclin B but exert contrasting functions on cyclin-dependent kinase 1 activities. Oncogene. 2008;27:6141–6150. doi: 10.1038/onc.2008.206. [DOI] [PubMed] [Google Scholar]
- 9.Schnieders F, Dörk T, Arnemann J, Vogel T, Werner M, Schmidtke J. Testis-specific protein, Y-encoded (TSPY) expression in testicular tissues. Hum Mol Genet. 1996;5:1801–1807. doi: 10.1093/hmg/5.11.1801. [DOI] [PubMed] [Google Scholar]
- 10.Honecker F, Stoop H, de Krijger RR, Lau YF Chris, Bokemeyer C, Looijenga LH. Pathobiological implications of the expression of markers of testicular carcinoma in situ by fetal germ cells. J Pathol. 2004;203:849–857. doi: 10.1002/path.1587. [DOI] [PubMed] [Google Scholar]
- 11.Lau YF, Lau HW, Kömüves LG. Expression pattern of a gonadoblastoma candidate gene suggests a role of the Y chromosome in prostate cancer. Cytogenet Genome Res. 2003;101:250–260. doi: 10.1159/000074345. [DOI] [PubMed] [Google Scholar]
- 12.Kido T, Lau YF. A Cre gene directed by a human TSPY promoter is specific for germ cells and neurons. Genesis. 2005;42:263–275. doi: 10.1002/gene.20147. [DOI] [PubMed] [Google Scholar]
- 13.Mathias N, Bayés M, Tyler-Smith C. Highly informative compound haplotypes for the human Y chromosome. Hum Mol Genet. 1994;3:115–123. doi: 10.1093/hmg/3.1.115. [DOI] [PubMed] [Google Scholar]
- 14.Repping S, van Daalen SK, Brown LG, Korver CM, Lange J, Marszalek JD, Pyntikova T, van der Veen F, Skaletsky H, Page DC, Rozen S. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- 15.Smith GP. Evolution of repeated DNA sequences by unequal crossover. Science. 1976;191:528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- 16.Guttenbach M, Müller U, Schmid M. A human moderately repeated Y-specific DNA sequence is evolutionarily conserved in the Y chromosome of the great apes. Genomics. 1992;13:363–367. doi: 10.1016/0888-7543(92)90254-p. [DOI] [PubMed] [Google Scholar]
- 17.Murphy WJ, Pearks Wilkerson AJ, Raudsepp T, Agarwala R, Schäffer AA, Stanyon R, Chowdhary BP. Novel gene acquisition on carnivore Y chromosomes. PLoS Genet. 2006;2:e43. doi: 10.1371/journal.pgen.0020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyler-Smith C. An evolutionary perspective on Y-chromosomal variation and male infertility. Int J Androl. 2008;31:376–382. doi: 10.1111/j.1365-2605.2008.00889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vodicka R, Vrtel R, Dusek L, Singh AR, Krizova K, Svacinova V, Horinova V, Dostal J, Oborna I, Brezinova J, Sobek A, Santavy J. TSPY gene copy number as a potential new risk factor for male infertility. Reprod Biomed Online. 2007;14:579––587. doi: 10.1016/s1472-6483(10)61049-8. [DOI] [PubMed] [Google Scholar]
- 20.World Heath Organization . WHO Laboratory Manual for the Examination of Human Semen and Sperm-Cervical Mucus Interaction. 3rd ed Cambridge University Press; Cambridge, UK: 1999. pp. 4–33. [Google Scholar]
- 21.Jobling MA, Lo IC, Turner DJ, Bowden GR, Lee AC, Xue Y, Carvalho-Silva D, Hurles ME, Adams SM, Chang YM, Kraaijenbrink T, Henke J, Guanti G, McKeown B, van Oorschot RA, Mitchell RJ, de Knijff P, Tyler-Smith C, Parkin EJ. Structural variation on the short arm of the human Y chromosome: recurrent multigene deletions encompassing amelogenin Y. Hum Mol Genet. 2007;16:307–316. doi: 10.1093/hmg/ddl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oakey R, Tyler-Smith C. Y chromosome DNA haplotyping suggests that most European and Asian men are descended from one of two males. Genomics. 1990;7:325–330. doi: 10.1016/0888-7543(90)90165-q. [DOI] [PubMed] [Google Scholar]
- 23.Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, Armenteros M, Arroyo E, Barbujani G, Beckman G, Beckman L, Bertranpetit J, Bosch E, Bradley DG, Brede G, Cooper G, Côrte-Real HB, de Knijff P, Decorte R, Dubrova YE, Evgrafov O, Gilissen A, Glisic S, Gölge M, Hill EW, Jeziorowska A, Kalaydjieva L, Kayser M, Kivisild T, Kravchenko SA, Krumina A, Kucinskas V, Lavinha J, Livshits LA, Malaspina P, Maria S, McElreavey K, et al. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet. 2000;67:1526–1543. doi: 10.1086/316890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krausz C, Degl’Innocenti S. Y chromosome and male infertility: update, 2006. Front Biosci. 2006;11:3049–3061. doi: 10.2741/2032. [DOI] [PubMed] [Google Scholar]
- 25.Giachini C, Laface I, Guarducci E, Balercia G, Forti G, Krausz C. Partial AZFc deletions and duplications: clinical correlates in the Italian population. Hum Genet. 2008;124:399–410. doi: 10.1007/s00439-008-0561-1. [DOI] [PubMed] [Google Scholar]
- 26.Repping S, Skaletsky H, Brown L, van Daalen SK, Korver CM, Pyntikova T, Kuroda-Kawaguchi T, de Vries JW, Oates RD, Silber S, van der Veen F, Page DC, Rozen S. Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet. 2003;35:247–251. doi: 10.1038/ng1250. [DOI] [PubMed] [Google Scholar]
- 27.Krausz C, Quintana-Murci L, Forti G. Y chromosome polymorphisms in medicine. Ann Med. 2004;36:573–583. doi: 10.1080/07853890410018853. [DOI] [PubMed] [Google Scholar]
- 28.Nuti F, Krausz C. Gene polymorphisms/mutations relevant to abnormal spermatogenesis. Reprod Biomed Online. 2008;16:504–513. doi: 10.1016/s1472-6483(10)60457-9. [DOI] [PubMed] [Google Scholar]
- 29.Tyler-Smith C, Krausz C. The will-o’-the-wisp of genetics— hunting for the azoospermia factor gene. N Engl J Med. 2009;360:925–927. doi: 10.1056/NEJMe0900301. [DOI] [PMC free article] [PubMed] [Google Scholar]