Abstract
Postnatal development of fast skeletal muscle is characterized by a transition in expression of myosin heavy chain (MHC) isoforms, from primarily neonatal MHC at birth to primarily IIb MHC in adults, in a tightly coordinated manner. These isoforms are encoded by distinct genes, which are separated by ∼17 kb on rat chromosome 10. The neonatal-to-IIb MHC transition is inhibited by a hypothyroid state. We examined RNA products [mRNA, pre-mRNA, and natural antisense transcript (NAT)] of developmental and adult-expressed MHC genes (embryonic, neonatal, I, IIa, IIx, and IIb) at 2, 10, 20, and 40 days after birth in normal and thyroid-deficient rat neonates treated with propylthiouracil. We found that a long noncoding antisense-oriented RNA transcript, termed bII NAT, is transcribed from a site within the IIb-Neo intergenic region and across most of the IIb MHC gene. NATs have previously been shown to mediate transcriptional repression of sense-oriented counterparts. The bII NAT is transcriptionally regulated during postnatal development and in response to hypothyroidism. Evidence for a regulatory mechanism is suggested by an inverse relationship between IIb MHC and bII NAT in normal and hypothyroid-treated muscle. Neonatal MHC transcription is coordinately expressed with bII NAT. A comparative phylogenetic analysis also suggests that bII NAT-mediated regulation has been a conserved trait of placental mammals for most of the eutherian evolutionary history. The evidence in support of the regulatory model implicates long noncoding antisense RNA as a mechanism to coordinate the transition between neonatal and IIb MHC during postnatal development.
Keywords: long noncoding antisense RNA, thyroid hormone, gene transcription, phylogenetic, RT-PCR
six myosin heavy chain (MHC) genes, each encoding a specific motor protein impacting contractile velocity and hence muscle performance, are expressed across the spectrum of skeletal muscle of mammals. In the rat the genes of the three fast MHC types, IIa, IIx, and IIb MHC, are clustered on chromosome 10 in a tandem alignment (IIa-IIx-IIb). These MHCs, along with the slow type I MHC, are the primary adult-specific isoforms. Also located on this genomic locus are the embryonic (Emb), situated 5′ to IIa, and the neonatal (Neo), located 3′ to IIb. The Emb and Neo are the predominant MHC isoforms expressed during fetal and early postnatal development (63). Postnatal development is characterized by replacement of these developmental isoforms with the adult isoforms of MHC, in a highly regulated and precisely coordinated manner, such that individual muscles attain various proportions of each MHC isoform to confer optimal function for a given usage pattern. Thyroid hormone, which is essential to normal development and differentiation of all cells, exerts a powerful influence on muscle function and MHC gene regulation during postnatal development (5).
The adult rat plantaris consists of ∼80–85% IIx/IIb fibers, with IIb being the dominantly expressed of the MHC isoforms (6, 17). At birth, however, the majority of MHC is represented by the Neo isoform in the plantaris (1, 2, 10). It was demonstrated previously that there is an inverse relationship in expression of the Neo and IIb MHCs, such that during the postnatal transition to the adult muscle phenotype, Neo MHC downregulation was coordinated with upregulation of the IIb MHC (1, 2, 10). This transition is also characterized by the presence of a significant number of single myofiber hybrids of varying combinations of Neo, IIx MHC, and IIb MHC in 20-day-old rats, indicating a progressive nature of the transition in a single cell (10). Furthermore, this developmental scheme is disrupted in a hypothyroid state, which delays attainment of the adult phenotype and prevents the normal Neo to IIb MHC transition (1, 2, 10, 34). Thus there appears to be a coupling in expression between the Neo and IIb MHC isoforms during postnatal development in plantaris myofibers. However, the mechanism by which these two genes are coordinated to achieve a precise Neo to IIb transition during development has remained unknown.
The genes encoding the IIa, IIx, IIb, and Neo MHC isoforms (MYH2, MYH1, MYH4, and MYH8, respectively) are tandemly linked in a head-to-tail fashion and are separated by relatively short intergenic distance on rat chromosome 10. For example, in the rat, IIa-IIx are separated by 2.7 kb, IIx-IIb by 14 kb, and IIb-Neo by 17 kb. The genomic order, relative spacing, and the coding sequence are highly conserved across a broad evolutionary span. We have previously shown that functional significance for this gene order and intergenic spacing is apparent by the transcriptional coordination of MHC genes with long noncoding RNA transcripts that are transcribed in the antisense orientation to the IIa, IIx, and IIb MHC genes during alterations to muscle loading state (42, 47). For example, muscle inactivity and unloading induce a coordinated IIa to IIx MHC shift in the soleus muscle whereby IIa decreases while IIx increases concurrently. Along with these shifts, an antisense transcript is also induced, the so-called aII natural antisense transcript (NAT), which is transcribed in reverse orientation across the entire length of the IIa MHC gene (42). The aII NAT appears to repress IIa MHC gene transcription (42). Sense-antisense pairs were also observed to regulate IIx and IIb MHC in both fast and slow skeletal muscle and in cardiac muscle in coordination of α- and β-MHC expression (18, 20, 42, 47). These reports are accompanied by other recent reports in nonmuscle cells describing mechanisms of cis-acting long noncoding antisense RNA-mediated transcriptional repression (23, 28, 37, 58, 61, 68). Thus a model of coordinated MHC gene switching mediated by cis-acting long noncoding antisense RNA has emerged as a mechanism to explain how precise stoichiometric relationships between MHC isoforms are maintained in response to altered environmental conditions.
Given the apparent coupling between Neo and IIb MHC previously observed during postnatal development and the previously defined role for antisense RNA in the coordination of other MHC genes, we hypothesized that a paralogous antisense IIb transcript is expressed strategically within the IIb-Neo MHC gene locus to coordinate the transition from Neo to IIb MHC gene expression during postnatal maturation of fast fiber-type skeletal muscle. We tested this hypothesis by examining sense and antisense RNA expression of the MHC genes in the rat plantaris muscle at various time points during the first 6 wk of muscle development. We also examined animals under a hypothyroid state, which arrests MHC conversion from Neo to IIb MHC, to determine whether a IIb antisense transcript would be responsive and regulated.
Herein we report that IIb MHC transcriptional regulation during muscle maturation to the adult state is consistent with our previously defined model of antisense-mediated coordination of genomically adjacent MHC genes, as is evident in the transcriptionally divergent euthyroid and hypothyroid states. Furthermore, the antisense transcript has a transcription start site in the rat IIb-Neo MHC intergenic region that is encompassed by DNA sequence that is highly conserved across most of the evolutionary history of placental mammals.
METHODS
Animal care and experimental design.
Pregnant Sprague-Dawley rats were obtained from Taconic Farms (Germantown, NY). Litters were equalized to eight neonates per dam, with six female and two male neonates per litter. Only tissue from female neonates were used for subsequent analyses. Litters were organized into control (CON) and hypothyroid (propylthiouracil, PTU). Both the dam and neonates from the PTU group received daily intraperitoneal injections of PTU (12 mg/kg) until the time of euthanasia. PTU treatment began at day 2 after birth. Animals in a particular litter were euthanized using Pentosol at the following number of postnatal days: 2, 10, 20, and 40 (P2, P10, P20 P40, respectively). To obtain sufficient muscle tissue to analyze both the RNA and protein, two litters were assigned for P2, P10, and P20 groups. For P10 (CON and PTU) and P20 (PTU), muscles from two neonates were pooled for each n which resulted in n = 6 for P10, P20 and P40. For P2 rats 4–5 muscles were pooled together for analysis to yield n = 4 for RNA analysis and n = 2 for protein analysis. The plantaris muscle was obtained from each neonate and weighed and frozen at −80° C for later analysis. All procedures were approved by the University of California, Irvine Institutional Animal Care and Use Committee.
Thyroid hormone analysis.
Plasma total thyroid hormone [triiodothyronine (T3) and l-thyroxine (T4)] concentrations were assayed using a commercially available RIA kit (MP Biomedical). Readings at P2 that were below the level of detection (T3 <5 ng/dl; and T4 <0.5 μg/dl) were assigned values of zero for statistical analysis.
RNA analysis.
Total RNA was extracted from frozen plantaris muscle using the Tri Reagent protocol (Molecular Research Center). Extracted RNA was DNase-treated using one unit of RQ1 RNase-free DNase (Promega) per microgram of total RNA and was incubated at 37°C for 10 min followed by a second RNA extraction using Tri Reagent LS (MRC).
Strand-specific RT-PCR used the one-step real-time reverse transcription polymerase chain reaction (RT-PCR) kit from Qiagen. These assays were utilized in the determination of the relative level of expression of pre-mRNA, antisense RNA, and mRNA in a known amount of total RNA in comparing various developmental stages in CON versus PTU states. These procedures were also utilized in the analyses of antisense RNA expression across the skeletal MHC gene locus between the IIb and Neo MHC. The manufacturer's protocol was followed with some modifications as described previously (42, 47). This protocol has been optimized to avoid amplification of nonspecific transcripts (21, 42). These one-step RT-PCR analyses were performed using 100 ng total RNA and 15 pmoles of specific primers in 25 μl total volume and were carried out on a Robocycler (Stratagene). For the same target RNA, all samples were run under similar conditions (template amounts, PCR cycle numbers). RT reactions were performed at 50°C for 30 min followed by 15 min of heating at 95°C and followed by PCR cycling for a varied number of cycles (19–33 cycles). The annealing temperature was based on the PCR primers optimal annealing temperature. PCR primers used for RNA analysis were reported previously (43) except for those shown in Table 1. The amount of RNA and the number of PCR cycles were adjusted so that the accumulated product was in the linear range of the exponential curve of the PCR amplifications. PCR products were separated by electrophoresis on agarose gels and stained with ethidium bromide. The ultraviolet light-induced fluorescence of stained DNA was captured by a digital camera, and band intensities were quantified by densitometry with ImageQuant software (GE Healthcare) on digitized images. RNA levels are reported as concentration in arbitrary units per unit RNA.
Table 1.
PCR primer sequences, their specific target, and PCR product size
| Target | RT-PCR primers: 5′→3′ | PCR Product Size, bp |
|---|---|---|
| Neonatal pre-mRNA | Fwd: GAAACAGGGCAGCGCTAAGTCTCCT | 226 |
| Rev: ATGGCTTTTGTGCAAGGGAAGGT | ||
| Neonatal mRNA | Fwd: TGAAGGGCGGCAAGAAGCAGAT | 458 |
| Rev: AAGCGACCCAAGGCAGCACATT | ||
| bII NAT | Fwd: CTAAAGGCGGTCTGAAGGGGTGAG | 220 |
| Rev: TTGGATGAGTAACCGCATGCTGTTTT | ||
| β-actin pre-mRNA | Fwd: CACGCCCTTTCTCAATTGTCTTTCT | 225 |
| Rev: GGCCATTTATCACCAGCCTCATTAG | ||
| β-actin mRNA | Fwd: GAGGCCCCTCTGAACCCTAAGG | 230 |
| Rev: CGGCCAGCCAGGTCCAGA |
NAT, natural antisense transcript.
To determine the approximate 5′ and 3′ boundaries of the bII NAT, we performed strand-specific RT-PCR across regular overlapping intervals to amplify antisense transcript from the IIb-Neo intergenic region to within the IIb MHC gene with sufficient controls to ensure accuracy and validity, i.e., no-primer RT and nonspecific PCR primer conditions, as described previously (21).
Pre-mRNAs are the nascent, unprocessed, transcriptional products. Pre-mRNA transcript abundance serves as a better marker of a gene's level of transcriptional activity than the mRNA because its half-life is much shorter. Assessing the transcriptional activity of other genes by measuring pre-mRNA with RT-PCR has been validated as an alternative to the nuclear run-on approach (12).
MHC mRNA isoform distribution.
The MHC mRNA isoform distribution was assessed by RT with oligo dT/random primers followed by PCR with primers targeting the embryonic, neonatal, I, IIa, IIx, and IIb MHC mRNAs, as described previously (9, 66). In these PCR reactions, each MHC mRNA signal was corrected to an externally added control DNA fragment that was coamplified with the MHC cDNAs using the same PCR primer pair. This approach provides a means to correct for any differences in the efficiency and/or pipetting of each PCR reaction. A correction factor was used for each control fragment band on the ethidium bromide-stained gel to account for the staining intensity of the variably sized fragments (224 to 324 bp), as reported previously (9).
SDS-PAGE MHC isoform separation.
A preweighed frozen muscle piece was homogenized in 20 volume PBS using a tight-fitting glass homogenizing tube and a pestle. Total protein concentration was determined using the Bio-Rad Protein Assay and gamma globin as a standard. Total muscle homogenate proteins were diluted to 1 mg/ml in a solution consisting of 50% glycerol, 50 mM Na4P2O7, 2.5 mM EGTA, and 1 mM β-mercaptoethanol; pH 8.8, and stored at −20°C. To quantify MHC protein isoform distribution we performed SDS-PAGE of 2.5 μg of the stored total protein (53). In addition to the four primary adult MHCs, this technique enabled us to separate both neonatal and embryonic MHCs. These procedures were performed as described previously (1). Quantification of relative proportions of MHC isoforms were obtained from these gels by densitometry of bands (Image Quant Software, GE Healthcare). Values per muscle were calculated based on protein concentration and muscle weight.
5′RACE: rapid amplification of cDNA ends.
These assays were conducted to identify the transcription start site (TSS) of the antisense IIb RNA (bII NAT). We adapted the method described by Invitrogen as supplied with the kit (5′RACE). The cDNA was synthesized using Superscript II at 50°C and an RT primer reverse complementary to a region of the target RNA near its 5′ end based on transcript mapping via one step RT-PCR. The specific cDNA was RNase H-treated and purified using the Qiaquick PCR Purification Kit (Qiagen). The pure cDNA was tailed with dCTP using terminal transferase (New England Biolab). The tailed cDNA was amplified using a gene-specific reverse primer that was nested to the RT primer and an adapter primer (5′RACE primer, Invitrogen). Two different nested gene-specific primers were utilized for the subsequent PCR reactions (see Table 2) for the RT and gene-specific primer sequence information. The PCR products were cloned into pGEMT easy (Promega), and the insert of individual clones was sequenced to determine the 5′ end corresponding to the end of the cDNA that is adjacent to the poly dC tail. Sequencing and data acquisition were obtained commercially (Retrogen, San Diego, CA).
Table 2.
Primers used for the 5′RACE targeting the bII NAT
| Primer ID | Position on IIb-Neo Intergenic DNA Versus IIb 3′ End | *Primer Sequence: 5′→3′ |
|---|---|---|
| RT | +4308 | GGCAGGCTACGGGCTTGGAGA |
| GSP PCR-1 | +4671 | AAGTTCGCTAATTTCTTTTAAGTTTGAGC |
| GSP PCR 2 | +4988 | CCCACCGGCCACCACCATC |
GSP: gene specific primer;
Note the primer sequence is in reference to the sense strand, but the primer is reverse complement to the target antisense RNA.
Phylogenetic analysis.
DNA sequence was obtained from NCBI. Sequence was masked for repetitive sequence with RepeatMasker (www.repeatmasker.org) so that repetitive sequences are ignored in evolutionary conservation analysis. Rat sequence was used as the base sequence for comparison to other species. The IIb-Neo (MYH4-MYH8) MHC intergenic DNA sequence between the IIb MHC poly-A signal and the Neo MHC TSS were utilized as the respective 5′ to 3′ boundaries for the rat. Nucleotide sequence encompassing these same boundaries were obtained from NCBI GenBank databases for the following species provided with their corresponding accession numbers: human (Homo sapiens; NC_000017.10), chimpanzee (Pan troglodytes; NC_006484.3), rhesus monkey (Macaca mulatta; NC_006484.3NC_007873.1), mouse (Mus musculus; NC_000077.5), rat (Rattus norvegicus; NC_005109.2), dog (Canis familiaris; NC_006587.2), cow (Bos taurus; NC_007317.4), pig (Sus scrofa; NC_010454.2), rabbit (Oryctolagus cuniculus; NC_013687.1), horse (Equus caballus; NC_009154.2), opossum (Monodelphis domestica; NC_008802.1), and chicken (Gallus gallus; NC_006105.2). Draft assembly of the elephant (Loxodonta Africana) was provided by The Broad Institute. Local multiple DNA sequence alignments were implemented on mulan.org for the IIb-Neo intergenic region. Methods for generating alignments to identify evolutionarily conserved regions (ECR) and the display design for alignment visualization of divergent species are described by Ovcharenko et al. (40, 41). Regions of at least 100 bases and 70% similarity was utilized for ECR parameters.
Statistical analyses.
Data are reported as means ± SE. Significant differences between groups were determined by one-way analyses of variance (ANOVA); when differences were detected for a given variable, a Newman-Keuls as a post hoc test was used (GraphPad Software). Relationships between two variables were assessed using linear regression and Pearson correlation analyses (GraphPad Software). Statistical significance was set at P < 0.05, or lower, as noted in the figure legends.
RESULTS
Gross effects of hypothyroid state.
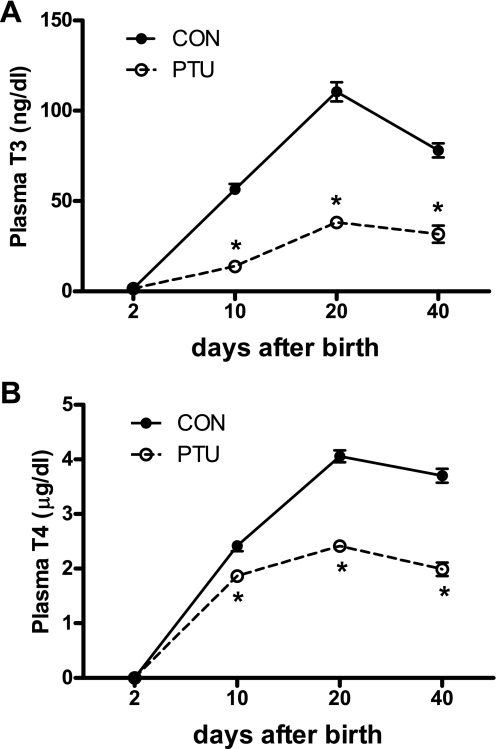
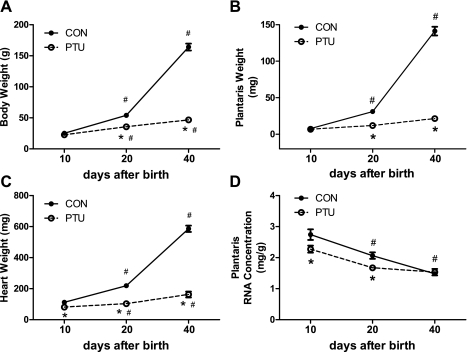
The typical thyroid hormone surge was observed in CON rats during the first 20 days of postnatal life (Fig. 1). Daily injections of PTU successfully inhibited synthesis of T3 at all time points analyzed (Fig. 1A). Plasma T4 levels were also markedly suppressed by PTU treatment (Fig. 1B). During postnatal development, from 10 to 40 days after birth, both body weight and plantaris muscle weight were markedly increased in CON (Fig. 2, A and B). As the euthyroid (CON) animal matured from P10 to P40, the body weight increased 6.5-fold, whereas the muscle weight increased 18.5-fold. With PTU treatment, both body and muscle growth were largely blunted in the hypothyroid state (Fig. 2, A and B), demonstrating the powerful influence of thyroid hormone in normal postnatal growth. Heart weights were significantly repressed at every time point in the PTU group (Fig. 2C), which confirms a hypothyroid state and effectiveness of the treatment. Total RNA concentration in the plantaris significantly decreased as the CON animals aged from P10 to P40 (Fig. 2C). In the PTU-treated group, total RNA concentration decreased as the animal aged from P10 to P20 but remained constant after that at P40 (Fig. 2D). The RNA concentration was also significantly lower in PTU compared with CON at P10 and P20.
Fig. 1.
Hypothyroid (propylthiouracil, PTU) treatment reduces thyroid hormone levels during postnatal development. A: plasma triiodothyronine (T3); B: plasma l-thyroxine (T4) concentrations. Data points are means ± SE. *Significantly different from control (CON) (P < 0.01).
Fig. 2.
PTU treatment alters normal growth in developing rats. A: body weight; B: plantaris muscle weight; C: heart weight; D: plantaris total RNA concentration. Data points are means ± SE. *Significantly different from CON at same time point (P < 0.05). #Significantly different from previous intratreatment time point (P < 0.05).
Effects of hypothyroid on MHC expression.
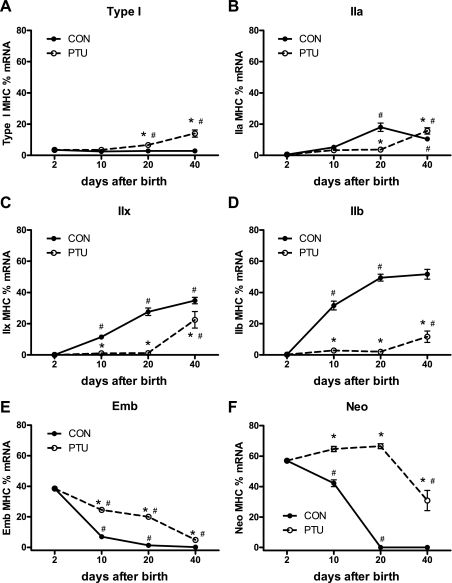
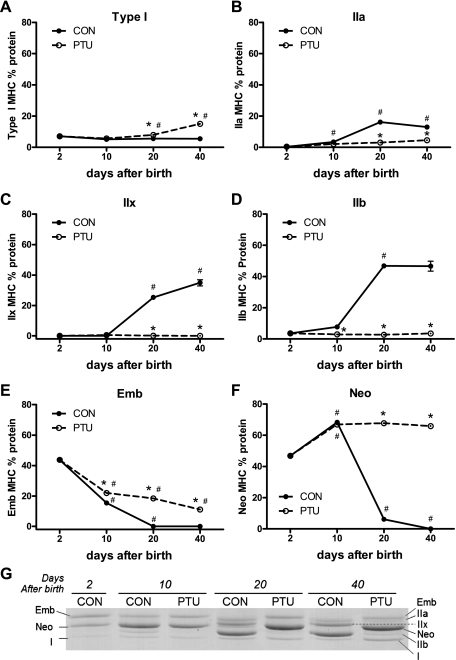
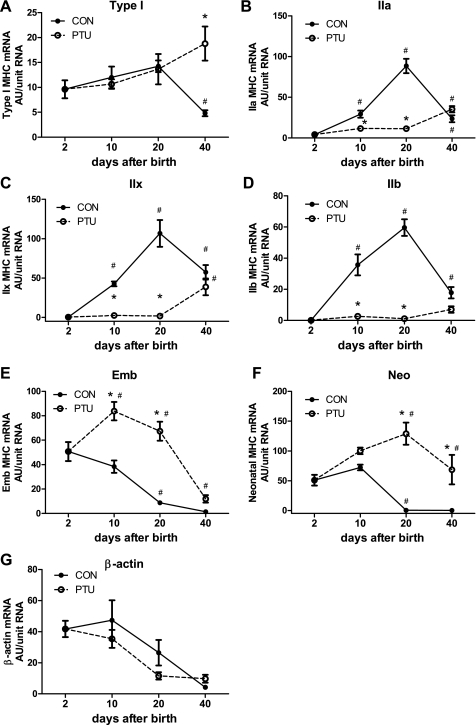
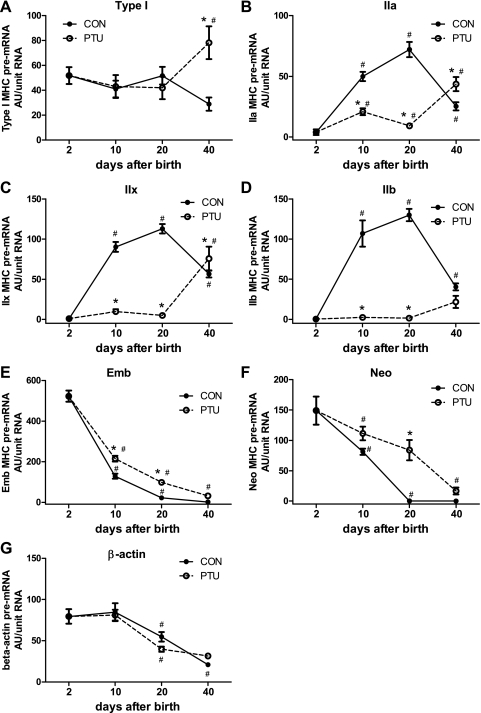
Figure 3 shows the MHC mRNA distribution in the plantaris muscle across all groups. Each individual MHC mRNA isoform is expressed as a percentage of the total MHC mRNA. These data demonstrate the gradual replacement of developmental MHC (Emb and Neo) with the adult isoforms (IIa, IIx, and IIb) during postnatal development in CON rats. By 20 days of age, the developmental MHC mRNAs were not detected in the CON plantaris. Analysis of the PTU-treated muscle demonstrates that expression of each of the six MHC isoforms is altered by the hypothyroid state during postnatal development. By maturity, at P40, the composition of the different MHCs remains significantly altered by PTU administration compared with CON, and significant amounts of developmental MHC isoforms remains expressed. However, with the exception of the type I MHC, the PTU influence on the percentage of each MHC isoforms' mRNA content appears to have been mitigated at P40 compared with earlier time points, suggesting some normalization of MHC isoform mRNA expression independent of thyroid hormone as the animal reaches adult age. The MHC isoform composition at the level of protein expression is shown in Fig. 4. These changes in percent protein isoform expression are similar to the percent mRNA shown in Fig. 3, with differences between the mRNA and protein during the analysis reflecting the different half-lives of the two gene products and the kinetics for the accumulation of transcriptional products (mRNA) versus translated products (protein). Each MHC mRNA isoform was also analyzed independently of the other isoforms, and these are expressed as arbitrary units (AU)/unit of total RNA (Fig. 5), and the corresponding levels of pre-mRNA are shown in Fig. 6. Levels of β-actin pre-mRNA and mRNA are provided for comparison (Figs. 5G and 6G, respectively). In general, there is broad correspondence in relative levels of pre-mRNA and mRNA for each age and treatment (compare Figs. 5 and 6). There are also similar alterations in these absolute transcript levels compared with MHCs expressed as a percentage of total MHC (compare with Fig. 3), though differences are apparent at P40 for IIx (Fig. 3C vs. Fig. 5C) and IIb MHC (Fig. 3D vs. Fig. 5D) (see discussion for interpretation).
Fig. 3.
Myosin heavy chain (MHC) mRNA proportions are developmentally regulated and development are delayed by altered thyroid state. The relative percent expression of individual MHC isoforms as a percentage of the total MHC mRNA expression is shown and determined by competitive RT-PCR. A: type I; B: IIa; C: IIx; D: IIb; E: embryonic (Emb); F: neonatal (Neo) MHCs. Data points are means ± SE. *Significantly different from CON at same time point (P < 0.05). #Significantly different from previous intratreatment time point (P < 0.05).
Fig. 4.
MHC protein proportions are developmentally regulated and MHC expression is altered by thyroid state. Relative percent expression of individual MHC isoforms as a percentage of the total MHC protein expression is shown. A: type I; B: IIa; C: IIx; D: IIb; E: Emb; F: Neo MHCs; G: representative SDS-PAGE separation of MHC isoforms at 2, 10, 20, and 40 days after birth in CON and PTU, as labeled. Data points are means ± SE. *Significantly different from CON at same time point (P < 0.05). #Significantly different from previous intratreatment time point (P < 0.05).
Fig. 5.
mRNA abundance of individual MHCs and β-actin. The dynamic changes in expression of the individual MHC genes during development in normal and hypothyroid states are demonstrated with altered mRNA levels. A: type I; B: IIa; C: IIx; D: IIb; E: Emb; F: Neo MHCs; G: β-actin. Data points are means ± SE of RT-PCR analyses. AU, arbitrary units. *Significantly different from CON at same time point (P < 0.05). #Significantly different from previous intratreatment time point (P < 0.05).
Fig. 6.
Pre-mRNA abundance of individual MHCs and β-actin. The dynamic changes in transcription of the individual MHC genes during development in normal and hypothyroid states are indicated with altered pre-mRNA levels. A: type I; B: IIa; C: IIx; D: IIb; E: Emb; F: Neo MHCs; G: β-actin. Data points are means ± SE of RT-PCR analyses. *Significantly different from CON at same time point (P < 0.05). #Significantly different from previous intratreatment time point (P < 0.05).
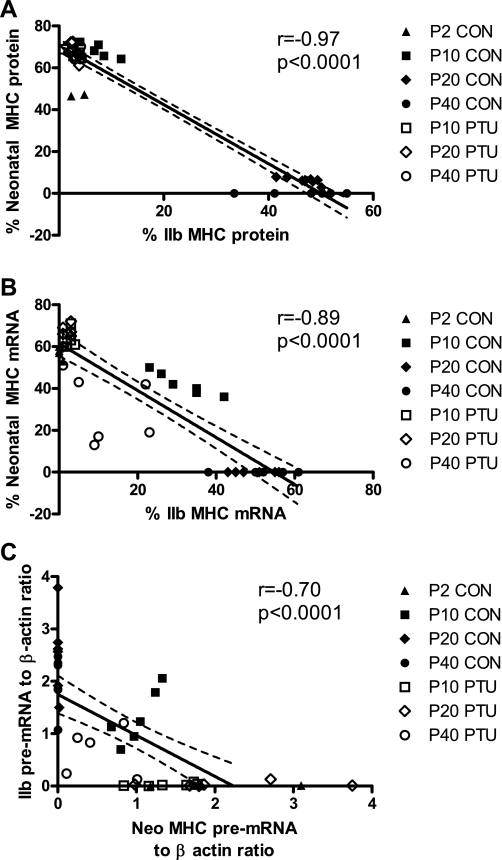
The relationship of Neo MHC and IIb MHC is illustrated by correlation analysis, and this was analyzed at the protein, mRNA, and pre-mRNA levels (Fig. 7). At the protein level the inverse relationship is especially striking, where Pearson coefficient r = −0.97 (P < 0.0001; Fig. 7A), with the input data represented by the proportion of each MHC as a percentage of the total MHC pool. When MHCs are similarly analyzed by the proportion of MHC mRNA, the correlation between Neo and IIb MHC is −0.89 (P < 0.0001; Fig. 7B). Analysis of absolute transcript abundance is difficult to interpret, so Neo and IIb primary transcripts were normalized to β-actin levels. Normalizing the data to β-actin, Neo, and IIb pre-mRNA are similarly inversely correlated (r = −0.70, P < 0.0001, Fig. 7C; using nonnormalized data, r = −0.41, P = 0.01). These data suggest a strong coordinated regulatory mechanism that tightly links the expression of IIb to the repression of the Neo, and this coordination is significant at the transcriptional (pre-mRNA) as well as posttranscriptional (mRNA) and translational (protein) levels.
Fig. 7.
There is an inverse relationship between IIb and Neo MHC expression. The normal neonatal to IIb MHC transition during development is altered in the hypothyroid state, whereas a significantly correlated inverse relationship is maintained. A: relationship between IIb and Neo MHC at the protein level each expressed as % total MHC. B: relationship between IIb and Neo MHC at the mRNA level, each expressed as % total MHC. C: relationship between IIb MHC pre-mRNA and Neon MHC pre-mRNA. Pre-mRNA values are shown relative to individual β-actin pre-mRNA levels. Dotted lines indicate 95% confidence interval of linear regression.
Antisense RNA of IIb MHC gene: bII NAT.
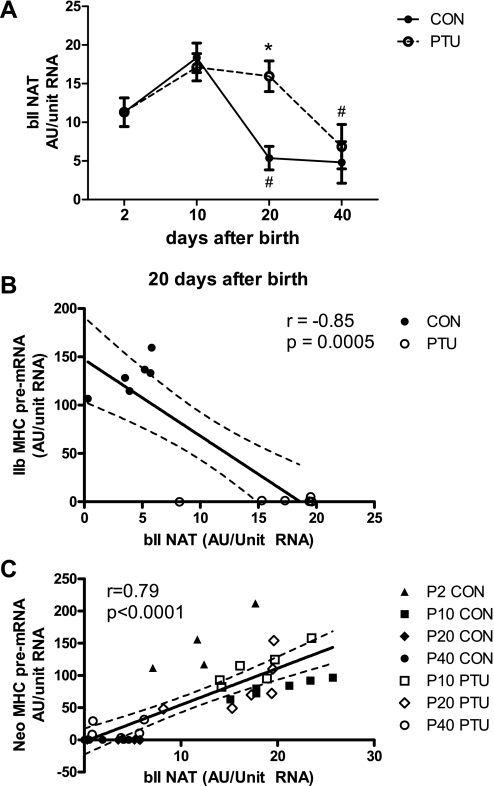
Long noncoding antisense RNA, also referred to as natural antisense transcripts (NAT), have been previously shown to act in an inhibitory manner to overlapping genes (23, 28, 37, 58, 61, 68). To determine whether long noncoding antisense RNA could be involved in the Neo to IIb MHC coordinated shift during postnatal development, we first performed analysis of the antisense strand-specific RNA transcripts in the coding region of the IIb MHC gene at the 3′ end of the IIb gene (at ∼20 kb from its TSS). Antisense IIb (bII NAT) expression in the plantaris of CON and PTU rats in postnatal development is shown in Fig. 8A. bII NAT RNA expression was observed at P2 and P10 in the plantaris of both CON and PTU groups. In CON, bII NAT significantly decreased at P20 compared with P10. This downregulation in bII NAT expression was not observed in the PTU-treated group (Fig. 8A). Thus, at P20, bII NAT was significantly elevated in PTU compared with CON. The level of bII NAT was downregulated at P40 in the PTU group so that at P40, there is no differential expression between CON and PTU. There was no difference in bII NAT levels between CON and PTU at either P10 or P40. In CON plantaris muscle the levels of bII NAT after birth (P2) and at P10 are significantly elevated relative to P20 and P40.
Fig. 8.
bII natural antisense transcript (NAT) is regulated as a function of age and thyroid state and is coordinately regulated. A: DNAse-treated RNA analyzed on antisense strand at the 3′ end of the IIb MHC coding region with one-step RT-PCR. Data points are means ± SE of RT-PCR analyses. *Significantly different from CON at same time point (P < 0.05). #Significantly different from previous intratreatment time point (P < 0.05). B: correlation analysis between bII NAT and IIb MHC pre-mRNA at P20 shows inverse relationship. C: correlation analysis of bII NAT and Neo MHC pre-mRNA shows positive correlation. Lines were drawn using linear regression analyses.
The relationships between IIb pre-mRNA and bII NAT and between Neo MHC pre-mRNA and bII-NAT were evaluated using correlation analyses and linear regression (Fig. 8, B and C). A significant negative correlation between IIb pre-mRNA and bII NAT was observed (r = −0.85, P < 0.0005) at P20. In contrast, a significant positive correlation was found between Neo MHC pre-mRNA and bII NAT (r = 0.79, P < 0.0001) with all groups.
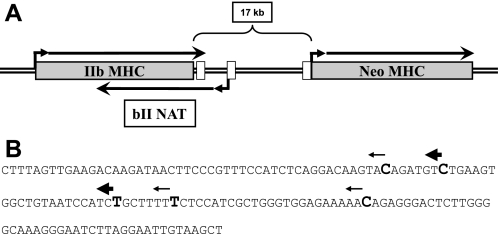
We next sought to determine the transcription start site (TSS) of the bII NAT. Strand-specific RT-PCR across regular overlapping intervals showed that the bII NAT was detected at intergenic regions ∼5 kb from the IIb poly(A) signal at the 3′ end of the IIb gene, but the NAT was not detectable in regions further downstream from this site with the primer sets utilized (locations of detectable transcription is diagramed in Fig. 9). With the location of TSS of the bII NAT narrowed, we mapped the precise start site with 5′RACE procedures. Primer extension and cloning of the 5′ ends of this transcript indicated several TSSs clustered between 5148 and 5207 bp from the 3′ end of the IIb MHC gene, with two major sites located at 5155 and 5176 bp (Sequence shown in Fig. 9).
Fig. 9.
The bII NAT starts in the intergenic region between the IIb and Neo MHC genes. A: schematic diagram of the IIb-Neo MHC genomic locus. The diagram is drawn approximately to scale indicating the size of the intergenic region and the start and termination sites of the bII NAT relative to the IIb and Neo MHC gene coding regions. White boxes in the 17-kb intergenic region correspond to the location of evolutionarily conserved regions, as described for Fig 10. B: sequence shown is from 5100 to 5250 from IIb 3′ end. Larger bold font nucleotides mark the TSS of the bII NAT. Thicker arrows mark the more frequent TSSs according to 5′RACE sequencing results.
Phylogenetic analysis.
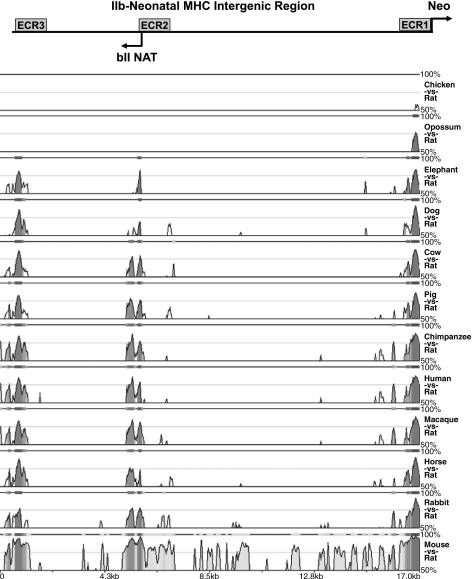
Noncoding intergenic sequences that contain an ECR are typically indicative of the presence of regulatory elements with essential functions. To determine whether potential regulatory elements of the bII NAT are evolutionarily conserved, local multiple DNA sequence alignments were performed. The ∼17-kb IIb-Neo intergenic region of the rat was compared with a diverse array of eutherian (placental mammal) species representing broad evolutionary time scales: primates (human, chimpanzee, and Rhesus macaque), artiodactyls (cow and pig), perissodactyl (horse), carnivore (dog), lagomorph (rabbit), another rodent (mouse), and a proboscidean (African Elephant) as well as a representative of marsupials (short-tailed opossum) and a bird (domestic chicken).
As illustrated in Fig. 10, there are three primary ECRs (ECR1, -2, and -3) that are common to all 11 of the placental mammals. ECR1, indicates conserved DNA sequence common to all 13 of the species examined, including the more evolutionarily distant opossum and chicken. ECR1 corresponds to the proximal promoter region of the Neo MHC gene. This is the only ECR in the IIb-Neo intergenic region that the opossum and chicken share with the placental mammals examined.
Fig. 10.
Three regions are evolutionarily conserved in the rat IIb-Neo MHC intergenic region. The evolutionary conserved regions (ECR) correspond to the Neo MHC transcription start site (ECR1), the bII NAT promoter region (ECR2), and the IIb MHC transcription termination site (ECR3). For each pairwise alignment visualization the rat reference sequence is linear along the horizontal axis and the percent identity is plotted along the vertical axis. Input criteria utilized for identifying and displaying evolutionarily conserved regions is as follows, ECR length of least 100 bp and ECR similarity of at least 70%. Pairwise alignment of rat sequence to particular species are as labeled on the right side of each visualization. The IIb-Neo intergenic region is diagramed above the alignment display, with the three ECRs indicated at the corresponding locations of conservation. The amplitude of graphed peaks indicates the percent identity for each animal vs. rat. The top of each graph indicates 100% identity, whereas the bottom cut-off of each graph is 50% identity. Intensity of grey-scale shading of graphed peaks indicates interspecies conservation, where the most common conserved regions among different species are shown with the darkest shading.
A region corresponding to ∼5.1 to 5.7 kb downstream from the 3′ end of the IIb MHC gene in the rat is highly conserved in each of the 11 placental mammals examined and designated as ECR2 (Fig. 10). This ECR is located immediately upstream of the bII NAT TSS. ECR2 is ∼645 bp in size and presumed to encode regulatory elements of the bII NAT. It also encodes numerous ultra-conserved sequences that are predicted transcription factor binding sites that have been shown to regulate muscle specific genes. One of these sites is an MCAT element to which binds the TEF-1/TEAD-1 family of transcription factors, which regulate MHC genes (57, 67). This element, 5′-CAATTCCTAA-3′ on the antisense strand, is perfectly conserved in all placentals except the elephant. Also of note, the sequence, 5′-CCACCTGTTAA-3′ on the antisense strand, is perfectly conserved in each of the 11 placental mammals studied. This conforms to the E box consensus sequence 5′-CANNTG-3′ to which the myogenic basic helix-loop-helix (bHLH) family of transcription factors interact (36). This family of transcription factors includes MyoD, myogenin, myf5, and MRF4 that determine the myogenic phenotype during skeletal muscle development (36). There was also a highly conserved TATA box, 5′-TTAATTA-3′ on the antisense strand in this region. The cardiac muscle-specific homeobox transcription factor nkx2.5, known to be required for early heart development and morphogenesis (25), binds a consensus sequence 5′-CTTAATTG-3′ that is perfectly conserved in eight of the eutherians examined. Also encompassing this sequence is 5′CTTAATTGTAA-3′, with the same species conservation, that may have ability to bind the MADS box of the MEF2 family of transcription factors that dimerize and bind to AT-rich sites (36). The functional significance of these conserved sequences will have to be confirmed experimentally.
ECR3 is positioned at +549 to +1237 relative to the IIb poly(A) signal site located at the 3′ end of the IIb MHC gene (Fig. 10). The functional significance of the sequence conservation in this region is likely a consequence of elements required for transcriptional termination of the IIb MHC. Termination elements located at similar distances downstream of the poly(A) signal have been shown to be required for efficient transcription termination of mammalian genes (11, 44, 45, 54, 62). The 3′ end of the IIb pre-mRNA was detected by RT-PCR with primers amplifying a product ending at +1857. A large repetitive sequence (∼1.5kb) follows this site, which precludes primer design. At sites further downstream the IIb pre-mRNA was not detectable by RT-PCR, indicating that transcriptional termination of the rat IIb MHC occurs relatively close to ECR3.
Transcription termination of the bII NAT within the IIb MHC gene.
To determine the length of the bII NAT and its approximate site of transcriptional termination, we also performed antisense strand-specific RT-PCR along the length of the IIb MHC gene. The bII NAT terminates in the proximity of intron 15, corresponding to ∼8.8 kb from the IIb TSS (see schematic diagram in Fig. 9). The bII NAT could be detected by RT-PCR through the remaining length of the IIb MHC gene, i.e., from the intron 15 site through to the 5′ end of bII NAT within the IIb-Neo MHC intergenic region (as indicated in Fig. 9). Thus these data indicate that the bII NAT is a long noncoding antisense RNA transcript of ∼19 kb in length.
DISCUSSION
We reported herein that a long noncoding antisense-oriented RNA, termed bII NAT, is transcribed from within the IIb-Neo intergenic region; it is regulated during postnatal development and in response to hypothyroidism. Evidence for a regulatory mechanism is reported that suggests that mediation of the coordinated shift from Neo to IIb MHC involves bII NAT. Neo MHC expression was associated with bII NAT transcription, which may act in an inhibitory manner on transcription of the IIb MHC. From a teleological perspective, coregulation of bII NAT and Neo MHC would ensure that transcription of Neo MHC is coordinated with inhibition of IIb MHC transcription. In addition, the coregulation observed between bII NAT and Neo MHC transcription supports our previously reported model of MHC gene transcriptional coordination, which entails a functional linkage between two neighboring MHC genes mediated by cotranscription of a NAT with one MHC gene that negatively regulates the other MHC gene (19, 42, 47). However, the relatively large distance between the TSS of the bII NAT and Neo MHC stands out compared with the previously reported MHC NATs, which are much more closely spaced to their coregulated MHC gene. Other factors, including thyroid hormone, likely contribute to the complexity of regulating the coordination between the IIb and Neo MHC expression. A balance between positive and negative regulatory influence on IIb MHC gene expression, as reported herein with T3 and bII NAT, respectively, seems to play a key role in development of the fast-fiber contractile phenotype. The evidence in support of this NAT-centered model provides a mechanism for the coordinated transition between Neo and IIb MHC during postnatal development.
Shifts in MHC expression.
Dynamic alterations in MHC gene expression and composition in the plantaris muscle are acutely evident with the interventions studied. Though the percentage of IIx and IIb MHC mRNA (Fig. 3, C and D) and protein (Fig. 4, C and D) reached a peak by P40, unexpectedly, it appears that absolute mRNA and pre-mRNA levels of the IIx (Figs. 5C and 6C) IIb (Figs. 5D and 6D) MHCs in P40 CON muscles markedly fell relative to the P20 time point, such that CON levels are similar to PTU. This apparent decrease in transcriptional activity of the fast type II MHCs that occurred at P40 may be attributable to the animals having reached maturity and thus a steady state with regard to muscle size and therefore gene transcription. In support, the mRNA and pre-mRNA levels of the housekeeping gene β-actin steadily decreased from P10 to P40 (Figs. 5G and 6G). Furthermore, when IIx and IIb MHC expression are normalized to β-actin, CON levels of IIx and IIb MHC mRNA and pre-mRNA are increased or unchanged from P20 to P40 (data not shown), indicating that relative to β-actin, the IIx and IIb MHC transcription was not curtailed at P40 to the degree suggested by Figs. 5 and 6.
Mechanism and function of antisense transcripts.
Overlapping transcripts, or sense/anti-sense pairs, such as the IIb MHC/bII NAT, are prevalent across whole mammalian genomes (7, 8, 13, 14, 25a). Analyses based on full-length mouse cDNAs provided evidence of at least 66% of protein-coding genes with antisense transcription (25a). Analysis of a randomly selected subset of human transcripts determined that 61% of them were transcribed on both genomic strands (8). Long noncoding antisense RNAs, such as the bII NAT described in this report, have been shown to be key regulatory molecules in many types of cells (35, 60, 64). Many of these RNAs have been shown to negatively affect expression of overlapping or nearby genes (reviewed in Refs. 30, 38, 39, 60, and 64). Though prevalent and shown to be associated with regulatory roles, the underlying regulatory mechanisms and functions of most of the long noncoding RNAs previously identified are poorly characterized. Regulatory mechanisms that have recently emerged for nonimprinted genes indicate that long noncoding antisense RNAs negatively regulate gene transcription by targeting epigenetic modifications to the overlapping gene and/or its promoter region (23, 28, 37, 58, 61, 68). The function of the bII NAT is delineated in the present report. Further research will be required to determine the underlying regulatory mechanism by which bII NAT exerts its influence.
Regulatory role of bII NAT.
In hypothyroid rats bII NAT levels were elevated at all time points up to P20, before falling at P40 (Fig. 8A). From P2 to P20, IIb MHC levels remained undetectable, at the pre-mRNA, mRNA, and protein levels in hypothyroid rats. Thus, provided what is known about regulatory relationships of sense/anti-sense pairs, these data suggest that the sense/antisense pairs IIb MHC and bII NAT function in such a way that bII NAT could inhibit transcription of IIb MHC. In normal euthyroid muscle bII NAT levels were elevated at P2 and P10 before falling at P20 and P40 (Fig. 8A). Since bII NAT is presumed to be inhibitory of IIb MHC expression, it was somewhat surprising that the bII NAT remains at high levels of transcription at P10, while IIb MHC pre-mRNA levels are also high in the CON muscle. This indicates that the bII NAT has little regulatory influence at this particular stage of development in the euthyroid state and that other factors can override any negative regulatory influence of bII NAT. We speculate that IIb MHC expression is elevated at P10 despite bII NAT expression due to a strong transcriptional influence on IIb MHC by T3. The T3 surge that occurs during the first 20 days (see Fig. 1) activates thyroid response elements on the IIb promoter to engage transcriptional activity and thus may override any negative influence of the bII NAT. The strength of this T3 influence on IIb MHC activation is demonstrated when a slow muscle is impacted by hindlimb suspension, which creates a transcriptionally permissive environment for IIb MHC expression and results in a sevenfold increase in IIb MHC mRNA in the soleus (22). If rats were induced to be hyperthyroid, by administration of T3, in addition to hindlimb suspension, then IIb mRNA levels are increased 21-fold above normal conditions, demonstrating the powerful synergistic effect of T3 with unloading (22). In contrast, postnatal development during a hypothyroid state attenuates the T3-induced transactivation of the IIb MHC gene. Thus, while elevated transcription levels of bII NAT during the first 20 days of life in PTU-treated rats may directly repress IIb MHC transcription, in CON rats surging T3 levels will transactivate the IIb MHC gene, leading to increased accumulation of IIb mRNA, despite the presence of elevated bII NAT levels, at 10 days of age.
Conversely, a posttranscriptional, rather than transcriptional, type of regulatory mechanism could provide a rationale for concurrently elevated levels of bII NAT and IIb MHC mRNA at P10 in CON muscle. For example, bII NAT could inhibit translation via sense-antisense pairing or through formation of secondary structures generated by interactions between the sense and antisense RNAs (52), though this is not well supported mechanistically. Alternatively, the possibility of the bII NAT encoding an unknown micro-RNA that regulates posttranscriptional processes should also be considered. However, the regulatory mechanism of the majority of previously described long antisense noncoding RNAs are consistent with transcription-level regulation of its sense counterpart via epigenetic mechanisms (e.g., see Refs. 27 and 60).
One question raised by these experiments is whether bII NAT plays a role in inhibiting expression of IIb MHC, and potentially the other adult MHCs in the same genomic locus, during embryonic development. In the fetal stage, Neo, along with Emb MHC, are the primary MHCs expressed in developing fetal limb muscle, whereas IIb MHC remains essentially repressed (32, 63). T3 is not synthesized or circulated in prenatal embryos. During embryonic development and in the first ∼5 days of life rats are effectively hypothyroid. Thus a potent transactivator of IIb MHC is absent. A bII NAT inhibitory role during prenatal development is suggested by the data at P2, when bII NAT is elevated, relative to levels at P20 and P40 in the CON group, and P2 transcriptional activity of IIb MHC is undetectable (see Fig. 6D).
With data indicating that long noncoding antisense RNA is a regulator of IIb MHC gene expression under certain conditions during development, the question arises regarding any regulatory role in the adult muscle. It appears that the bII NAT is only expressed in concurrence with the Neo MHC, which is not normally expressed in most adult skeletal muscles. However, we previously reported on the presence of the bII NAT and its regulation in response to motoneuron silencing in adult soleus and vastus intermedius muscle (42). Under these conditions there was also a small increase in expression of Neo MHC mRNA in the slow fiber type. Under normal control conditions bII NAT is not expressed, and the IIb MHC gene remains essentially repressed in a slow muscle such as the rat soleus (42), indicating that additional regulatory mechanisms exist to inhibit IIb MHC gene transcription.
Our previous analyses of the MHC NATs, which appear to regulate IIx, IIa, and type I (β) MHC were found to have TSSs within 1,000 bp of the IIb, IIx, and α-MHC TSSs, respectively (19, 42, 47). The sense and antisense transcripts are suggested to be regulated by bidirectional promoters, such that the MHC gene promoter and antisense promoter are coregulated. In the case of the bII NAT, it is ∼11.8 kb from the Neo MHC TSS in the rat, which is generally considered to be outside of the range of cooperative regulation via bidirectional promoters. However, the bII NAT and Neo pre-mRNA levels were strongly positively correlated (r = 0.79 across all groups), suggesting these sense/antisense genes are coregulated functionally to maintain the embryonic/early developmental state in which Neo MHC is expressed while IIb MHC is repressed. Therefore, is seems likely that despite the relatively large distance between the bII NAT and Neo TSSs, they may be coregulated in an analogous manner to a bidirectional promoter, such as through intrachromosomal interaction (48).
Evolutionary conservation of bII NAT start site.
Local multiple DNA sequence alignments revealed that there are three ECRs in the rat IIb-Neo intergenic region common to the 11 placental mammals examined, but only one (ECR2) is uniquely situated far from the typically conserved regulatory regions of the IIb or Neo MHC genes (see Fig. 10). This unique site is likely the conserved promoter region of bII NAT; it contains the rat bII NAT TSS. The described regulation of the bII NAT during rat development and in altered thyroid hormone states, together with an evolutionarily conserved region at the TSS, provides convincing evidence of an important regulatory function.
A small marsupial, the short-tailed opossum, did not have any conserved sequence in the IIb-Neo intergenic region, except for the proximal promoter region of the Neo MHC gene. Marsupials diverged from placental mammals ∼180 million years ago, suggesting that the antisense-mediated regulation of IIb MHC must have evolved after this divergence (65). Comparing the IIb-Neo intergenic region in 12 diverse eutherians confirms that the origin of the bII NAT promoter region dates back to at least 105 million years ago when the proboscidean (e.g., elephant) ancestors diverged from other placentals (29, 51). Our comparative analysis also suggests that bII NAT-mediated regulation has been a conserved trait of placental mammals for most of the eutherian evolutionary history. Confirmation of the presence of bII NAT in adult human skeletal muscle has been achieved in recent experiments in our laboratory, providing further evidence that the evolutionary conservation of the rat bII NAT promoter region is of functional consequence (Pandorf CE, Haddad F, Baldwin KM; unpublished data).
Regulation of IIb MHC in large vs. small mammals.
A. V. Hill (24) originally observed that maximum speed of locomotion is largely independent of body size in different mammalian species. Hill hypothesized, and others have since largely confirmed, that differences in maximum shortening velocity (V0) of skeletal muscle is primarily responsible for this relationship. Thus limb muscles of small mammals such as rodents have a greater V0, which provides for greater relative stride frequency, than larger mammals. The primary means to achieve greater shortening velocities in muscle is with a bias toward the IIx and IIb MHC isoforms in a myofiber. The IIb MHC has the fastest ATPase activity of all MHC isoforms (4). The scaling of body size to muscle speed of contraction is illustrated by the fact that IIb MHC is abundantly expressed in small mammals such as rats, mouses, guinea pigs, and rabbits (15, 59), whereas it is not expressed at all in skeletal muscles of most large mammals, such as humans, baboons, horses, cows, goats, dogs, and cats (3, 26, 31, 33, 49, 50, 55).
The question remains as to the molecular mechanism that inhibits expression of the IIb MHC gene in large mammals. It is therefore curious that the rat promoter region of bII NAT, a potential negative regulator of IIb gene transcription, remains evolutionarily conserved in a broad range of placental mammals. In contrast, the rat promoter region of bII NAT was not conserved in the opossum IIb-Neo IG region. Zhong et. al. (69) determined that the IIb MHC is highly expressed in seven different marsupial species, including the opossum and some large kangaroo species. The question arises then as to whether there is a mechanistic linkage between the absence of the conserved bII NAT promoter (and therefore absence of the bII NAT inhibitory transcript) and the presence of the IIb MHC in large mammals such as the kangaroo and opossum. In contrast to all other previously examined large body-size eutherians, the domesticated pig and llama, both of the Artiodactyla order, expresses the IIb MHC in apparently abundant proportions in certain skeletal muscles (16, 56). Future research will be required to determine whether the bII NAT plays a role in inhibiting expression of IIb MHC in certain mammals, such as humans, and if a lack of bII NAT expression plays a mechanistic role in permitting IIb MHC transcription in other mammalian species.
In conclusion, we report that transcription of long noncoding antisense RNA, termed bII NAT, is positively correlated with Neo MHC expression during postnatal development in normal and hypothyroid states and is negatively correlated with IIb MHC expression during postnatal development, particularly under hypothyroid conditions that reduces the normally strong transactivating potential of thyroid hormone at the IIb MHC promoter. Thus, based on this evidence, and the previously reported evidence of a regulatory role for other NATs to the IIa and IIx MHCs in skeletal muscle and β-MHC in cardiac muscle, and the mechanistic evidence of the regulatory roles of other long noncoding antisense RNAs, we conclude that the bII NAT is likely a negative regulator of IIb MHC transcription.
Perspectives and Significance
Long noncoding antisense RNAs transcribed in reverse orientation to individual MHC genes appear to have functional importance and likely play a role in coordinating shifts in MHC during a variety of adaptive processes, including those enunciated herein. The bII NAT is transcribed from a region in the IIb-Neo intergenic region that is highly conserved across most of the evolutionary history of eutherians. The data presented suggest that coregulation of Neo MHC and bII NAT ensures that under conditions where Neo MHC is highly expressed, such as during early developmental states, particularly under hypothyroid conditions, the IIb MHC gene receives direct negative feedback from the neighboring Neo MHC gene, via the bII NAT, so that a reciprocal relationship is maintained between the developmental and adult MHC isoforms. Further research is needed to develop insight into mechanisms utilized by the MHC antisense transcripts and should include epigenetic approaches. Future research should also investigate whether bII NAT plays a role in limiting the expression of IIb MHC in certain mammals, which has implications for understanding how variations in the speed of muscle contraction of small compared with large mammals, including humans, is controlled.
GRANTS
This work was supported by National Institutes of Health AR-30346 (to K. M. Baldwin) and by the National Space Biomedical Research Institute through National Aeronautics and Space Administration NCC 9-58 (to C. E. Pandorf).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: C.E.P., K.M.B., and F.H. conception and design of research; C.E.P., W.J., A.X.Q., P.W.B., and F.H. performed experiments; C.E.P., K.M.B., and F.H. analyzed data; C.E.P., K.M.B., and F.H. interpreted results of experiments; C.E.P. prepared figures; C.E.P. drafted manuscript; C.E.P., K.M.B., and F.H. edited and revised manuscript; C.E.P., W.J., A.X.Q., P.W.B., K.M.B., and F.H. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Ming Zeng, LiYing Zhang, Michele Kunde, Peter Van Kha, Patrice Jang, Roberto Flores, Brian Chen, Nancy Nguyen, Janelle Sauz, and Cindy Ngo for excellent technical assistance and Tomasz Owerkowicz for critical reading of the manuscript.
REFERENCES
- 1. Adams GR, Haddad F, McCue SA, Bodell PW, Zeng M, Qin L, Qin AX, Baldwin KM. Effects of spaceflight and thyroid deficiency on rat hindlimb development. II. Expression of MHC isoforms. J Appl Physiol 88: 904– 916, 2000 [DOI] [PubMed] [Google Scholar]
- 2. Adams GR, McCue SA, Zeng M, Baldwin KM. Time course of myosin heavy chain transitions in neonatal rats: importance of innervation and thyroid state. Am J Physiol Regul Integr Comp Physiol 276: R954–R961, 1999 [DOI] [PubMed] [Google Scholar]
- 3. Arguello A, Lopez-Fernandez JL, Rivero JL. Limb myosin heavy chain isoproteins and muscle fiber types in the adult goat (Capra hircus). Anatomic Rec 264: 284–293, 2001 [DOI] [PubMed] [Google Scholar]
- 4. Bottinelli R, Canepari M, Reggiani C, Stienen GJ. Myofibrillar ATPase activity during isometric contraction and isomyosin composition in rat single skinned muscle fibres. J Physiol 481: 663–675, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caiozzo VJ, Haddad F. Thyroid hormone: modulation of muscle structure, function, and adaptive responses to mechanical loading. Exerc Sport Sci Rev 24: 321–361, 1996 [PubMed] [Google Scholar]
- 6. Caiozzo VJ, Haddad F, Baker M, McCue S, Baldwin KM. MHC polymorphism in rodent plantaris muscle: effects of mechanical overload and hypothyroidism. Am J Physiol Cell Physiol 278: C709–C717, 2000 [DOI] [PubMed] [Google Scholar]
- 7. Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res 32: 4812–4820, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, Sementchenko V, Piccolboni A, Bekiranov S, Bailey DK, Ganesh M, Ghosh S, Bell I, Gerhard DS, Gingeras TR. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308: 1149–1154, 2005 [DOI] [PubMed] [Google Scholar]
- 9. di Maso NA, Haddad F, Zeng M, McCue SA, Baldwin KM. Role of denervation in modulating IIb MHC gene expression in response to T(3) plus unloading state. J Appl Physiol 88: 682–689, 2000 [DOI] [PubMed] [Google Scholar]
- 10. di Maso NA, Caiozzo VJ, Baldwin KM. Single-fiber myosin heavy chain polymorphism during postnatal development: modulation by hypothyroidism. Am J Physiol Regul Integr Comp Physiol 278: R1099–R1106, 2000 [DOI] [PubMed] [Google Scholar]
- 11. Dye MJ, Proudfoot NJ. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell 105: 669–681, 2001 [DOI] [PubMed] [Google Scholar]
- 12. Elferink CJ, Reiners JJ., Jr Quantitative RT-PCR on CYP1A1 heterogeneous nuclear RNA: a surrogate for the in vitro transcription run-on assay. Biotechniques 20: 470–477, 1996 [DOI] [PubMed] [Google Scholar]
- 13. Engstrom PG, Suzuki H, Ninomiya N, Akalin A, Sessa L, Lavorgna G, Brozzi A, Luzi L, Tan SL, Yang L, Kunarso G, Ng EL, Batalov S, Wahlestedt C, Kai C, Kawai J, Carninci P, Hayashizaki Y, Wells C, Bajic VB, Orlando V, Reid JF, Lenhard B, Lipovich L. Complex Loci in human and mouse genomes. PLoS Genet 2: e47, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ge X, Rubinstein W, Jung Yc, Wu Q. Genome-wide analysis of antisense transcription with Affymetrix exon array. BMC Genomics 9: 27, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gorza L. Identification of a novel type 2 fiber population in mammalian skeletal muscle by combined use of histochemical myosin ATPase and anti-myosin monoclonal antibodies. J Histochem Cytochem 38: 257–265, 1990 [DOI] [PubMed] [Google Scholar]
- 16. Graziotti GH, Rios CM, Rivero JL. Evidence for three fast myosin heavy chain isoforms in type ii skeletal muscle fibers in the adult llama (Lama glama). J Histochem Cytochem 49: 1033–1044, 2001 [DOI] [PubMed] [Google Scholar]
- 17. Haddad F, Arnold C, Zeng M, Baldwin K. Interaction of thyroid state and denervation on skeletal myosin heavy chain expression. Muscle Nerve 20: 1487–1496, 1997 [DOI] [PubMed] [Google Scholar]
- 18. Haddad F, Bodell PW, Qin AX, Giger JM, Baldwin KM. Role of antisense RNA in coordinating cardiac myosin heavy chain gene switching. J Biol Chem 278: 37132–37138, 2003 [DOI] [PubMed] [Google Scholar]
- 19. Haddad F, Qin AX, Bodell PW, Jiang W, Giger JM, Baldwin KM. Intergenic transcription and developmental regulation of cardiac myosin heavy chain genes. Am J Physiol Heart Circ Physiol 294: H29–H40, 2008 [DOI] [PubMed] [Google Scholar]
- 20. Haddad F, Qin AX, Bodell PW, Zhang LY, Guo H, Giger JM, Baldwin KM. Regulation of antisense RNA expression during cardiac MHC gene switching in response to pressure overload. Am J Physiol Heart Circ Physiol 290: H2351–H2361, 2006 [DOI] [PubMed] [Google Scholar]
- 21. Haddad F, Qin AX, Giger JM, Guo H, Baldwin KM. Potential pitfalls in the accuracy of analysis of natural sense-antisense RNA pairs by reverse transcription-PCR. BMC Biotechnol 7: 21, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haddad F, Qin AX, Zeng M, McCue SA, Baldwin KM. Interaction of hyperthyroidism and hindlimb suspension on skeletal myosin heavy chain expression. J Appl Physiol 85: 2227–2236, 1998 [DOI] [PubMed] [Google Scholar]
- 23. Hawkins PG, Morris KV. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription 1: 165–175, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hill AV. The dimensions of animals and their muscular dynamics. Sci Prog 38: 209–230, 1950 [Google Scholar]
- 25. Kasahara H, Usheva A, Ueyama T, Aoki H, Horikoshi N, Izumo S. Characterization of homo- and heterodimerization of cardiac Csx/Nkx2.5 homeoprotein. J Biol Chem 276: 4570–4580, 2001 [DOI] [PubMed] [Google Scholar]
- 25a. Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. RIKEN. Genome Exploration Research Group, and Genome Science Group (Genome Network Project Core Group) FANTOM Consortium Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566, 2005 [DOI] [PubMed] [Google Scholar]
- 26. Kawai M, Minami Y, Sayama Y, Kuwano A, Hiraga A, Miyata H. Muscle fiber population and biochemical properties of whole body muscles in thoroughbred horses. Anat Rec 292: 1663–1669, 2009 [DOI] [PubMed] [Google Scholar]
- 27. Knowling S, Morris KV. Non-coding RNA and antisense RNA. Nature's trash or treasure? Biochimie 93: 1922–1927, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene 30: 1956–1962, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature 392: 917–920, 1998 [DOI] [PubMed] [Google Scholar]
- 30. Lipovich L, Johnson R, Lin CY. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta 1799: 597–615, 2010 [DOI] [PubMed] [Google Scholar]
- 31. Lucas CA, Kang LHD, Hoh JFY. Monospecific antibodies against the three mammalian fast limb myosin heavy chains. Biochem Biophys Res Commun 272: 303–308, 2000 [DOI] [PubMed] [Google Scholar]
- 32. Lyons GE, Ontell M, Cox R, Sassoon D, Buckingham M. The expression of myosin genes in developing skeletal muscle in the mouse embryo. J Cell Biol 111: 1465–1476, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maccatrozzo L, Patruno M, Toniolo L, Reggiani C, Mascarello F. Myosin heavy chain 2B isoform is expressed in specialized eye muscles but not in trunk and limb muscles of cattle. Eur J Histochem 48: 357–366, 2004 [PubMed] [Google Scholar]
- 34. Mahdavi V, Izumo S, Nadal-Ginard B. Developmental and hormonal regulation of sarcomeric myosin heavy chain gene family. Circ Res 60: 804–814, 1987 [DOI] [PubMed] [Google Scholar]
- 35. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155–159, 2009 [DOI] [PubMed] [Google Scholar]
- 36. Molkentin JD, Olson EN. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci USA 93: 9366–9373, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet 4: e1000258, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morris KV. Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics 4: 296–301, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morris KV. The emerging role of RNA in the regulation of gene transcription in human cells. Semin Cell Develop Biol 22: 351–358, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ovcharenko I, Loots GG, Giardine BM, Hou M, Ma J, Hardison RC, Stubbs L, Miller W. Mulan: multiple-sequence local alignment and visualization for studying function and evolution. Genome Res 15: 184–194, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ovcharenko I, Loots GG, Hardison RC, Miller W, Stubbs L. zPicture: dynamic alignment and visualization tool for analyzing conservation profiles. Genome Res 14: 472–477, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pandorf CE, Haddad F, Roy RR, Qin AX, Edgerton VR, Baldwin KM. Dynamics of myosin heavy chain gene regulation in slow skeletal muscle: role of natural antisense RNA. J Biol Chem 281: 38330–38342, 2006 [DOI] [PubMed] [Google Scholar]
- 43. Pandorf CE, Jiang WH, Qin AX, Bodell PW, Baldwin KM, Haddad F. Calcineurin plays a modulatory role in loading-induced regulation of type I myosin heavy chain gene expression in slow skeletal muscle. Am J Physiol Regul Integr Comp Physiol 297: R1037–R1048, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Proudfoot NJ. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem Si 14: 105–110, 1989 [DOI] [PubMed] [Google Scholar]
- 45. Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Develop 23: 1247–1269, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rinaldi C, Haddad F, Bodell PW, Qin AX, Jiang W, Baldwin KM. Intergenic bidirectional promoter and cooperative regulation of the IIx and IIb MHC genes in fast skeletal muscle. Am J Physiol Regul Integr Comp Physiol 295: R208–R218, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 42: 53–61, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Serrano AL, Petrie JL, Rivero JL, Hermanson JW. Myosin isoforms and muscle fiber characteristics in equine gluteus medius muscle. Anatom Rec 244: 444–451, 1996 [DOI] [PubMed] [Google Scholar]
- 50. Smerdu V, Karsch-Mizrachi I, Campione M, Leinwand L, Schiaffino S. Type IIx myosin heavy chain transcripts are expressed in type IIb fibers of human skeletal muscle. Am J Physiol Cell Physiol 267: C1723–C1728, 1994 [DOI] [PubMed] [Google Scholar]
- 51. Springer MS, Murphy WJ, Eizirik E, O'Brien SJ. Placental mammal diversification and the CretaceousΓÇôTertiary boundary. Proc Natl Acad Sci USA 100: 1056–1061, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stolt P, Zillig W. Antisense RNA mediates transcriptional processing in an archaebacterium, indicating a novel kind of RNase activity. Mol Microbiol 7: 875–882, 1993 [DOI] [PubMed] [Google Scholar]
- 53. Talmadge RJ, Roy RR. Electrophoretic separation of rat skeletal muscle myosin heavy-chain isoforms. J Appl Physiol 75: 2337–2340, 1993 [DOI] [PubMed] [Google Scholar]
- 54. Tantravahi J, Alvira M, Falck-Pedersen E. Characterization of the mouse beta maj globin transcription termination region: a spacing sequence is required between the poly(A) signal sequence and multiple downstream termination elements. Mol Cell Biol 13: 578–587, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Toniolo L, Maccatrozzo L, Patruno M, Pavan E, Caliaro F, Rossi R, Rinaldi C, Canepari M, Reggiani C, Mascarello F. Fiber types in canine muscles: myosin isoform expression and functional characterization. Am J Physiol Cell Physiol 292: C1915–C1926, 2007 [DOI] [PubMed] [Google Scholar]
- 56. Toniolo L, Patruno M, Maccatrozzo L, Pellegrino MA, Canepari M, Rossi R, D'Antona G, Bottinelli R, Reggiani C, Mascarello F. Fast fibres in a large animal: fibre types, contractile properties and myosin expression in pig skeletal muscles. J Exp Biol 207: 1875–1886, 2004 [DOI] [PubMed] [Google Scholar]
- 57. Tsika RW, Schramm C, Simmer G, Fitzsimons DP, Moss RL, Ji J. Overexpression of TEAD-1 in transgenic mouse striated muscles produces a slower skeletal muscle contractile phenotype. J Biol Chem 283: 36154–36167, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. van Dijk EL, Chen CL, 'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Ne P, Loeillet S, Nicolas A, Thermes C, Morillon A. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475: 114–117, 2011 [DOI] [PubMed] [Google Scholar]
- 59. Wada M, Hamalainen N, Pette D. Isomyosin patterns of single type IIB, IID and IIA fibres from rabbit skeletal muscle. J Muscle Res Cell Motil 16: 237–242, 1995 [DOI] [PubMed] [Google Scholar]
- 60. Wang X, Song X, Glass CK, Rosenfeld MG. The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harbor Perspect Biol 3: a003756, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Watanabe Y, Numata K, Murata S, Osada Y, Saito R, Nakaoka H, Yamamoto N, Watanabe K, Kato H, Abe K, Kiyosawa H. Genome-wide analysis of expression modes and DNA methylation status at sense-antisense transcript loci in mouse. Genomics 96: 333–341, 2010 [DOI] [PubMed] [Google Scholar]
- 62. West S, Proudfoot NJ, Dye MJ. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol Cell 29: 600–610, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weydert A, Barton P, Harris AJ, Pinset C, Buckingham M. Developmental pattern of mouse skeletal myosin heavy chain gene transcripts in vivo and in vitro. Cell 49: 121–129, 1987 [DOI] [PubMed] [Google Scholar]
- 64. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Develop 23: 1494–1504, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Woodburne MO, Rich TH, Springer MS. The evolution of tribospheny and the antiquity of mammalian clades. Mol Phylogenet Evol 28: 360–385, 2003 [DOI] [PubMed] [Google Scholar]
- 66. Wright C, Haddad F, Qin AX, Baldwin KM. Analysis of myosin heavy chain mRNA expression by RT-PCR. J Appl Physiol 83: 1389–1396, 1997 [DOI] [PubMed] [Google Scholar]
- 67. Yoshida T. MCAT Elements and the TEF-1 family of transcription factors in muscle development and disease. Arterioscler Thromb Vasc Biol 28: 8–17, 2008 [DOI] [PubMed] [Google Scholar]
- 68. Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451: 202–206, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhong WW, Lucas CA, Kang LH, Hoh JF. Electrophoretic and immunochemical evidence showing that marsupial limb muscles express the same fast and slow myosin heavy chains as eutherians. Electrophoresis 22: 1016–1020, 2001 [DOI] [PubMed] [Google Scholar]