Abstract
Systematic biological screens typically identify many genes or proteins that are implicated in a specific phenotype. However, deriving mechanistic insight from these screens typically involves focusing upon one or a few genes within the set in order to elucidate their precise role in producing the phenotype. To find these critical genes, researchers use a variety of tools to query the set of genes to uncover underlying common genetic or physical interactions or common functional annotations (e.g. gene ontology terms). Not only it is necessary to find previous screens containing genes in common with the new set, but also useful to easily access the individual manuscript or study that classified those genes. Unfortunately, no tool currently exists to facilitate this task. We have developed a web-based tool (ScreenTroll) that queries one or more genes against a database of systematic yeast screens. The software determines which genome-wide yeast screens also identified the queried gene(s) and the resulting screens are listed in an order based on the extent of the overlap between the queried gene(s) and the open reading frames (ORFs) characterized in each individual yeast screen. In a separate list, the corresponding ORFs that are found in both the queried set of genes and each individual genome-wide screen are displayed along with links to the relevant manuscript via NIH’s PubMed database. ScreenTroll is useful for comparing a list of ORFs with genes identified in a wide array of published genome-wide screens. This comparison informs users whether any of their queried ORFs overlaps a previous study in the ScreenTroll database. By listing the manuscript of the published screen, users can read more about the phenotype associated with that study. Together, this information provides insight into the function of the queried genes and helps the user focus on a subset of them.
Background
The creation of a comprehensive collection of non-essential open reading frame (ORF) deletions in the yeast Saccharomyces cerevisiae has made this organism a primary model for genomics and high-throughput biology (1). The genomics data generated using the gene deletion collection has been central in driving the development of systems biology (2). When analyzing the ORFs identified in a genome-wide yeast screen, it is possible to determine the genetic and physical interactions between them using relatively sophisticated approaches [for example, Biopixie, (3)]. Additionally, using gene ontology term enrichment analysis, it is possible to determine if functional categories are enriched within a set of ORFs [reviewed in (4)]. There are also tools that provide an overview of multiple phenotypic properties (interactions, localization, etc.) for a given list of genes [for example, FunSpec (5)]. However, what is lacking is a database and search tool that (i) identifies common ORFs between a queried set and ORFs characterized by individual genome-wide studies, (ii) orders the results based upon the likelihood of the overlap and (iii) lists the manuscripts associated with the studies.
Construction and content
We have assembled a database from published manuscripts of hundreds of groups of ORFs identified in, or derived from, large-scale yeast screens. We have focused our attention upon datasets that have systematically utilized the non-essential gene deletion collection to assay a specific phenotype. However, there are an increasing number of screens included in ScreenTroll using collections of mutant alleles of essential genes. There are two types of screens that are commonly reported: first, those that list a set of ORFs as affected. For example, Alvaro et al. (6) screened each non-essential gene deletion for its ability to increase the frequency of nuclear foci of Rad52, a key DNA repair protein. That study produced a list of 86 ORF deletions with this phenotype. The database entry for that study includes a short description of the screen phenotype: Elevated Rad52 foci, a summary of the manuscript describing the screen: Alvaro et al. 2007. Plos Genetics. 3; e228 with its PubMed ID number and finally, a list of the ORFs that were identified in the study. Much of this type of data is not included in other databases and these data are the core of the ScreenTroll database.
The second type is genome-wide screens that report quantitative data for each deletion, but do not necessarily provide a cutoff value or a defined list of affected ORFs. For example, a growth ratio on the experimental condition versus the control condition is listed for each deletion. To assemble a list of ORF deletions from these quantitative screens, we chose a specific cutoff value to generate a list of ORFs with the strongest phenotype and have indicated this cutoff value in the screen description. For example, the description ‘1.5 M sorbitol sensitive at 15 generations (competition assay >100 fitness defect)’, indicates that the selection of strains chosen for the ScreenTroll database from the study of Giaever et al. (7) are sensitive to 1.5 M sorbitol after 15 generations and showed a ‘fitness defect’ greater than 100. Further details of the definition of ‘fitness defect’ are clearly explained in the manuscript describing the screen, whose link is accessible directly from the ScreenTroll output.
The ScreenTroll database includes most of the significant interactions from the Costanzo et al. (8) large-scale synthetic genetic array (SGA) study, where more than 1700 different query gene deletions were assayed against the entire library. The data from these screens were reported quantitatively and we have included ORFs within an intermediate cutoff value defined by the authors (|ε| > 0.08, P < 0.05). Thus, the description of the database entry for the synthetic interactions with cln2Δ is labeled ‘Costanzo SGA Screen, Intermediate Cutoff (|ε| > 0.08 & p-value < 0.05) - Query: YPL256C (CLN2)’.
Utility and discussion
To query the ScreenTroll database for commonalities, we have built a web-based search tool that enables users to enter one or more yeast ORFs (the ‘query set’) into a single search window. The screens that most closely match the query set are listed in a rank order based upon a ‘rank score’ (a description of the statistical methods used to evaluate this score is provided on the website and in Supplementary Data). The rank order is not a precise statistical ranking, but allows the user to focus on screens with extensive overlap as well as screens that identify mutually exclusive sets of ORFs (highlighted in blue). This latter group also provides functional insight since mutual exclusivity likely indicates that the two different phenotypes result from separate molecular pathways. Additionally, when ScreenTroll identifies an overlap, a list of ORFs in common with the query set is provided along with a link to the PubMed reference for the manuscript describing the particular screen. This feature facilitates access to the details of each screen enabling users to evaluate the potential biological significance of the individual ORFs identified.
The ScreenTroll output is ordered by ‘rank score’, which is a calculation of the hypergeometric P-value of each comparison. A simple adjustment for multiple comparison testing using the Bonferroni method (9) can be readily applied by multiplying the rank score by the number of screens tested (provided at the top of the results screen). We purposely use the term ‘rank score’ as opposed to ‘P-value’ for a number of reasons. First, the rank score assumes that both the user’s query set and each screen in the database are derived from the same set of 4800 strains in the viable yeast deletion collection. However, this may not be the case. Second, each published yeast screen has its own, often unknown or unreported, false positive and negative discovery rates, which directly affects the likelihood of an overlap. Further, we cannot predict the user’s own false discovery rates. Third, we feel that the biological importance of an overlap between two groups of ORFs is best determined by carefully examining the manuscript describing how the ORF list was derived.
Some screens in the ScreenTroll database are the result of characterizing each deletion strain individually, however, many screens use a competition method. For this latter approach, the entire deletion collection is pooled together and exposed to experimental conditions (7). Subsequent microarray hybridization analysis of ‘bar code’ sequences specific for each yeast deletion reveals the relative levels of each strain in the pooled population. In this way, strains affected by the experimental condition are identified. In one such study, an exhaustive list of hundreds of different conditions and compounds were tested in both homozygous and heterozygous diploid strains (10) and much of this data is included in the ScreenTroll database. However, competition assays do not directly test each strain separately and some users may prefer to exclude this type of data from their analysis. Consequently, ScreenTroll includes the option of excluding data from competition assays in each search.
ScreenTroll was initially designed to highlight screens that include significant analysis of the associated phenotype. These screens, although genome wide, generally focus on a specific mechanism or phenotype (e.g. Rad52 focus formation, methyl methanesulfonate (MMS) sensitivity, chromosome instability, etc.) and the manuscripts associated with them provide considerable detail about their findings. Nevertheless, as noted above, we have also included most of the very large scale screening data from the Costanzo et al. (8) SGA screens. However, some users may prefer to restrict their search to the core ScreenTroll data set, since the massive amount of data from the SGA screens may overwhelm the output and mask overlaps with the more focused screens. Hence, we have included an option to exclude the data from these large-scale synthetic genetic array experiments.
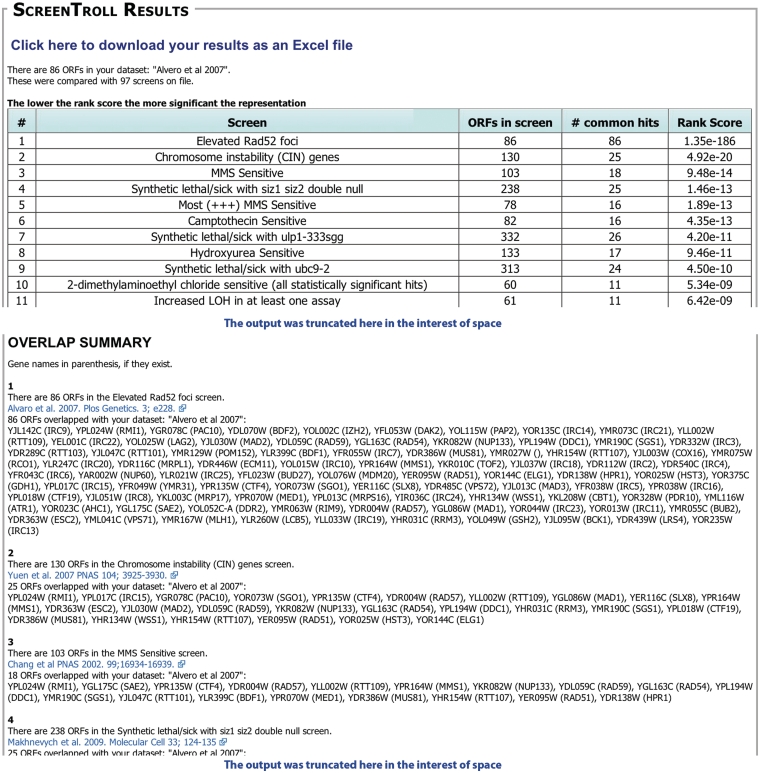
Primarily, users will enter a set of ORFs identified in a new screen, ‘the query set,’ to compare with those identified from other screens. If a strong match is found, it suggests that both the user’s screen and the published screen share a common feature. For example, we entered a set of ORFs identified by our laboratory as being important to prevent high levels of Rad52 foci (6). ScreenTroll identifies screens that assay for chromosomal instability (11), sensitivity to methyl methanesulfonate (12) and the sumoylation pathway (13), as those that most closely match the query set (a portion of the ScreenTroll output is shown in Figure 1). These matches confirm the shared pathway of DNA damage repair for all of these screens and highlight potentially new insights into the role of sumoylation in regulating the DNA damage response. In addition, having the complete list of overlapping screens is useful since some of the individual ORFs further down the list, which are common to a particular screen, may be of interest to the user. For example, a screen for propanol sensitivity identified IRC15 and IRC25, two previously uncharacterized ORFs from the Alvaro screen (14).
Figure 1.
Example of a ScreenTroll Output. Portions of the output from a ScreenTroll search using the set of 86 ORFs identified by Alvaro et al. (6) are shown. ScreenTroll displays a table of the screens in the database ordered by a rank score of the overlap with the query set (top). Below the table, the ‘overlap summary’ includes more details of each screen including the specific ORFs that overlap with the query set and a link to the PubMed entry for the manuscript that describes the screen (bottom). Both the competition and the Costanzo et al. (8) SGA screens were excluded from this search.
If the user is interested in exploring a new or existing pathway, ScreenTroll can be used to query the ORFs that encode that pathway to determine whether they were enriched in previous screens. For example, the spindle assembly checkpoint (SAC) is a key regulator of mitosis and it is possible to query the database with MAD1, MAD2, MAD3, BUB1 and BUB3, each of which encode key non-essential components of the SAC [see (15) and references therein]. The ScreenTroll output from this query can be viewed by selecting the example provided on the ScreenTroll website. At the time of publication, the first four screens [excluding the Costanzo et al. (8) SGA data] that most closely match this query set are (i) gene deletions that are synthetic lethal with kinetochore mutants (16), (ii) deletions that fail to maintain an ‘originless’ plasmid (17), (iii) deletions that are sensitive to the microtubule poison benomyl (18) and (iv) deletions that result in chromosome instability (11). Since the SAC proteins are located at the kinetochore and help to direct chromosome segregation, these data are consistent with the known mechanism of the SAC. However, the fifth screen listed is a screen for increased Rad52 DNA repair centers (6), reinforcing a role for the SAC in preventing DNA damage (19,20).
If a user is interested in a single gene, ScreenTroll can list all of the screens that identified it. For example, if a user enters RAD50, the results show that this gene was identified in numerous genome-wide screens for DNA damage sensitivity, consistent with its known role in DNA repair.
Finally, there are a wealth of gene-gene and protein-protein interaction data available for yeast (8,16,18,21–25) and excellent tools to query these data [for examples, see (26,27)]. The ScreenTroll webpage provides a link to access many of these tools.
Conclusions
Using this simple tool, similarities between screens are revealed and listed in rank order. The results of ScreenTroll are useful for deciding which ORFs identified in a new screen are of specific interest due to a shared phenotype. Moreover, identifying the ‘screen phenotype’ conferred by deletion of a specific ORF, or set of ORFs, can illuminate the biological function of the encoded protein(s) and aid in the design of new assays to test its function. We envision that ScreenTroll will be of use to anyone interested in analyzing the results of yeast genomic data. The package with documentation is available at http://www.rothsteinlab.com/tools/apps/screenTroll. This website includes the option to separately download the entire database, the source code for the application and information about the statistics used to generate the rank score. There are no access restrictions. In addition, ScreenTroll is available through individual ORF pages on the Saccharomyces Genome Database (yeastgenome.org).
Supplementary Data
Supplementary data are available at Database Online.
Funding
National Institutes of Health (GM50237 and GM67055 to R.R. and GM008798 and CA009503 to J.C.D.). Funding for open access charge: National Institutes of Health (GM50237 and GM67055).
Conflict of interest. None declared.
Acknowledgments
We would like to thank our colleagues Michael Chang, Bob Reid, Kara Bernstein and Steven Pierce for helpful discussion. J.C.D. and P.H.T. assembled the database from the published literature and created the search software. J.C.D., P.H.T. and R.R. wrote the manuscript.
References
- 1.Winzeler EA, Shoemaker DD, Astromoff A, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 2.Snyder M, Gallagher JE. Systems biology from a yeast omics perspective. FEBS Lett. 2009;583:3895–3899. doi: 10.1016/j.febslet.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Myers CL, Robson D, Wible A, et al. Discovery of biological networks from diverse functional genomic data. Genome Biol. 2005;6:R114. doi: 10.1186/gb-2005-6-13-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson MD, Grigull J, Mohammad N, Hughes TR. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics. 2002;3:35. doi: 10.1186/1471-2105-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alvaro D, Lisby M, Rothstein R. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 2007;3:e228. doi: 10.1371/journal.pgen.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giaever G, Chu AM, Ni L, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 8.Costanzo M, Baryshnikova A, Bellay J, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonferroni CE. Teoria statistica delle classi e calcolo delle probabilità. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze. 1936;8:3–62. [Google Scholar]
- 10.Hillenmeyer ME, Fung E, Wildenhain J, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuen KW, Warren CD, Chen O, et al. Systematic genome instability screens in yeast and their potential relevance to cancer. Proc. Natl Acad. Sci. USA. 2007;104:3925–3930. doi: 10.1073/pnas.0610642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang M, Bellaoui M, Boone C, Brown GW. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc. Natl Acad. Sci. USA. 2002;99:16934–16939. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makhnevych T, Sydorskyy Y, Xin X, et al. Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol. Cell. 2009;33:124–135. doi: 10.1016/j.molcel.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 14.Auesukaree C, Damnernsawad A, Kruatrachue M, et al. Genome-wide identification of genes involved in tolerance to various environmental stresses in Saccharomyces cerevisiae. J. Appl. Genet. 2009;50:301–310. doi: 10.1007/BF03195688. [DOI] [PubMed] [Google Scholar]
- 15.Skibbens RV, Hieter P. Kinetochores and the checkpoint mechanism that monitors for defects in the chromosome segregation machinery. Annu. Rev. Genet. 1998;32:307–337. doi: 10.1146/annurev.genet.32.1.307. [DOI] [PubMed] [Google Scholar]
- 16.Measday V, Baetz K, Guzzo J, et al. Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation. Proc. Natl Acad. Sci. USA. 2005;102:13956–13961. doi: 10.1073/pnas.0503504102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Theis JF, Irene C, Dershowitz A, et al. The DNA damage response pathway contributes to the stability of chromosome III derivatives lacking efficient replicators. PLoS Genet. 2010;6:e1001227. doi: 10.1371/journal.pgen.1001227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parsons AB, Brost RL, Ding H, et al. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 19.Garber PM, Rine J. Overlapping roles of the spindle assembly and DNA damage checkpoints in the cell-cycle response to altered chromosomes in Saccharomyces cerevisiae. Genetics. 2002;161:521–534. doi: 10.1093/genetics/161.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim EM, Burke DJ. DNA damage activates the SAC in an ATM/ATR-dependent manner, independently of the kinetochore. PLoS Genet. 2008;4:e1000015. doi: 10.1371/journal.pgen.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito T, Chiba T, Ozawa R, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uetz P, Giot L, Cagney G, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 23.Venkatesan K, Rual JF, Vazquez A, et al. An empirical framework for binary interactome mapping. Nat. Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Kakaradov B, Collins SR, et al. A complex-based reconstruction of the Saccharomyces cerevisiae interactome. Mol. Cell Proteomics. 2009;8:1361–1381. doi: 10.1074/mcp.M800490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valente AX, Roberts SB, Buck GA, Gao Y. Functional organization of the yeast proteome by a yeast interactome map. Proc. Natl Acad. Sci. USA. 2009;106:1490–1495. doi: 10.1073/pnas.0808624106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Z, Hung JH, Wang Y, et al. VisANT 3.5: multi-scale network visualization, analysis and inference based on the gene ontology. Nucleic Acids Res. 2009;37:W115–W121. doi: 10.1093/nar/gkp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopes CT, Franz M, Kazi F, et al. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]